Abstract
The serine protease HtrA2/Omi is released from the mitochondrial intermembrane space following apoptotic stimuli. Once in the cytosol, HtrA2/Omi has been implicated in promoting cell death by binding to inhibitor of apoptosis proteins (IAPs) via its amino-terminal Reaper-related motif, thus inducing caspase activity, and also in mediating caspase-independent death through its own protease activity. We report here the phenotype of mice entirely lacking expression of HtrA2/Omi due to targeted deletion of its gene, Prss25. These animals, or cells derived from them, show no evidence of reduced rates of cell death but on the contrary suffer loss of a population of neurons in the striatum, resulting in a neurodegenerative disorder with a parkinsonian phenotype that leads to death of the mice around 30 days after birth. The phenotype of these mice suggests that it is the protease function of this protein and not its IAP binding motif that is critical. This conclusion is reinforced by the finding that simultaneous deletion of the other major IAP binding protein, Smac/DIABLO, does not obviously alter the phenotype of HtrA2/Omi knockout mice or cells derived from them. Mammalian HtrA2/Omi is therefore likely to function in vivo in a manner similar to that of its bacterial homologues DegS and DegP, which are involved in protection against cell stress, and not like the proapoptotic Reaper family proteins in Drosophila melanogaster.
The serine protease HtrA2/Omi was identified as a mammalian homologue of the Escherichia coli protein HtrA, also known as DegP (3, 5). Like the bacterial protein, HtrA2/Omi has a PDZ domain in addition to the serine protease domain. HtrA2/Omi is localized to the mitochondrial intermembrane space by a mitochondrion-targeting sequence. A proteolytic-processing event reveals an amino-terminal sequence motif related to that of the critical Drosophila melanogaster proapoptotic proteins Reaper, Grim, and Hid. Following apoptotic stimuli, HtrA2/Omi is released from the mitochondria into the cytosol, where it uses this motif to bind to inhibitor of apoptosis proteins (IAPs) (8, 17, 27, 35). Binding of HtrA2/Omi to IAPs relieves their inhibitory activities towards caspases. This binding results in the increase of the proteolytic activity of HtrA2/Omi (18) and has also been reported to cause proteolytic degradation of bound IAPs (26, 40). HtrA2/Omi has therefore been proposed to be a proapoptotic protein analogous to Reaper that removes the protective effect of IAPs and thus potentiates the ability of the cytochrome c that is released from the mitochondria at the same time to trigger caspase activation (16, 29, 30, 38). In addition, HtrA2/Omi has also been implicated in mediating caspase-independent cell death through its own serine protease activity (8, 27).
Smac/DIABLO is another mammalian mitochondrial protein containing an amino-terminal Reaper motif that has been identified due to its ability to interact with and antagonize IAPs (2, 34). When overexpressed, Smac/DIABLO can sensitize cells to apoptotic stimuli. However, there is relatively little evidence that endogenous Smac/DIABLO plays an important physiological role in the regulation of apoptosis (33) and mice with the gene for this protein deleted show no detectable abnormalities (19). Cells derived from these Smac/DIABLO knockout mice also display normal apoptosis regulation. Similarly, mice with a deletion of the gene for the broadly expressed IAP family member XIAP show no abnormal phenotype (6). It is possible that IAPs and their antagonists play relatively minor roles in the regulation of apoptosis in mammals, unlike in flies. Alternatively, the high degree of redundancy among IAPs, and possibly also among Reaper motif-containing proteins, may make their in vivo function hard to analyze by single-gene deletion experiments.
While Smac/DIABLO has no obvious catalytic function and no clear homologues (outside of the 4-amino-acid Reaper motif) in invertebrates, HtrA2/Omi is a member of a well-conserved family of PDZ domain-containing serine proteases that are found in most eukaryotes and prokaryotes. Well-characterized members of the family include DegS and DegP in E. coli. DegP is a protein that protects against heat stress and that acts as a chaperone for unfolded proteins at low temperatures, while acting to degrade them at elevated temperatures (25). DegS detects unfolded outer membrane porins in the periplasm, triggering a signaling protease cascade leading to adaptive changes in gene expression (36). The known protective stress response activities of bacterial HtrA2/Omi homologues thus appear to be very different from the proposed proapoptotic, Reaper-like action of mammalian HtrA2/Omi. However, it has been reported recently that the neurodegenerative phenotype leading to juvenile mortality in the Mnd2 strain of mice is caused by a protease-inactivating point mutation in the gene encoding HtrA2/Omi (12). These mice show loss of a population of striatal neurons (11), suggesting possible failure of a protective mechanism and providing circumstantial evidence that the protease activity of HtrA2/Omi might have a function in protein quality control akin to that of its bacterial homologues. On the other hand, the Mnd2 mice still express HtrA2/Omi protein with a correctly processed amino-terminal Reaper motif, so they are not expected to show the effect of removing the IAP-antagonizing activity of HtrA2/Omi.
In this paper we report the phenotype of mice with a homozygous deletion of the gene encoding HtrA2/Omi. Unlike Mnd2 mice, these animals lack all expression of HtrA2/Omi and hence have neither its Reaper motif nor its protease activity. The phenotype of HtrA2/Omi knockout mice is extremely similar to that of Mnd2 mice, with early death resulting from the loss of a population of striatal neurons, suggesting that while the serine protease activity of HtrA2/Omi has a protective function, the amino-terminal Reaper motif does not have an obvious nonredundant proapoptotic role. To address whether the removal of both the major XIAP binding and the Reaper motif-containing proteins would reveal a physiological proapoptotic function for mammalian Reaper-related proteins, HtrA2/Omi knockout mice were crossed with animals in which the gene encoding Smac/DIABLO had been deleted. Loss of Smac/DIABLO did not further exacerbate the phenotype of the HtrA2/Omi knockout mice. We conclude that if Reaper motif-containing proteins play any physiological role in apoptosis regulation in mice, it is very highly redundant and must be mediated in addition by proteins other than the major reported IAP interactors. HtrA2/Omi, on the other hand, has a significant protective function that requires its protease activity and may be related to that of the homologous bacterial stress-adaptive proteins DegP and DegS.
MATERIALS AND METHODS
Generation of HtrA2/Omi-deficient and HtrA2/Omi Smac/DIABLO double-knockout mice.
The HtrA2/Omi locus was targeted in mouse embryonic day 14.5 (E14.5) (129/Ola) embryonic stem (ES) cells using standard ES cell culture and gene-targeting techniques (10, 13). The targeting vector was generated using DNA fragments from a 129/sv mouse Bac genomic clone (Invitrogen). The 5′ arm of homology was isolated as a 2.7-kb XhoI-BglII fragment, and the 3′ arm was isolated as a 2.1-kb HindIII-XhoI fragment. Targeting the HtrA2/Omi locus with this vector inserts the neomycin resistance gene, under the control of the PGK promoter (PGK-neo), in the first coding HtrA2/Omi exon at a position 140 bp downstream of the translational start codon. The PGK-neo gene replaces part of exon 1, exons 2 to 6, and part of exon 7 as well as intervening intronic sequences. This process removes 83% of the mouse HtrA2 coding sequence, including the proteolytic catalytic site of HtrA2/Omi. The targeting construct was linearized with PmeI and transfected into ES cells (Bio-Rad GenePulser; 230 kV, 500 μF). Transfected clones were selected in 180 μg of G418 (Sigma) per ml. A total of 300 G418-resistant ES cell colonies were isolated and screened for homologous recombination by Southern blot analysis. Three correctly targeted clones were identified using the screening strategies and probes indicated in the legend to Fig. 1. Two targeted clones were injected into C57BL6/J-derived blastocysts. Male chimeras from both clones were crossed with C57BL6/J females to produce F0 offspring. Heterozygous HtrA2/Omi offspring derived from one of the targeted ES cell clones were backcrossed for at least five generations onto the C57BL6/J strain. Mutant heterozygote F1 crosses were set up to generate study populations of animals containing all three possible genotypes (wild-type and heterozygous and homozygous mutants). Animal husbandry and experimental procedures were performed in full compliance with the United Kingdom Animal (Scientific Procedures) Act of 1986 and were approved by the GlaxoSmithKline UK procedures panel. Genotypes of mutant mice were determined by Southern blotting and confirmed by PCR as shown in Fig. 1B. Primers NeoF (5′-CCGGCCGCTTGGGTGGAGAGG-3′) and NeoR (5′-TCGGCAGGAGCAAGGTGAGATGACA-3′) specific for neo were used to detect the mutant allele. Primers AM04230701 (5′-CCCCGGATCTCTGGGCACGATTGAAT-3′) and AM05230701 (5′-ATCCCCGCTAGGCAGCCTCACTCGTA-3′), which are specific to sequences within exons 1 and 2, respectively, were used to detect the wild-type allele.
FIG. 1.
Targeting the HtrA2/Omi gene by homologous recombination. (A) Schematic representations of the wild-type mouse HtrA2/Omi (WT mHtrA2) locus, the targeting construct used in this study, and the targeted HtrA2/Omi allele. (B) Genotyping of the wild-type and mutant HtrA2/Omi loci by PCR. (C) Western blot analysis of protein lysates obtained from wild-type, HtrA2/Omi heterozygous, and HtrA2/Omi knockout MEFs.
To obtain double-knockout (HtrA2/Omi−/− Smac/DIABLO−/−) animals, we first generated compound heterozygous animals (HtrA2/Omi+/− Smac/DIABLO+/−). This F1 generation was intercrossed to generate double-knockout mice. Genotypes of mutant mice were determined by PCR. Primers AM04230701 and AM05230701 were used to detect the HtrA2/Omi wild-type locus, primers H2F_1-21 (5′-ATGGCTGCGCTGAAAGCGGGG-3′) and NeoR were used to detect the HtrA2/Omi mutant locus, and primers Smac-a, Smac-b, and Smac-c were used to detect the Smac/DIABLO wild-type and mutant loci as previously described (19).
Cell culture.
Primary mouse embryo fibroblasts (MEFs) were established from E14.5 embryos according to standard procedures (10). The cells were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum, l-glutamine, and antibiotics. To establish immortalized MEFs overexpressing the simian virus 40 large T antigen, primary MEFs were cultured in the presence of viral supernatants prepared by transfecting the Phoenix ecotropic retrovirus packaging cell line with pBabe Puro SV40LT. Cells that integrated the virus were selected in medium containing 2 μg of puromycin per ml (Sigma). Thymocytes were prepared from HtrA2/Omi+/+ and HtrA2/Omi−/− mice by standard procedures (22) and plated at 5 × 106/ml in 24-well plates in RPMI medium containing 10% fetal calf serum. Primary cortical neurons were isolated from E14.5 embryos and seeded at a density of 5 × 105 per well in 12-well plates coated with poly-l-lysine. Cells were maintained in Neurobasal medium (Invitrogen) supplemented with B27, Glutamax, and penicillin-streptomycin (Invitrogen). Following 3 days in culture, contamination from fibroblasts and glial cells was reduced by supplementing culture media with 10 μM cytosine arabinoside (Sigma). Cells were cultured for 10 days, with exchange of 50% of the medium volume with fresh medium every 2 days before treatment with toxic stimuli.
Inclined-platform test.
Mice were placed in the middle portion of a 60o-inclined platform, and evaluation was performed as previously described (23).
Histology and immunohistochemistry.
Brain tissues were harvested from 20- and 30-day-old mice and fixed in 10% formalin, embedded in paraffin, cut into 3-μm-thick sections, and processed for hematoxylin-eosin staining. In addition, immunostaining with the following stains was carried out according to the manufacturer's instructions: glial fibrillary acidic protein (GFAP; rabbit polyclonal antibody; dilution, 1:300; DAKO), NeuN (antineuronal nuclei; mouse monoclonal antibody; dilution, 1:4,000; Chemicon), MAP2 (microtubule-associated protein-2 mouse monoclonal antibody; dilution, 1:500; Chemicon), calbindin (rabbit polyclonal antiserum; dilution, 1:200; Chemicon), and tyrosine hydroxylase (TH; rabbit polyclonal antiserum; dilution, 1:1,000; Chemicon).
Biotinylated secondary antibodies were used for all staining, and visualization was with a horseradish peroxidase-conjugated streptavidin complex and diaminobenzidine as a chromogen. All immunostaining was carried out with the automated Nexus staining apparatus (Ventana Medical Systems) according to the manufacturer's guidelines. Photographs were obtained on a ColorView II digital camera (Soft Imaging System) mounted on a Zeiss Axioplan microscope and composed in Adobe Photoshop.
Stereological cell counts.
Immunohistochemistry was performed on serial sections (50 μm thick) of the striatum after the tissue had been fixed with paraformaldehyde cryoprotected in 30% (wt/vol) sucrose in phosphate buffer and frozen rapidly in liquid isopentane. Counts were performed by using the optical fractionator method as described previously (39). In agreement with this method, NeuN-positive neurons were counted in the left striatum of every third section throughout the entire extent of the striatum. Each section was viewed at low power (×2.5 objective), and the striatum was outlined. Then the number of NeuN-positive cells in the various groups of animals were counted at high power (×63 oil immersion lens). In another set, the first and fourth sections from the striatum, just lateral of the thalamus, in a relatively posterio-medial portion of the basal ganglia were outlined and cells in the lower half were counted with a stereological program (StereoInvestigator; Microbrightfield, Williston, Vt.). TH immunostaining was carried out on striatal and midbrain sections, and the TH-stained substantia nigra pars compacta neurons were counted by stereology using the optical fractionator method. The striatal density of TH immunoreactivity was determined as described previously (28).
Determination of mitochondrial enzyme activities.
A 4-mm-thick coronal slice of each mouse brain containing the medial striatum was isolated with a razor blade. The striatum was microdissected, placed in dry ice, and stored at −80°C until analysis. Prior to analysis, samples were homogenized at a concentration of 10% (wt/vol) in 10 mM Trizma base-1 mM EDTA and subjected to three cycles of freezing and thawing to lyse membranes. All activities were determined at 30°C. Enzyme activities were assessed using a Uvikon 940 spectrophotometer (Kontron Instruments Ltd., Watford, United Kingdom). Complex I (NADH ubiquinone reductase) activity was measured according to the method of Ragan and colleagues (20). Complexes II and III (succinate cytochrome c reductase) were measured according to the method of King (14). Complex IV (cytochrome c oxidase) was measured according to the method of Wharton and Tzagoloff (37). Citrate synthase was measured as described by Shepherd and Garland (24).
Morphological analysis of mitochondria.
Wild-type and HtrA2/Omi knockout MEFs were cultivated in Dulbecco modified Eagle medium containing 10% fetal bovine serum until confluence. Cells were then subjected to the following treatments for 30 min: 25 μM m-chlorocarbonyl cyanide phenylhydrazone (CCCP; Calbiochem) or 25 μM rotenone (Calbiochem). Following treatment, the medium was replaced and cells were further incubated at 37°C for 2 h. Following incubation, cells were processed for electron microscopy by gently replacing the culture medium with fixative solution (4% paraformaldehyde, 5% glutaraldehyde, and 5 mM CaCl2 in 0.2 M cacodylate buffer at pH 7.4) and incubating the mixture for 1 h at room temperature. Cells were postfixed with 1% osmium tetroxide-1% potassium ferrocyanide for 1 h at room temperature. Fixed cells were stained with 5% aqueous uranyl acetate overnight at room temperature and harvested by scraping. The resulting pellet was dehydrated and embedded in TAAB embedding resin. Ultrathin sections were stained with lead citrate and examined in a Jeol 100-CXII electron microscope equipped with a rotating-stage and eucentric goniometer. All quantitative assessments of mitochondrial morphology were based on scoring a minimum of 25 cells for each treatment. For each treatment, the number of mitochondria showing specific changes was determined.
Induction of cell death in MEFs, thymocytes, and primary neurons.
For induction of cell death in MEFs, cells (5 × 104 per well) were plated in 12-well plates and treated 24 h later with UV, etoposide (Sigma), or tumor necrosis factor alpha (TNF-α) (Roche) in the presence of 5 μg of cycloheximide (Sigma) per ml. Cells were harvested after 16 h. Cell viability was determined by staining cells with propidium iodide (Molecular Probes) according to the manufacturer's instructions, followed by flow cytometric analysis. For thymocyte cell death analysis, cells were incubated for 16 h in the presence or absence of anti-Fas antibody (Jo2; BD-Pharmingen), etoposide (Sigma), CCCP (Calbiochem), or rotenone (Calbiochem). Cells were harvested after 16 h, and viability was determined as described above. Immortalized MEFs were plated (2 × 105 per well) in six-well plates and treated 16 h later with CCCP, rotenone, tunicamycin (Sigma), or hydrogen peroxide (Sigma) for 27 h. DNA content was determined by staining cells with propidium iodide, followed by flow cytometric analysis as previously described (1). For neuron cell death analysis, cells were incubated in the presence or absence of glutamate (Sigma) for 4 h, fixed in 4% paraformaldehyde, and stained with Hoechst 33342 dye (Molecular Probes) according to the manufacturer's instructions, and nuclear chromatin was scored as being condensed or not condensed by fluorescence microscopy.
Immunoprecipitation of Smac/DIABLO complexes.
MEFs derived from HtrA2/Omi+/+ and HtrA2/Omi−/− animals were lysed in phosphate-buffered saline containing 1% Triton X-100 and a 1-μg/ml concentration (each) of chymostatin, leupeptin, antipain, and pepstatin A. Cell lysates were cleared by centrifugation at 10,000 × g and incubated in the presence of anti-Smac antibody (567360, 50 ng/ml; Calbiochem) in phosphate-buffered saline-1% Triton X-100. Antigen-antibody complexes were allowed to form at 4°C for 1 h and then precipitated using protein G-Sepharose beads. Proteins were resolved in a sodium dodecyl sulfate-12% polyacrylamide gel and transferred to an Immobilon-P membrane (Millipore). Immunoblot analysis was performed with the following antibodies: anti-Smac (567360; Calbiochem), anti-HtrA2/Omi as previously described (17), and anti-XIAP (AAM-050; Stressgen).
RESULTS
Deletion of HtrA2/Omi results in a neurological phenotype leading to death at 1 month postbirth.
In order to address the physiological role of HtrA2/Omi in vivo, we used homologous recombination in ES cells to disrupt the HtrA2/Omi gene and generate HtrA2/Omi knockout mice (Fig. 1A). The targeting vector was designed such that 83% of the coding region, encompassing part of exon 1, all of exons 2 to 6, and part of exon 7 were replaced with the PGK-neo cassette. We obtained three independent ES cell clones carrying the mutated HtrA2/Omi allele. Two targeted clones were injected into C57BL6/J-derived blastocysts. Male chimeras with high similarity percentages from both clones were crossed with C57BL6/J females to produce F0 offspring. Heterozygous HtrA2/Omi offspring derived from one of the targeted ES cell clones were backcrossed for at least five generations onto the C57BL6/J strain. Backcrossed HtrA2/Omi heterozygotes were then intercrossed to produce the F1 progeny. Analysis of genomic DNA prepared from tail biopsy samples by PCR (Fig. 1B) and Southern blotting confirmed the successful disruption of the HtrA2/Omi gene. To confirm that the knockout allele was a null mutation, MEFs were prepared from HtrA2/Omi+/+, HtrA2/Omi+/−, and HtrA2Omi−/− littermates and lysates were subjected to Western blotting to detect HtrA2/Omi expression. This analysis revealed the absence of HtrA2/Omi protein in cells derived from knockout animals and a partial reduction of protein levels in cells derived from embryos heterozygous for the HtrA2/Omi locus (Fig. 1C). Genotypic analysis of the F1 progeny by PCR revealed that out of 99 postbirth day 0 (P0) pups, 26 (26%) were wild type, 50 (50%) were heterozygous for the mutation, and 23 (23%) were homozygous mutants, consistent with normal Mendelian inheritance and indicated that deletion of the HtrA2/Omi gene does not result in embryonic lethality. Analysis of the resulting HtrA2/Omi−/− animals revealed that from P18, they displayed striking phenotypic alterations compared with wild-type or heterozygous littermates. The most dramatic alteration observed was failure to gain weight following weaning at P18 (Fig. 2A), resulting in small runted animals (Fig. 2B). Animals lacking HtrA2/Omi also show a general decrease in organ size. The degree of decrease in organ size varied, being particularly dramatic in the cases of thymus, heart, and spleen (Fig. 2C). Hair loss was occasionally seen in the HtrA2/Omi knockout mice.
FIG. 2.
Phenotypic alterations of HtrA2/Omi knockout mice. (A) Body weights of wild-type (HtrA2/Omi+/+), heterozygous (HtrA2/Omi+/−), and knockout (HtrA2/Omi−/−) littermates. Data shown are means ± standard deviations of results for at least six animals for each point. (B) Appearance of day 30 HtrA2/Omi knockout and littermate control wild-type animals. (C) HtrA2/Omi knockout mice show a general decrease in organ size. This decrease is more dramatic with the spleen, thymus, and heart; the brain is approximately 75% of the weight of wild-type control brains.
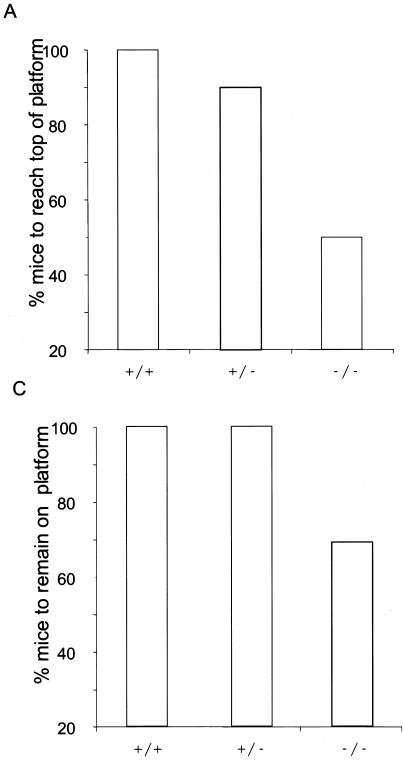
Examination of the behavior of HtrA2/Omi−/− animals revealed several neurological abnormalities. While wild-type and heterozygous mice extended their hind limbs and digits when suspended by their tails, from P24, mice lacking HtrA2/Omi retracted their hind feet and clenched their digits (Table 1). Animals lacking HtrA2/Omi are also characterized by a progressive movement disorder beginning at P18. They display a progressive akinetic, rigid syndrome, showing lack of coordination, decreased mobility, bended posture, and tremor (data available on request). Such features are characteristic of a parkinsonian syndrome and resulted in decreased performance in an inclined-platform test (Fig. 3A and B). The fact that HtrA2/Omi knockout mice took longer than control mice to climb to the top of inclined platform suggests that deletion of HtrA2/Omi results in deficiencies in coordination and/or strength. Although a subset of the knockout animals failed to remain on the platform for the 60-s trial period (Fig. 3C), 63% of knockout animals remained on the platform for the entire trial, suggesting that they were not grossly impaired in muscle strength and that disturbances in coordination were principally responsible for their poor performance.
TABLE 1.
Assessment of neuromuscular abnormalities in wild-type and HtrA2/Omi knockout mice by determining hind leg posture following suspension by the tail
| Mouse HtrA2/Omi genotype | Posturea on day:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | |
| +/+ and +/− | N | N | N | N | N | N | N | N |
| −/− | N | N | A | A | A | A | A | A |
N, normal posture with extended hind limbs and digits; A, abnormal with retracted hind feed and clenched digits. Each data point is the result of analysis of at least three animals.
FIG. 3.
Sensorimotor tests on knockout mice. HtrA2+/+, HtrA2+/−, and HtrA2−/− mice were placed on a 60o-inclined platform. (A) Percentages of mice that climbed to the top of the platform. Wild-type (+/+; n = 8); heterozygote (+/−; n = 20), and knockout (−/−; n = 16) littermates were compared. (B) Average elapsed times for mice to climb to the top of the platform. Data are means ± standard deviations of results from a minimum of eight trials. (C) Percentages of mice that remained on the platform for the full 60-s trial period. Wild-type (+/+; n = 8), heterozygote (+/−; n = 20), and knockout (−/−; n = 16) littermates were compared.
HtrA2/Omi deletion results in the selective loss of a population of striatal neurons.
The observation that HtrA2/Omi knockout animals displayed features of a parkinsonian syndrome led us to investigate whether these animals possessed either presynaptic deficits by analyzing the substantia nigra or postsynaptic deficits by analyzing the striatum. Analysis of TH-positive neurons in the substantia nigra of HtrA2/Omi knockout animals revealed no loss of dopaminergic neurons compared with those of wild-type littermates (Fig. 4G).
FIG. 4.
Morphological features of localized striatal degeneration in HtrA2/Omi knockout mice. (A) Hematoxylin and eosin staining showing focal loss of neurons and occasional ballooning in the striata of HtrA2/Omi knockout animals (right) compared to controls (+/+) (left). (B) GFAP immunohistochemistry revealing the reaction of astrocytes in knockout animals (right). (C) MAP2 immunohistochemistry revealing the loss of this protein in sections from knockout animals (right). (D) Diagram of a mouse brain in sagital orientation indicating the area of striatal degeneration (red box) in the striatum. (E and F) Density of NeuN-positive neurons in the striata (Str) of wild-type and HtrA2/Omi knockout mice scored for both the whole striatum (total basal ganglia) and the area where localized degeneration was observed (posterior portion of the basal ganglia). Analysis was performed with both P20 and P30 animals. (G and H) Average optical density of TH staining in both the total basal ganglia and the area where localized degeneration was observed (posterior portion of the basal ganglia). (I) Average number of TH-positive neurons in the substantia nigra pars compacta of wild-type and HtrA2/Omi knockout animals.
We next examined the neuronal circuits in the striata of HtrA2/Omi knockout animals. P30 knockout animals were analyzed, as they displayed the most severe akinetic phenotypes. Analysis of brains from HtrA2/Omi knockout animals revealed a selective loss of a neuronal population in the striatum, just lateral of the thalamus in a relatively posteriomedial portion of the basal ganglia (Fig. 4D). In the hematoxylin- and eosin-stained sections (Fig. 4A), we identified focal loss of neurons, with occasional ballooned cells and concomitant reaction of the astrocytes, as highlighted in a GFAP immunostain (Fig. 4B). This loss of neurons is more clearly seen in sections immunostained for the microtubule-associated protein MAP2, a neuronal marker (Fig. 4C). We could detect occasional apoptotic cell bodies (data not shown). In order to rule out a general and widespread loss of neurons in the striatum, we performed stereological cell counts of striatal neurons. These results indicated that there is no significant change in the overall neuronal density in the striata of HtrA2/Omi knockout animals (Fig. 4E). On the other hand, when we performed a similar analysis of the posterior portion of the basal ganglia, we detected a decrease of neuronal density in brains from HtrA2/Omi knockout animals both at P20, when no phenotypic alterations are detectable, and at P30 (Fig. 4F). Concomitant with this neuronal loss, we detected a selective loss of terminals of the nigrostriatal pathway in this area (Fig. 4I) compared to the terminals of the whole striatum (Fig. 4H). At present, we do not know if this loss of terminals is a cause or a consequence of the neuronal loss detected.
Deletion of HtrA2/Omi results in a mitochondrial dysfunction.
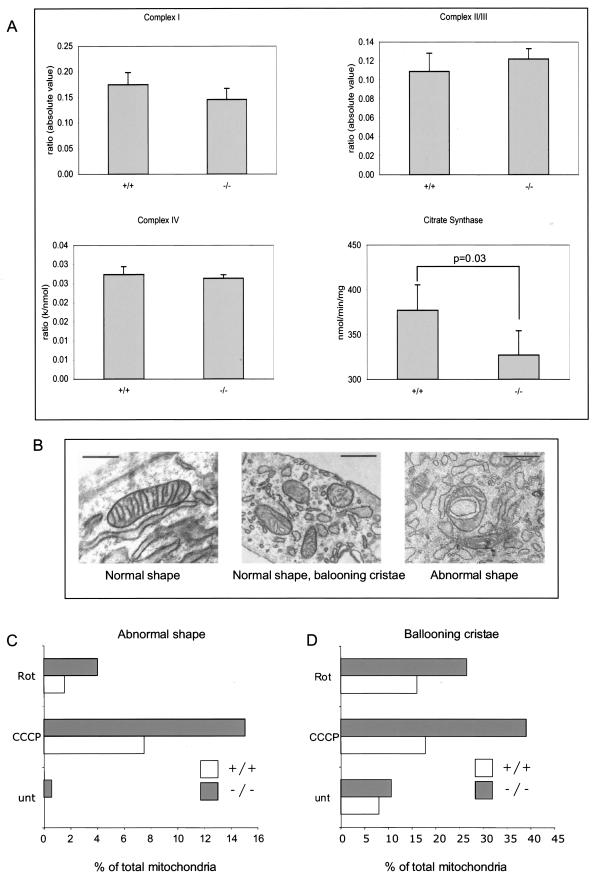
The serine protease HtrA2/Omi is localized to the intermembrane space of mitochondria of healthy cells. In order to determine if HtrA2/Omi's absence resulted in a compromise of mitochondrial metabolism, we analyzed at the biochemical level the components of the mitochondrial electron transport chain in extracts prepared from the striata of HtrA2/Omi+/+ and HtrA2/Omi−/− animals. This analysis failed to reveal any significant compromise of the function of complex I, II, III, or IV (Fig. 5A) when enzymatic activities were normalized against the activity of the mitochondrial matrix enzyme citrate synthase, to account for differences in mitochondrial density in the samples. However, extracts prepared from HtrA2/Omi knockout animals showed a significant decrease of the yield of mitochondrial citrate synthase (Fig. 5A), indicating that mitochondrial density is likely to be reduced in tissues derived from knockout animals.
FIG. 5.
Mitochondrial function in wild-type and HtrA2/Omi knockout animals. (A) Activities of mitochondrial complexes I, II and III, and IV corrected for mitochondrial enrichment by normalizing the activity against the mitochondrial matrix enzyme citrate synthase; values for detected citrate synthase activities are also shown. Extracts were prepared from striata of P30 control (+/+) and HtrA2/Omi knockout (−/−) animals. A statistically significant (t test analysis) decrease in the amount of citrate synthase activity was detected in samples from HtrA2/Omi−/− animals. (B) Ultrastructural features of mitochondria present in cells from wild-type and HtrA2/Omi knockout animals. Scale bars correspond to 500 nm. (C and D) Quantification of mitochondrial morphological abnormalities detected in cells before and after treatment with mitochondrial stress stimuli. Results are representative of three independent experiments performed on three to five different cell lines isolated from different animals. Rot, rotenone; unt, untreated.
The decrease in mitochondrial enrichment in tissues derived from HtrA2/Omi-deficient animals suggested that cells lacking this protease might be more susceptible to cellular stresses that affect these organelles. To test this possibility, we cultured cells derived from control and HtrA2/Omi knockout animals and compared mitochondrial morphologies at the ultrastructural level following treatment with mitochondrial-stress-inducing agents. Using transmission electron microscopy, we observed three types of previously described mitochondria (4) in untreated cells (Fig. 5B): normal mitochondria, mitochondria displaying ballooning of cristae (intracristal swelling), and mitochondria with abnormal forms (the last were detected only in cells from knockout animals). When cells were subjected to mitochondrial stresses in the form of the complex I inhibitor rotenone and the mitochondrial membrane potential uncoupler CCCP, we found that cells lacking HtrA2/Omi contained an increased percentage of mitochondria characterized by abnormal morphological alterations (Fig. 5C and D). We therefore conclude that deletion of HtrA2/Omi may confer on mitochondria an increased sensitivity to stress.
Deletion of HtrA2/Omi results in increased susceptibility to cell death stimuli.
The observation that mitochondria in cells lacking HtrA2/Omi displayed an increased susceptibility to cellular stresses led us to ask whether this increased sensitivity could affect cell viability. We therefore subjected thymocytes from control and HtrA2/Omi knockout animals to classical apoptosis-inducing agents that activate both the intrinsic and extrinsic pathways, as well as drugs that specifically target mitochondria. Our results indicate that although cells lacking HtrA2/Omi do not show an altered sensitivity to agents that trigger the extrinsic cell death pathway, such as Fas agonistic antibodies (Fig. 6A), they display increased sensitivity to agents that trigger apoptosis through the intrinsic pathway, such as the DNA-damaging agent etoposide (Fig. 6B), and agents that perturb mitochondrial function, such as CCCP and rotenone (Fig. 6C and D). Analysis of immortalized MEFs derived from either wild-type or HtrA2/Omi knockout animals was consistent with an increased sensitivity of cells lacking HtrA2/Omi to mitochondrial stress stimuli as well as agents that cause both oxidative stress (hydrogen peroxide) and endoplasmic reticulum stress (tunicamycin) (Fig. 6E).
FIG. 6.
Deletion of HtrA2/Omi results in increased sensitivity to mitochondrion-damaging agents. Thymocytes were isolated from control and HtrA2/Omi knockout animals and incubated with increasing concentrations of anti-Fas antibody (A), etoposide (B), or the mitochondrion-damaging agents CCCP (C) and rotenone (D). Viability was determined 16 h after treatment by flow cytometry using propidium iodide. Results are representative of three independent experiments. (E) Viability of simian virus 40 large-T-antigen-immortalized MEFs derived from wild-type (+/+) and HtrA2/Omi knockout animals, determined by measurement of sub-G1 cell populations by flow cytometry. Cells were incubated in the presence of CCCP (25 μM for 27 h), rotenone (25 μM for 27 h), tunicamycin (2.5 μg/ml for 27 h), or hydrogen peroxide (3 μM for 4.5 h) and compared to untreated control cells. Results show the means ± standard deviations of results of three independent experiments. (F) The neuronalresponse to glutamate-induced cytotoxicity was determined in primary neurons isolated from E14.5 embryos cultured in vitro for 10 days. Following incubation with the indicated concentrations of glutamate for 4 h, cells were stained with Hoechst 33342 and nuclear morphology was determined. Results show the means ± standard deviations of results from two independent experiments where six fields with at least 40 cells were scored for each data point.
The observation that neuronal cells showed clear signs of degeneration in HtrA2/Omi knockout animals led us to ask whether cultured neurons isolated from knockout mice were more sensitive than wild-type controls to excitotoxic stimuli. We isolated primary cortical neurons from control and HtrA2/Omi knockout animals and subjected these to glutamate-mediated excitotoxicity. These experiments revealed a significant increase in the susceptibility of neurons obtained from HtrA2/Omi knockout animals to excitotoxic stimuli (Fig. 6E).
Endogenous Smac/DIABLO is capable of sequestering all XIAP in cells lacking HtrA2/Omi.
The results presented so far indicate that, rather than protecting cells from apoptosis, the lack of HtrA2/Omi results in increased sensitivity to some death stimuli. It is possible that the presence of the other major Reaper motif-containing protein, Smac/DIABLO, might compensate for the removal of HtrA2/Omi in terms of its ability to neutralize IAPs. To explore this possibility, we immunoprecipitated Smac/DIABLO from lysates of MEFs from wild-type or HtrA2/Omi knockout mice with close to 100% efficiency (Fig. 7). Removal of Smac/DIABLO resulted in the depletion of the major portion of the caspase inhibitor XIAP from wild-type cell lysates and almost all of XIAP from HtrA2/Omi knockout cell lysates (Fig. 7, upper panel, middle section, which shows the amount of XIAP in cell lysates from which Smac/DIABLO has been immunodepleted). This depletion indicates that there is sufficient Smac/DIABLO present in cells to compensate for the loss of HtrA2/Omi in terms of its potential ability once released from mitochondria to bind XIAP and neutralize its caspase-inhibitory activity.
FIG. 7.
Immunoprecipitation of Smac/DIABLO from wild-type and HtrA2/Omi knockout cells effectively depletes endogenous XIAP. Lysates were prepared from MEFs and incubated with an anti-Smac/DIABLO monoclonal antibody. Following separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blot analysis was performed using antibodies to detect endogenous XIAP, Smac/DIABLO, and HtrA2/Omi. “I” indicates the input lysates (10% of the total immunoprecipitated proteins loaded), “ID” indicates the equivalent amount of lysate following immunodepletion, and “IP” indicates the immunopurified eluate (50% of the total protein equivalent).
Mice with both the Smac/DIABLO and the HtrA2/Omi gene deleted display a phenotype identical to that of HtrA2/Omi knockout animals.
Since cells lacking HtrA2/Omi failed to confer enhanced resistance to apoptotic stimuli, we decided to investigate the consequence of removal of both HtrA2/Omi and Smac/DIABLO. In order to delete the genes for both HtrA2/Omi and Smac/DIABLO from mice, we first crossed animals heterozygous for HtrA2/Omi (HtrA2/Omi+/−) with animals heterozygous for Smac/DIABLO (Smac/DIABLO+/−) (19). From these crosses we identified F1 compound heterozygotes for both Smac/DIABLO and HtrA2/Omi mutant alleles (Smac/DIABLO+/− HtrA2/Omi+/−). We then proceeded to intercross these compound heterozygotes to generate F2 progeny. Genotypic analysis of the F2 progeny by PCR (Fig. 8A) revealed that out of 173 P0 pups, 9 (5.2%) were homozygous for both the Smac/DIABLO and HtrA2/Omi mutations. These numbers are consistent with the expected Mendelian inheritance (6.25%) and indicate that deletion of both genes does not result in embryonic lethality. Analysis of F2 animals lacking both Smac/DIABLO and HtrA2/Omi indicated that these are again characterized by a failure to gain weight (Fig. 8B), resulting in small runted animals and a progressive movement disorder beginning at P18, identical to the one found in HtrA2/Omi knockout animals. Postmortem analysis of double-knockout animals failed to reveal any alterations specific to the animals with both genes deleted. We therefore conclude that deletion of Smac/DIABLO and HtrA2/Omi confers a phenotype similar to the one found in mice lacking only HtrA2/Omi.
FIG. 8.
Analysis of HtrA2/Omi Smac/DIABLO double-knockout animals. (A) Detection of the wild-type and mutant HtrA2/Omi and Smac/DIABLO loci in the offspring of animals with a heterozygous deletion of the genes for both HtrA2/Omi and Smac/DIABLO. PCR was performed on genomic tail DNA templates. (B) Average body weights of Smac/DIABLO+/+ HtrA2/Omi+/+ (wild-type [wt]) and Smac/DIABLO−/− HtrA2/Omi−/− (double-knockout [KO]) mice. (C and D) The sensitivities of wild-type and double-knockout MEFs to TNF-α (C), and etoposide (D) are shown. Viability was determined 16 h following treatment by flow cytometry using propidium iodide. Results show the means ± standard deviations of results from three independent experiments.
Deletion of both Smac/DIABLO and HtrA2/Omi fails to confer enhanced resistance to apoptotic stimuli.
In order to characterize the cellular response to apoptotic stimuli in animals with both HtrA2/Omi and Smac/DIABLO deleted, we isolated mouse embryonic fibroblasts from these animals and compared the levels of apoptosis with that of wild-type controls. Treatment of cells with apoptosis-inducing agents acting via death receptors, such as TNF-α, revealed that the levels of cell death in wild-type and double-knockout cells were identical (Fig. 8C). However, as was observed in cells lacking solely HtrA2/Omi, cells lacking both HtrA2/Omi and Smac/DIABLO displayed increased sensitivity to apoptosis induction by the DNA-damaging agent etoposide (Fig. 8D). We conclude that deletion of both genes for the mammalian IAP interactors HtrA2/Omi and Smac/DIABLO does not result in resistance to apoptosis in the cell types analyzed but rather in an increased sensitivity to some death-inducing agents.
DISCUSSION
When the mammalian Reaper motif-containing proteins Smac/DIABLO and HtrA2/Omi were first identified through their ability to interact with XIAP (2, 8, 17, 27, 34, 35), it appeared likely that these proteins were released from mitochondria following apoptotic insult along with cytochrome c and then acted together to block the caspase-inhibitory function of IAPs, thus promoting caspase activation and apoptosis. While overexpression of Smac/DIABLO or HtrA2/Omi certainly promotes caspase-mediated apoptotic death and also, in the case of HtrA2/Omi, caspase-independent cell death, the significance of the endogenous proteins in these pathways has been much less clear. The data presented here indicate that the complete loss of expression of HtrA2/Omi protein, along with its Reaper motif, does not obviously perturb mouse development and health beyond the effects seen due to the loss resulting from a point mutation of only the serine protease activity of HtrA2/Omi: the phenotype of the HtrA2/Omi knockout mouse is very similar to that of the Mnd2 mouse (11, 12, 21). Similarly, a total loss of Smac/DIABLO expression has no effect on mouse development or function (19). Mice completely lacking both HtrA2/Omi and Smac/DIABLO expression have a phenotype similar to that of Mnd2 mice lacking only HtrA2/Omi protease activity. Therefore, the loss of the two major XIAP binding Reaper motifs does not have any obvious effect on the phenotype of mice that can be detected within the limited life span of animals lacking HtrA2/Omi serine protease activity. Similarly, the loss of both HtrA2/Omi and Smac/DIABLO expression from cultured cells taken from the double-knockout mice does not protect them from cell death but rather sensitizes them to some death stimuli.
Why might the loss of these two Reaper motifs have so little effect in vivo? The possibility that the Reaper/IAP system is not a major regulator of cell death in mammals cannot be ruled out. While clearly of great importance in flies, the system does not exist in worms. Although at least seven mammalian IAPs have been identified, it could be argued that no true Reaper, Hid, or Grim analogues have been found. On the other hand, it is also possible that an effect might become apparent later in life in mice with a healthy genetic background whose life spans are not restricted to approximately 4 weeks due to the loss of HtrA2/Omi protease activity. In order to address this possibility rigorously, it will be necessary to create mice in which the Reaper motif (AVP) of HtrA2/Omi has been replaced by a gene with a sequence unable to bind to IAPs knocked in, such as GVP. A third possibility is that, even though Smac/DIABLO and HtrA2/Omi are the major IAP binding proteins in most cells studied, there may exist a much greater degree of redundancy in Reaper motif-containing proteins than previously suspected. It has been suggested that caspase cleavage of proteins may frequently generate novel amino termini with alanine residues that may be able to bind to IAPs in a Reaper-like manner (9). This possible ability to bind IAPs might provide feed forward amplification of caspase activation once activation is under way, although it seems unlikely to be able to promote the initiation of caspase activation. It is also possible that many mitochondrial proteins might have some form of amino-terminal Reaper motif (32). In this model, severe mitochondrial damage would cause the release of many proteins with the ability to antagonize IAPs, not just Smac/DIABLO and HtrA2/Omi, so the loss of the Reaper motifs of these two may have little impact. The reason for the possible frequent appearance of amino-terminal Reaper motifs in mitochondrial proteins may relate to the N end rule (31), under which amino-terminal alanine residues are destabilizing. Thus, a low-level release of mitochondrial proteins into the cytosol during normal cell function may be mopped up effectively by the proteasome machinery.
Data presented here in Fig. 7 show that Smac/DIABLO is clearly the most efficient XIAP binding protein in detergent lysates of cells: in the absence of HtrA2/Omi, all XIAP is complexed to Smac/DIABLO. In the presence of HtrA2/Omi, a minor fraction of XIAP is not complexed to Smac/DIABLO, presumably because it is bound to HtrA2/Omi. Thus, if there are other physiological Reaper motif-containing XIAP binding proteins in cells, it is unlikely that they will be found readily in the presence of competing Smac/DIABLO. However, it should be noted that all the XIAP complexes seen in coimmunoprecipitations from detergent cell lysates must have been formed after cell and organelle disruption. Another Reaper motif-containing XIAP binding protein has been identified, GSPT1/eRF3 (7), although this protein is not localized to the mitochondria and so would be unlikely to contribute to caspase regulation in the manner proposed for HtrA2/Omi and Smac/DIABLO.
If the Reaper motif at the processed amino terminus of HtrA2/Omi does not play a major nonredundant role in cell death regulation, what can be concluded about the function of the protease domain of HtrA2/Omi? As in Mnd2 mice, a population of neurons in the striata of HtrA2/Omi knockout mice can be seen to be lost from about 20 days of age. In the case of the Mnd2 mice, this cell death, and the mobility defect, is not blocked by Bcl2 overexpression, suggesting that apoptosis is not the sole cell death mechanism at work (21). Astrogliosis is detected by GFAP staining in this area. Neuronal cell populations isolated from the knockout mice are more sensitive than wild-type controls to excitotoxicity induced by glutamate. Nonneuronal cell types, such as fibroblasts and T lymphocytes, from the HtrA2/Omi knockout mice also show increased sensitivity to induction of cell death by DNA-damaging agents and inhibitors that target the mitochondria, such as the complex I inhibitor rotenone and the uncoupler CCCP.
Since HtrA2/Omi knockout mice develop a phenotype with parkinsonian features starting at about day 25 (unpublished data), it will be interesting to analyze patients with Parkinson's disease and parkinsonian syndromes for HtrA2/Omi deficits. We did not detect a degeneration of TH-positive dopaminergic neurons in the substantia nigra pars compacta, as occurs in Parkinson's disease patients. However, our data show a presynaptic and postsynaptic defect of the nigrostriatal pathway in the striatum: in a frontal area of the striatum there is a loss of neurons as well as a decrease of TH-positive fibers. This finding suggests similarity to some parkinsonian syndromes.
While HtrA2/Omi is ubiquitously expressed, it seems that a localized population of striatal neurons are uniquely dependent on its protease function. It is possible that HtrA2/Omi acts to protect these cells against some forms of pathogenic stress. Stresses that have been implicated in the death of neurons in vivo include oxidative stress, excitotoxic stress, and the accumulation of denatured proteins, including those with expanded polyglutamine repeats. Due to its mitochondrial localization, HtrA2/Omi probably acts to protect these organelles, and in particular the mitochondrial intermembrane compartment, from damage resulting from the stresses to which striatal neurons are normally exposed. In this regard, the similarity to the bacterial HtrA homologues, especially DegS, is particularly striking. This protein has exactly the same domain structure as HtrA2. It is localized in the periplasmic space between the inner and outer bacterial cell membranes, with its amino-terminal region inserted into the inner membrane: this localization is clearly analogous to that of full-length HtrA2/Omi within the mitochondria. The appearance of the exposed C termini of unfolded porins, outer membrane channel proteins, in the periplasmic space results in activation of the protease domain of DegS due their binding to its PDZ domain (36). Similarly, engagement of the PDZ domain of HtrA2/Omi results in activation of its protease activity (15, 18), although in the case of HtrA2/Omi its natural PDZ binding partners within the mitochondria remain to be identified. Once activated, DegS cleaves RseA, an inner membrane protein which, together with RseB, sequesters the transcriptional regulator σE at the cytoplasmic face of the inner membrane. It is interesting that DegS cleaves RseA between a valine and a serine (36), similar to the optimal peptide substrate cleavage for human HtrA2/Omi (18). Cleavage of RseA by DegS leads to further proteolysis by the intramembrane protease YaeL, resulting in the release of σE into the cytosol to induce the expression of periplasmic stress response genes (36). While the similarity between HtrA2/Omi and DegS is evident, it is not possible to identify obvious mammalian homologues of other downstream components of the bacterial periplasmic stress response system.
If HtrA2/Omi does act in a manner similar to that of DegS, it is at present unclear exactly what cellular stresses might stimulate the protease activity of HtrA2/Omi. The exact nature of the cell damage caused in striatal neurons lacking HtrA2/Omi is also unknown: there is no obvious defect in total brain tissue in the functions of the different mitochondrial electron transport complexes or the generation of reduced glutathione. However, it is possible that these could be altered in a small population of neurons that would be difficult to detect by biochemical assay of whole tissue extracts. The main effect on mitochondrial function seen in liver and in cultured fibroblasts from the HtrA2/Omi knockout mice is the reduction in mitochondrial membrane potential relative to that of controls, as was also reported for Mnd2 mice (12). If it acts like DegS, the loss of HtrA2/Omi may result in an inability to mount an adaptive transcriptional response under stress conditions that cause protein unfolding, leaving the mitochondria of striatal neurons unable to maintain their membrane potential and rendering the cells prone to either necrotic or apoptotic death. Alternatively, if HtrA2/Omi functions like another E. coli homologue, DegP (25), mitochondria from knockout mice may be unable to degrade unfolded proteins in the intermembrane space under conditions of cell stress, and these could form aggregates that would directly compromise the maintenance of the mitochondrial membrane potential. Which of these different models of HtrA2/Omi's neuroprotective function is correct may be clarified once its mitochondrial binding partners are identified and once differences in stress-induced transcriptional responses are determined.
Acknowledgments
We thank members of P. Nicotera's laboratory for help with neuronal cell culture techniques and Greg Gasic for advice.
Kristina Klupsch was funded by a studentship from the Boehringer Ingelheim Fonds.
REFERENCES
- 1.Darzynkiewicz, Z., and E. Bedner. 2000. Analysis of apoptotic cells by flow and laser scanning cytometry. Methods Enzymol. 322:18-39. [DOI] [PubMed] [Google Scholar]
- 2.Du, C., M. Fang, Y. Li, L. Li, and X. Wang. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102:33-42. [DOI] [PubMed] [Google Scholar]
- 3.Faccio, L., C. Fusco, A. Chen, S. Martinotti, J. V. Bonventre, and A. S. Zervos. 2000. Characterization of a novel human serine protease that has extensive homology to bacterial heat shock endoprotease HtrA and is regulated by kidney ischemia. J. Biol. Chem. 275:2581-2588. [DOI] [PubMed] [Google Scholar]
- 4.Ghadially, F. N. 1982. Ultrastructural pathology of the cell and matrix, 2nd ed. Butterworths, London, United Kingdom.
- 5.Gray, C. W., R. V. Ward, E. Karran, S. Turconi, A. Rowles, D. Viglienghi, C. Southan, A. Barton, K. G. Fantom, A. West, J. Savopoulos, N. J. Hassan, H. Clinkenbeard, C. Hanning, B. Amegadzie, J. B. Davis, C. Dingwall, G. P. Livi, and C. L. Creasy. 2000. Characterization of human HtrA2, a novel serine protease involved in the mammalian cellular stress response. Eur. J. Biochem. 267:5699-5710. [DOI] [PubMed] [Google Scholar]
- 6.Harlin, H., S. B. Reffey, C. S. Duckett, T. Lindsten, and C. B. Thompson. 2001. Characterization of XIAP-deficient mice. Mol. Cell. Biol. 21:3604-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hegde, R., S. M. Srinivasula, P. Datta, M. Madesh, R. Wassell, Z. Zhang, N. Cheong, J. Nejmeh, T. Fernandes-Alnemri, S. Hoshino, and E. S. Alnemri. 2003. The polypeptide chain-releasing factor GSPT1/eRF3 is proteolytically processed into an IAP-binding protein. J. Biol. Chem. 278:38699-38706. [DOI] [PubMed] [Google Scholar]
- 8.Hegde, R., S. M. Srinivasula, Z. Zhang, R. Wassell, R. Mukattash, L. Cilenti, G. DuBois, Y. Lazebnik, A. S. Zervos, T. Fernandes-Alnemri, and E. S. Alnemri. 2002. Identification of Omi/HtrA2 as a mitochondrial apoptotic serine protease that disrupts inhibitor of apoptosis protein-caspase interaction. J. Biol. Chem. 277:432-438. [DOI] [PubMed] [Google Scholar]
- 9.Hell, K., M. Saleh, G. D. Crescenzo, M. D. O'Connor-McCourt, and D. W. Nicholson. 2003. Substrate cleavage by caspases generates protein fragments with Smac/Diablo-like activities. Cell Death Differ. 10:1234-1239. [DOI] [PubMed] [Google Scholar]
- 10.Hogan, B., R. Beddington, F. Costantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 11.Jones, J. M., R. L. Albin, E. L. Feldman, K. Simin, T. G. Schuster, W. A. Dunnick, J. T. Collins, C. E. Chrisp, B. A. Taylor, and M. H. Meisler. 1993. mnd2: a new mouse model of inherited motor neuron disease. Genomics 16:669-677. [DOI] [PubMed] [Google Scholar]
- 12.Jones, J. M., P. Datta, S. M. Srinivasula, W. Ji, S. Gupta, Z. Zhang, E. Davies, G. Hajnoczky, T. L. Saunders, M. L. Van Keuren, T. Fernandes-Alnemri, M. H. Meisler, and E. S. Alnemri. 2003. Loss of Omi mitochondrial protease activity causes the neuromuscular disorder of mnd2 mutant mice. Nature 425:721-727. [DOI] [PubMed] [Google Scholar]
- 13.Joyner, A. L. 1999. Gene targeting: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 14.King, T. E. 1967. Preparations of succinate cytochrome c reductase and cytochrome b-c1 particle and reconstruction of succinate cytochrome c reductase. Methods Enzymol. 10:446-451. [Google Scholar]
- 15.Li, W., S. M. Srinivasula, J. Chai, P. Li, J. W. Wu, Z. Zhang, E. S. Alnemri, and Y. Shi. 2002. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9:436-441. [DOI] [PubMed] [Google Scholar]
- 16.Martins, L. M. 2002. The serine protease Omi/HtrA2: a second mammalian protein with a Reaper-like function. Cell Death Differ. 9:699-701. [DOI] [PubMed] [Google Scholar]
- 17.Martins, L. M., I. Iaccarino, T. Tenev, S. Gschmeissner, N. F. Totty, N. R. Lemoine, J. Savopoulos, C. W. Gray, C. L. Creasy, C. Dingwall, and J. Downward. 2002. The serine protease Omi/HtrA2 regulates apoptosis by binding XIAP through a reaper-like motif. J. Biol. Chem. 277:439-444. [DOI] [PubMed] [Google Scholar]
- 18.Martins, L. M., B. E. Turk, V. Cowling, A. Borg, E. T. Jarrell, L. C. Cantley, and J. Downward. 2003. Binding specificity and regulation of the serine protease and PDZ domains of HtrA2/Omi. J. Biol. Chem. 278:49417-49427. [DOI] [PubMed] [Google Scholar]
- 19.Okada, H., W. K. Suh, J. Jin, M. Woo, C. Du, A. Elia, G. S. Duncan, A. Wakeham, A. Itie, S. W. Lowe, X. Wang, and T. W. Mak. 2002. Generation and characterization of Smac/DIABLO-deficient mice. Mol. Cell. Biol. 22:3509-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragan, C. I., M. Y. Wilson, and V. M. Darley-Usmar. 1998. Subfractionation of mitochondria and isolation of proteins of oxidative phosphorylation, p. 79-113. In D. Rickwood and M. T. Wilson (ed.), Mitochondria: a practical approach. Oxford IRL Press, Oxford, United Kingdom.
- 21.Rathke-Hartlieb, S., U. Schlomann, P. Heimann, M. H. Meisler, H. Jockusch, and J. W. Bartsch. 2002. Progressive loss of striatal neurons causes motor dysfunction in MND2 mutant mice and is not prevented by Bcl-2. Exp. Neurol. 175:87-97. [DOI] [PubMed] [Google Scholar]
- 22.Ruland, J., G. S. Duncan, A. Elia, I. del Barco Barrantes, L. Nguyen, S. Plyte, D. G. Millar, D. Bouchard, A. Wakeham, P. S. Ohashi, and T. W. Mak. 2001. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell 104:33-42. [DOI] [PubMed] [Google Scholar]
- 23.Schaefer, M. L., S. T. Wong, D. F. Wozniak, L. M. Muglia, J. A. Liauw, M. Zhuo, A. Nardi, R. E. Hartman, S. K. Vogt, C. E. Luedke, D. R. Storm, and L. J. Muglia. 2000. Altered stress-induced anxiety in adenylyl cyclase type VIII-deficient mice. J. Neurosci. 20:4809-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shepherd, J. A., and P. B. Garland. 1969. Citrate synthase activity from rat liver. Methods Enzymol. 13:11-19. [Google Scholar]
- 25.Spiess, C., A. Beil, and M. Ehrmann. 1999. A temperature-dependent switch from chaperone to protease in a widely conserved heat shock protein. Cell 97:339-347. [DOI] [PubMed] [Google Scholar]
- 26.Srinivasula, S. M., S. Gupta, P. Datta, Z. Zhang, R. Hegde, N. Cheong, T. Fernandes-Alnemri, and E. S. Alnemri. 2003. Inhibitor of apoptosis proteins are substrates for the mitochondrial serine protease Omi/HtrA2. J. Biol. Chem. 278:31469-31472. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki, Y., Y. Imai, H. Nakayama, K. Takahashi, K. Takio, and R. Takahashi. 2001. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol. Cell 8:613-621. [DOI] [PubMed] [Google Scholar]
- 28.Teismann, P., K. Tieu, D. K. Choi, D. C. Wu, A. Naini, S. Hunot, M. Vila, V. Jackson-Lewis, and S. Przedborski. 2003. Cyclooxygenase-2 is instrumental in Parkinson's disease neurodegeneration. Proc. Natl. Acad. Sci. USA 100:5473-5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Gurp, M., N. Festjens, G. van Loo, X. Saelens, and P. Vandenabeele. 2003. Mitochondrial intermembrane proteins in cell death. Biochem. Biophys. Res. Commun. 304:487-497. [DOI] [PubMed] [Google Scholar]
- 30.van Loo, G., X. Saelens, M. van Gurp, M. MacFarlane, S. J. Martin, and P. Vandenabeele. 2002. The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet. Cell Death Differ. 9:1031-1042. [DOI] [PubMed] [Google Scholar]
- 31.Varshavsky, A. 2003. The N-end rule and regulation of apoptosis. Nat. Cell Biol. 5:373-376. [DOI] [PubMed] [Google Scholar]
- 32.Vaux, D. L., and J. Silke. 2003. HtrA2/Omi, a sheep in wolf's clothing. Cell 115:251-253. [DOI] [PubMed] [Google Scholar]
- 33.Vaux, D. L., and J. Silke. 2003. Mammalian mitochondrial IAP binding proteins. Biochem. Biophys. Res. Commun. 304:499-504. [DOI] [PubMed] [Google Scholar]
- 34.Verhagen, A. M., P. G. Ekert, M. Pakusch, J. Silke, L. M. Connolly, G. E. Reid, R. L. Moritz, R. J. Simpson, and D. L. Vaux. 2000. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 102:43-53. [DOI] [PubMed] [Google Scholar]
- 35.Verhagen, A. M., J. Silke, P. G. Ekert, M. Pakusch, H. Kaufmann, L. M. Connolly, C. L. Day, A. Tikoo, R. Burke, C. Wrobel, R. L. Moritz, R. J. Simpson, and D. L. Vaux. 2002. HtrA2 promotes cell death through its serine protease activity and its ability to antagonize inhibitor of apoptosis proteins. J. Biol. Chem. 277:445-454. [DOI] [PubMed] [Google Scholar]
- 36.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 37.Wharton, D. C., and A. Tzagoloff. 1967. Cytochrome oxidase from beef heart mitochondria. Methods Enzymol. 10:245-250. [Google Scholar]
- 38.Wolf, B. B., and D. R. Green. 2002. Apoptosis: letting slip the dogs of war. Curr. Biol. 12:R177-R179. [DOI] [PubMed] [Google Scholar]
- 39.Xia, X. G., T. Harding, M. Weller, A. Bieneman, J. B. Uney, and J. B. Schulz. 2001. Gene transfer of the JNK interacting protein-1 protects dopaminergic neurons in the MPTP model of Parkinson's disease. Proc. Natl. Acad. Sci. USA 98:10433-10438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, Q. H., R. Church-Hajduk, J. Ren, M. L. Newton, and C. Du. 2003. Omi/HtrA2 catalytic cleavage of inhibitor of apoptosis (IAP) irreversibly inactivates IAPs and facilitates caspase activity in apoptosis. Genes Dev. 17:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]