Abstract
Mitochondrial transcription factor A (TFAM), a transcription factor for mitochondrial DNA (mtDNA) that also possesses the property of nonspecific DNA binding, is essential for maintenance of mtDNA. To clarify the role of TFAM, we repressed the expression of endogenous TFAM in HeLa cells by RNA interference. The amount of TFAM decreased maximally to about 15% of the normal level at day 3 after RNA interference and then recovered gradually. The amount of mtDNA changed closely in parallel with the daily change in TFAM while in organello transcription of mtDNA at day 3 was maintained at about 50% of the normal level. TFAM lacking its C-terminal 25 amino acids (TFAM-ΔC) marginally activated transcription in vitro. When TFAM-ΔC was expressed at levels comparable to those of endogenous TFAM in HeLa cells, mtDNA increased twofold, suggesting that TFAM-ΔC is as competent in maintaining mtDNA as endogenous TFAM under these conditions. The in organello transcription of TFAM-ΔC-expressing cells was no more than that in the control. Thus, the mtDNA amount is finely correlated with the amount of TFAM but not with the transcription level. We discuss an architectural role for TFAM in the maintenance of mtDNA in addition to its role in transcription activation.
Human mitochondrial DNA (mtDNA) is a 16.5-kb double-stranded circular molecule that encodes 13 essential protein components of the mitochondrial oxidative phosphorylation complexes. The maintenance of mtDNA integrity is essential for normal function of the respiratory chain that is responsible for aerobic ATP production. There are hundreds to thousands of copies of mtDNA in one cell. Because the level of mtDNA transcripts largely depends on the copy number of mtDNA, the regulation of its copy number is important for maintaining mitochondrial ATP production. However, the regulation of mtDNA copy number is still poorly understood.
Mitochondrial transcription factor A (TFAM) (11, 30), a transcription factor for mtDNA, enhances mtDNA transcription in a promoter-specific fashion in the presence of mitochondrial RNA polymerase and transcription factor B (TFB1 M or TFB2 M) (10, 23). TFAM is a member of the high-mobility group (HMG) proteins because it contains two HMG boxes. TFAM possesses DNA-binding properties regardless of sequence specificity, although it shows a higher affinity for the light- and heavy-strand promoters (LSP and HSP, respectively) (11, 30). In addition to these two HMG boxes, human TFAM has a linker region between the two HMG boxes and a carboxyl-terminal tail region (C-tail) composed of 27 and 25 residues, respectively (6).
According to the strand-coupled model (4, 16, 38), replication of the L-strand, i.e., lagging-strand replication, occurs simultaneously with that of the H-strand. On the other hand, in another mtDNA replication model, the strand displacement model (34), replication of the nascent H-strand proceeds and displaces the parental H-strand until a replication origin of the L-strand, OL, is exposed on a single strand. The process of mtDNA replication begins with the initiation of transcription at LSP. The transcript initiated from LSP forms an RNA-DNA hybrid at a replication origin for the H-strand, OH. The RNA-DNA hybrid is processed to generate an RNA primer utilized by mitochondrial DNA polymerase γ (20). Thus, in the latter model, the replication of mammalian mtDNA is proposed to be coupled with transcription, and therefore TFAM is thought to be essential for replication of mtDNA (34). The role of transcription in the former model has not yet been clarified.
Abf2p, a TFAM homolog in Saccharomyces cerevisiae, has two HMG boxes and a short linker region between them, but unlike TFAM, it does not have a C-tail (12). Abf2p is abundant in mitochondria, with one Abf2p polypeptide present for every 15 bp of mtDNA (8). Disruption of the ABF2 gene leads to a loss of mtDNA and a resultant loss of respiratory competence when cells are grown in the presence of glucose. Expression of human TFAM in the S. cerevisiae abf2 strain rescued the phenotype, implying a potential functional homology between human TFAM and Abf2p (30). However, unlike mammalian TFAM, Abf2p is not required for the initiation of transcription in yeast mtDNA (8). An in vitro transcription assay demonstrated that Abf2p or C-tail-deleted TFAM does not activate transcription, whereas a chimeric Abf2p containing the C-tail does (6), suggesting that the C-tail of TFAM is necessary for transcriptional activation. This notion is also supported by a recent report that the C-tail of TFAM is necessary to bind mitochondrial transcription factor B (TFBM) and that this binding is required for transcription activation (24). In agreement with this, the import of full-length TFAM into isolated mitochondria increases transcription, but import of TFAM lacking the C-tail does not (14). Thus, the C-tail of TFAM is considered essential for the activation of transcription.
The mitochondrial nucleoids, protein-mtDNA complexes, have been studied extensively in the lower eukaryotes S. cerevisiae (17, 26) and Physarum polycephalum (32). In S. cerevisiae Abf2p is detected as a main component of the nucleoid and appears to function to maintain mtDNA and the nucleoid structure (17). In P. polycephalum, Glom, which also has two HMG boxes, shows a strong DNA-packaging activity (32). Both of these HMG family proteins can be functionally replaced by an Escherichia coli histone-like protein, HU (25, 32), implying that Abf2p and Glom package mtDNA.
There are several reports that mtDNA in higher eukaryotes is somewhat naked except for the D-loop region (2, 7, 29, 31), while the mtDNA of Xenopus laevis is reported to be packaged into regular beaded structures (3). These conflicting results on whether animal mtDNA takes on a higher nucleosome- or chromatin-like structure (mitochondrial nucleoid or mitochromatin/mitochondrial chromosome) are not fully resolved. Recent reports also suggest the existence of such a higher mtDNA structure in mammals (1, 13, 35). Because the amount of human TFAM is sufficient to cover the entire region of mtDNA (36) and because most TFAM molecules indeed bind to mtDNA (1), TFAM has been proposed to be one of the main components of the human mtDNA higher structure.
Homozygous gene disruption of Tfam is lethal in both mouse and chicken cells, at least in part due to mtDNA depletion and resultant loss of oxidative phosphorylation capacity (19, 22). In heterozygous cells, the expression of mouse and chicken TFAM was reduced by about 50% and the amount of mtDNA also decreased by about half (19, 22). These results suggest that TFAM is necessary to maintain mtDNA. There are two possibilities to explain mtDNA maintenance by TFAM. One is that, given that the replication of mtDNA is coupled to transcription (34), TFAM affects the replication of mtDNA directly. The other is that TFAM binds and stabilizes mtDNA, as do other HMG family proteins (5, 8, 12, 32, 39).
There have been no reports demonstrating overexpression of TFAM in mammalian cells. We established stable and inducible human cell lines overexpressing TFAM for the first time with a tetracycline-regulated gene expression system. In this study, with these TFAM-overexpressing cell lines and RNA interference (RNAi), we manipulated the amount of human TFAM in human HeLa cell lines and then analyzed mtDNA and mitochondrial transcription. We found that the amount of TFAM but not the transcription level is correlated to the amount of mtDNA.
MATERIALS AND METHODS
Antibodies.
Antibodies to human TFAM, prohibitin, and BAP37 were produced by immunizing rabbits with recombinant glutathione S-transferase-human TFAM (28), glutathione S-transferase-prohibitin, and glutathione S-transferase-BAP37 proteins, respectively. Antibodies to cytochrome b and complex II were produced by immunizing rabbits with peptides of the C-terminal eight amino acids of cytochrome b and the C-terminal nine amino acids of the complex II iron sulfur protein, respectively (1). An antibody to human mitochondrial single-stranded DNA-binding protein (mtSSB) was described previously (36) as was an antibody to P32 (27). Antibodies against the hemagglutinin (HA) HA.11 epitope tag, cytochrome c, and calnexin were obtained from Covance, Stressgen, and Santa Cruz Biotechnology, respectively.
Preparation of tetracycline-regulated TFAM-overexpressing cell lines.
To avoid suppression by RNAi, we introduced silent mutations in a human TFAM cDNA corresponding to the RNAi target in advance (from GTCTTGGCAAGTTGTCCAAAGAAACCTGTAAGTTCT to GTCTTAGCAAGTTGCCCTAAAAAGCCTGTAAGCTCT; the encoded amino acid sequence for both is VLASCPKKPVSS [amino acids 45 to 56]; sites where silent mutations have been introduced are in boldface) and named it pmod-TFAM. A DNA fragment encoding human TFAM lacking the C-terminal 25 amino acids (human TFAM-ΔC), i.e., it retained amino acids 1 to 221, was amplified with pMod-TFAM as a template. The sense primer contained a BamHI site, the Kozak sequence (18), and ATG for a first methionine in that order; the antisense primer contained an SpeI site. A DNA fragment specifying an HA tag was prepared by annealing the complementary synthesized oligonucleotides, which included, in order, the sequences for an SpeI restriction site, an HA epitope tag (11 amino acids), a stop codon, and an NheI site.
The DNA fragments for human TFAM and the HA tag were digested with appropriate restriction enzymes and inserted between the BamHI and NheI sites of vector pTRE2hyg (Clontech). This vector encodes the precursor human TFAM-ΔC with the HA tag at its C terminus and was named ph-TFAM-ΔC-HA. We also amplified a DNA fragment encoding precursor mouse TFAM (amino acids 1 to 243) by PCR, with a mouse cDNA library as a template with a sense primer (containing, in order, a BamHI site, the Kozak sequence [18], and ATG for a first methionine) and an antisense primer (containing an SpeI site). The mouse TFAM DNA fragment and the HA tag DNA fragment were inserted into the pTRE2hyg vector in the same way as the human construct (pm-TFAM-HA).
A HeLa Tet-Off cell line was obtained from Clontech. The cells were grown in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (Gibco) and 100 μg of G418 per ml (Sigma). We transfected HeLa Tet-Off cells with our plasmids with the FuGene 6 reagent (Roche Molecular Biochemicals) and selected cells bearing the transgenes in the presence of G418 (400 μg/ml) (Wako), hygromycin B (200 μg/ml) (Wako), and doxycycline, a tetracycline derivative (1 μg/ml) (ICN Biomedicals). We isolated hygromycin-resistant clones, cultured them with or without doxycycline for about 10 days, and then examined the expression of recombinant proteins by Western blotting. These mouse and human TFAM-overexpressing cell lines were designated MTF (mouse TFAM full-length) and HΔC (human C-tail-deleted TFAM), respectively.
Suppression of human TFAM by RNAi.
HeLa cells were grown in DMEM with 10% fetal bovine serum and antibiotics (100 μg of G418 per ml for control HeLa Tet-Off cells; 100 μg of G418 and 100 μg of hygromycin B per ml for MTF and HΔC cells) in 3.5-cm dishes. When the cells were about 50% confluent, the culture medium was replaced by 2 ml of DMEM containing 10% fetal bovine serum with and without doxycycline (1 μg/ml). The sequences of short interfering RNA (siRNA) duplexes were 5′-GUUGUCCAAAGAAACCUGUdTdT-3′ (sense siRNA) and 5′-ACAGGUUUCUUUGGACAACdTdT-3′ (antisense siRNA) (obtained from Qiagen); 12 μl of 20 μM siRNA duplexes was mixed with 200 μl of Opti-MEM (Gibco) in one tube; 12 μl of Oligofectamine (Invitrogen) was mixed with 48 μl of Opti-MEM in the other tube. The two mixtures were incubated for 10 min at room temperature and then combined and mixed gently by pipetting. The combined mixture was allowed to stand for another 20 min at room temperature. The siRNA-Oligofectamine mixture was then added to the cultured cells. After 24 h, the culture medium was replaced by new DMEM with 10% fetal bovine serum and appropriate antibiotics. When the cells were almost confluent, they were reseeded in two 3.5-cm dishes.
Western blotting.
The cells from a 3.5-cm dish were collected in 1 ml of phosphate-buffered saline (PBS), and half of them were solubilized with 100 μl of sodium dodecyl sulfate (SDS) denaturing buffer consisting of 0.5% SDS and 1% 2-mercaptoethanol (the other half was used for mtDNA quantification [see below]). The mixture was briefly sonicated immediately after solubilization. Proteins were separated on an SDS-12% polyacrylamide gel by polyacrylamide gel electrophoresis (PAGE) and subsequently detected by immunoblotting. The signals were visualized with horseradish peroxidase-labeled anti-rabbit immunoglobulin G (Biosource) and ECL reagents (Amersham Biosciences). The chemiluminescence was recorded and quantified with a chilled charge-coupled device camera, LAS1000plus (Fuji).
Quantification of mtDNA.
From the remaining half of the cells, total DNA was extracted with a DNeasy tissue kit (Qiagen). The total DNA was quantified with a PicoGreen double-stranded DNA quantitation kit (Molecular Probes). The relative amount of mtDNA was quantified by quantitative PCR with a LightCycler (Roche). The PCR mixture contained 2 ng of the total DNA (for a standard curve, 8, 4, 2, 1, and 0.5 ng of the total DNA were used), 10 pmol each of primers (5′-CCACCCAAGTATTGACTCACCC-3′ [nucleotides 16052 to 16073] and 5′-CGAGAAGGGATTTGACTGTAATG-3′ [nucleotides 16339 to 16361]) and 3 mM MgCl2 in 20 μl. To estimate the amount of genomic DNA as an internal standard, a genomic beta-globin gene was amplified in a 20-μl reaction mixture containing 16 ng of the total DNA (for a standard curve, 64, 32, 16, 8, and 4 ng of the total DNA were used), 10 pmol each of primers (5′-CTGCCCTGTGGGGCAAGGTGAACGTGGATG-3′ and 5′-CAGGTGAGCCAGGCCATCACTAAAGGCACC-3′), and 3 mM MgCl2. The amount of mtDNA was adjusted to the amount of genomic DNA.
Immunofluorescent imaging of HeLa cells.
Cells were incubated in the presence of 100 nM MitoTracker Red CMXRos (Molecular Probes) for 20 min. After washing with PBS three times, the cells were fixed with acetone-methanol (50:50, vol/vol) for 5 min. After washing with PBS three times, the fixed cells were blocked with bovine serum albumin-PBS (1% bovine serum albumin in PBS) for 30 min, then incubated with 250-fold-diluted anti-human TFAM antiserum or anti-HA antibody in bovine serum albumin-PBS for 1 h. After washing the cells with wash buffer (0.1% Tween 20 in PBS) three times, cells were incubated with 250-fold-diluted Alexa Fluor 488 goat anti-rabbit immunoglobulin G (Molecular Probes) for 30 min. Fluorescence images were taken with a confocal laser microscope (Bio-Rad Laboratories).
Isolation of mitochondria from HeLa cells.
All procedures were done at 4°C. HeLa cells cultured in 10-cm dishes were scraped with a cell lifter (Costar) into 3 ml of PBS, pelleted by centrifugation, and washed with homogenization buffer (10 mM HEPES-KOH, pH 7.4, and 0.25 M sucrose). The cells were suspended in 4 volumes of the buffer and homogenized with a Potter-Elvehjem homogenizer. The homogenate was centrifuged at 900 × g for 10 min to remove the unbroken cells and nuclei. The supernatant was centrifuged at 10,000 × g for 6 min. The pellet was collected as a crude mitochondrial fraction and used for in organello transcription assays or for preparing Nonidet P-40-insoluble fractions.
In vitro transcription assay.
The cDNA coding for mature human TFAM (amino acid residues 42 to 246) and mature human TFAM-ΔC-HA (amino acid residues 42 to 221 and C-terminal HA tag) were inserted into the pProEXHTb vector (Gibco). The recombinant His-human TFAM and His-human TFAM-ΔC-HA were expressed in Escherichia coli BL21 cells and purified Ni2+-bound chelating Sepharose resin (Amersham Biosciences) as described previously (28, 36). In vitro transcription reactions were performed with a cloned DNA fragments corresponding to nucleotides 1 to 477 of human mtDNA as previously described (10).
In organello transcription assay.
In organello transcription was measured by the method of Enriquez et al. with slight modifications (9). The mitochondrial fraction was resuspended in transcription buffer (10 mM Tris-HCl [pH 7.4], 25 mM sucrose, 75 mM sorbitol, 100 mM KCl, 10 mM K2HPO4, 50 μM EDTA, 5 mM MgCl2, 1 mM ATP, 1 mg of bovine serum albumin per ml). Mitochondria (200 μg of protein) were incubated in 300 μl of the transcription buffer containing 10 μCi of [α-32P]UTP (Amersham Biosciences) at 37°C for 30 min. After the incubation, the mitochondria were pelleted at 15,000 × g for 1 min and then washed with nuclease buffer (0.25 M sucrose, 10 mM Tris-HCl [pH 8.0], 1 mM CaCl2). The mitochondria were resuspended in 400 μl of nuclease buffer with 100 units of nuclease S7 (Roche Molecular Biochemicals) and incubated at room temperature for 20 min. The mitochondria were then pelleted at 15,000 × g for 1 min and washed with TES buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 0.25 M sucrose). The mitochondrial pellet was solubilized in 100 μl of lysis buffer (50 mM Tris-HCl [pH 8.0], 20 mM NaCl, 1 mM EDTA, 1% SDS) containing 20 μg of protease K (Gibco) and incubated at room temperature for 15 min. After addition of 100 μl of a phenol-chloroform-isoamyl alcohol mixture (25:25:1, vol/vol/vol) and vortexing for 1 min, the mixture was centrifuged at 20,000 × g for 5 min, and 80 μl of the aqueous phase was transferred to a clean tube. Then 1 μl of Pellet Paint (Novagen), 80 μl of 3 M sodium acetate (pH 5.2), and 300 μl of ethanol were added, and the mitochondrial nucleic acids were precipitated. The pellets were solubilized with sample buffer (99% formamide, 1 mM EDTA, and bromphenol blue) and incubated at 95°C for 3 min. DNA size makers (100-bp DNA ladder and 1-kb DNA ladder, New England BioLabs) were [γ-32P]ATP labeled with Ready-To-Go T4 polynucleotide kinase (Amersham Biosciences). Samples were separated on an 8 M urea-4% polyacrylamide gel by electrophoresis and analyzed with a BAS-2500 (Fuji). The isolated mitochondria (5 μg) were analyzed by Western blotting to check the amount of mitochondria and efficiency of RNAi.
Separation of mitochondrial NP-40-soluble and -insoluble fractions.
The mitochondria were resuspended in TES buffer containing 0.5% NP-40 and 1 mM dithiothreitol (0.5 mg of protein/ml) and incubated for 30 min on ice with intermittent mixing. The mitochondria were centrifuged at 20,000 × g for 30 min and separated into a pellet (P1) and a supernatant (S1). The P1 fraction (10 μg of protein) was resuspended with 20 μl of nuclease buffer (10 mM Tris-HCl [pH 7.4], 0.25 M sucrose, 1 mM dithiothreitol, 2.5 mM CaCl2, and 0.5% NP-40) and incubated for 30 min on ice with 2.5 U of nuclease S7 or 0.5 μg of RNase A (DNase free; Wako). After centrifugation at 20,000 × g for 30 min, the sample was separated into a pellet (P2) and a supernatant (S2). Proteins from each fraction were separated by SDS-PAGE and subsequently analyzed by Western blotting with appropriate primary antibodies.
mtDNA in the P1 and S1 fractions was detected by PCR as described previously (1). Briefly, total mitochondria, P1, and S1 (from 10 μg of mitochondria) were incubated at 95°C for 30 min with 0.5 μg of protease K (Gibco) and diluted in 200 μl of distilled water. With 1 μl each of the diluted samples, a fragment of mtDNA was amplified with PCR primers (nucleotides 16052 to 16073 and nucleotides 16339 to 16361) and Ex-taq DNA polymerase (Takara). The number of amplification cycles was 25, and the PCR products were separated on a 1% agarose gel.
RESULTS
Reduced expression of human TFAM by RNAi.
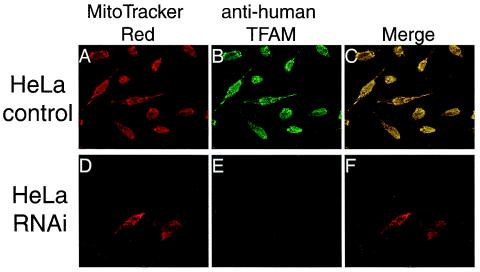
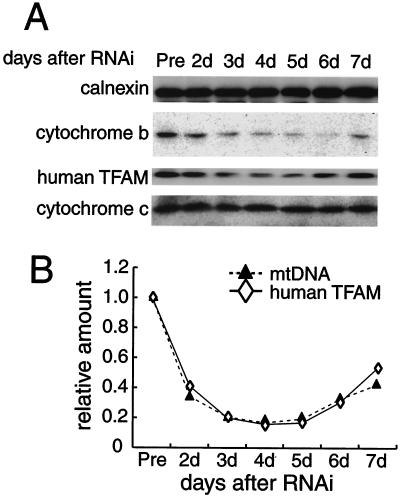
To observe a dose effect of TFAM, we downregulated the expression of human TFAM by RNAi in HeLa cells. Three days after RNAi treatment, human TFAM was barely detectable on immunofluorescent images with anti-human TFAM antibodies (compare Fig. 1B and E). To monitor the change in the levels of human TFAM and other mitochondrial proteins, cells were collected daily between days 2 and 7 and analyzed by Western blotting (Fig. 2A). The amount of human TFAM was decreased maximally at days 3 and 4 and then gradually increased again (Fig. 2A, third panel). The amounts of calnexin, a microsomal protein, and cytochrome c, a nucleus-encoded mitochondrial protein, did not change (Fig. 2A, top and bottom panels). The amounts of several other nucleus-encoded mitochondrial proteins (prohibitin, BAP37, and VDAC) were not affected by this RNAi (data not shown), confirming the specificity of the RNAi treatment. The reduction in cytochrome b, an mtDNA-encoded protein (Fig. 2A, second panel), probably resulted from a decrease in mtDNA (Fig. 2B). We measured the relative amount of mtDNA by quantitative PCR and found that the amount fell and rose in parallel with that of human TFAM (Fig. 2B). The levels of human TFAM and mtDNA were 0.141 ± 0.052 and 0.132 ± 0.061 (n = 4) of those of control cells at day 3. Thus, the amount of mtDNA was strongly correlated with the amount of TFAM in vivo.
FIG. 1.
Immunofluorescent images of HeLa cells treated by RNAi. Human TFAM in HeLa cells was identified with anti-human TFAM antibodies (B and E). Mitochondria were stained with MitoTracker Red (A and D). Panels C and F are merged images of panels A plus B and D plus E, respectively.
FIG. 2.
Parallel decreases and increases in human TFAM and mtDNA after RNAi. (A) Calnexin (as a standard of cell amount), cytochrome b, human TFAM, and cytochrome c in HeLa cells were analyzed by Western blotting 2 to 7 days after RNAi. The relative amount of mtDNA was measured by quantitative PCR. (B) Representative daily changes in the relative amounts of human TFAM (⋄) and mtDNA (▴).
Preparation of tetracycline-regulated TFAM-overexpressing cell lines.
Conversely, to see whether an increase in TFAM would affect the amount of mtDNA, we created TFAM-overexpressing cell lines. We made two kinds of Tet-Off gene expression vectors, ph-TFAM-ΔC-HA and pm-TFAM-HA, in order to express C-tail-deleted human TFAM with an HA tag and mouse full-length TFAM with an HA tag, respectively (Fig. 3A). To produce the stable TFAM-expressing cell lines, we transfected HeLa Tet-Off cells with each vector and cultured them with hygromycin for selection and with doxycycline to suppress expression. We isolated hygromycin-resistant colonies and checked the expression of TFAM by Western blotting after culturing for 10 days without doxycycline. Fourteen of 24 hygromycin-resistant ph-TFAM-ΔC-HA-transfected clones expressed high levels of human TFAM-ΔC-HA (about 50 to 120% compared with the amount of endogenous human TFAM). Thirteen of 24 hygromycin-resistant pm-TFAM-HA-transfected clones expressed high levels of mouse TFAM-HA (about 50 to 140% compared with the amount of endogenous human TFAM). These mouse TFAM- and human TFAM-ΔC-overexpressing cell lines were designated MTF (mouse TFAM full-length) and HΔC (human C-tail-deleted TFAM), respectively.
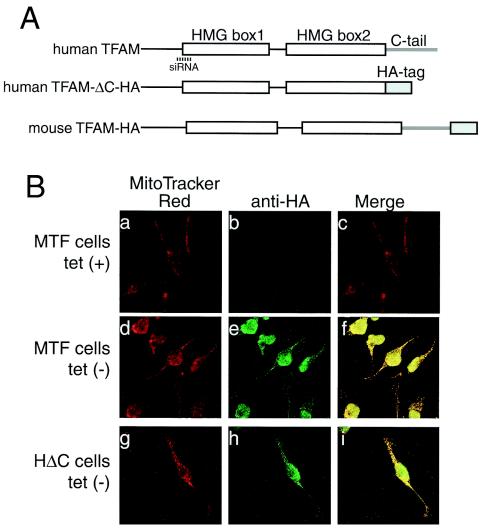
FIG.3.
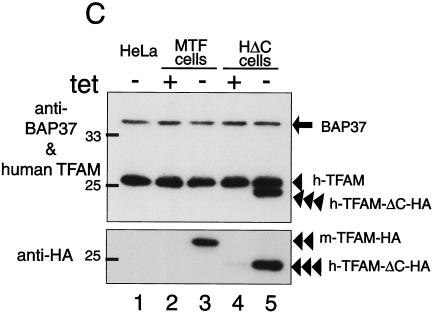
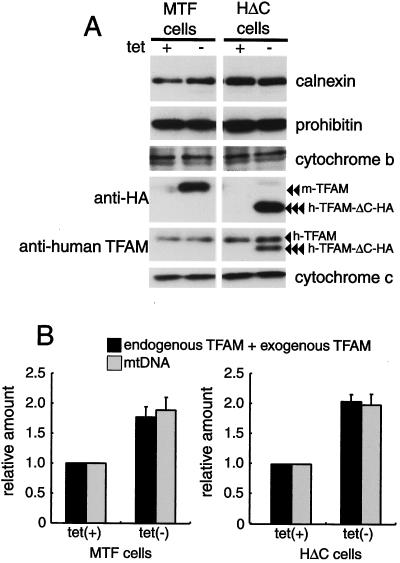
Tetracycline-regulated expression of TFAM. (A) The scheme of recombinant TFAM molecules. Human and mouse TFAM have two HMG boxes (white square) and a C-tail region (gray bar). An HA.11 epitope tag is indicated by a gray square. The position of the RNAi target is indicated by a dotted line. (B) MTF cells and HΔC cells were cultured with doxycycline [tet (+)] or without doxycycline [tet (−)]. Mitochondria were stained with MitoTracker Red (panels a, d, and g), and recombinant TFAM was stained with anti-HA antibodies (panels b, e, and h). The merged images are shown in panels c, f, and i. (C) Western blotting analysis of TFAM molecules. Total cell lysates were used for Western blotting. BAP37 (arrow) is shown as an internal standard for the amount of protein applied in the samples. Antibodies for BAP-37 and human TFAM were added together for detecting the two proteins (upper panel). Then the membrane was reprobed with anti-HA antibody (lower panel). Endogenous human TFAM (arrowhead), exogenous mouse TFAM-HA (double arrowheads), and exogenous human TFAM-ΔC-HA (triple arrowheads) are indicated.
When cultured without doxycycline, immunofluorescent imaging of MTF cells with anti-HA antibodies showed granular fluorescence. This pattern colocalized with that of MitoTracker Red, a mitochondrion-staining fluorescent dye (Fig. 3B, panels d to f), indicating that the expressed exogenous TFAM localized to mitochondria, while the cells cultured with doxycycline showed almost no fluorescence with anti-HA antibodies (Fig. 3B, panel b). HΔC cells showed the same pattern as the MTF cells (Fig. 3B, panels g to i). With Western blots, mouse TFAM-HA was detected by anti-HA antibodies in medium without doxycycline (double arrowheads), whereas there was no signal in medium containing doxycycline (Fig. 3C, compare lanes 2 and 3 in the lower panel). Anti-human TFAM did not react with mouse TFAM (Fig. 3C, lane 3; notice their molecular sizes). Similarly, human TFAM-ΔC-HA (triple arrowheads) was detected by both anti-human TFAM and anti-HA antibodies only in medium without doxycycline (Fig. 3C, lanes 4 and 5 in the upper and lower panels). Thus, expression of the recombinant proteins was completely regulated by doxycycline. Endogenous human TFAM (single arrowhead) was not affected by the overexpression of exogenous TFAM (Fig. 3C, upper panel). The amount of BAP37 (arrow), an inner membrane protein, was not altered (Fig. 3C, upper panel), indicating that the amount applied was essentially the same among all samples.
Overexpression of mouse TFAM-HA or human TFAM-ΔC-HA increases the amount of mtDNA.
Because it takes about 10 days to stably express exogenous mouse TFAM-HA or human TFAM-ΔC-HA after removal of doxycycline (data not shown), we did experiments at least 14 days after removal of doxycycline. In medium without doxycycline, the amounts of mouse TFAM-HA and human TFAM-ΔC-HA were 0.77 ± 0.17 (mean ± standard deviation) and 1.05 ± 0.11, respectively, of that of endogenous human TFAM; i.e., the total amount of TFAM was 1.77 and 2.05, respectively, that of cells grown in medium with doxycycline (Fig. 4B). Overexpression of total TFAM by a factor of 1.77 (in MTF cells) and 2.05 (in HΔC cells) increased the amount of mtDNA by a factor of 1.89 ± 0.22 and 1.99 ± 0.18, respectively (Fig. 4B). Neither nucleus-encoded mitochondrial proteins (prohibitin and cytochrome c) nor an mtDNA-encoded protein, cytochrome b, was affected by the overexpression of TFAM (Fig. 4A). Thus, the amount of TFAM led the change in mtDNA levels in parallel.
FIG. 4.
Amount of mitochondrial proteins and mtDNA in MTF and HΔC cells. (A) Total cell lysates of cells cultured with doxycycline [tet (+)] or without doxycycline [tet (−)] were used for Western blotting. Prohibitin and cytochrome c are mitochondrial proteins encoded by the nuclear genome. Cytochrome b is a mitochondrial protein encoded by mtDNA. Endogenous human TFAM (arrowhead), mouse TFAM-HA (double arrowheads), and human TFAM-ΔC-HA (triple arrowheads) are indicated. Calnexin, a microsomal protein, is shown as an internal standard. (B) Quantification of relative amounts of TFAM (black bar) and mtDNA (gray bar). Signal intensities on the Western blots were measured for estimating the number of TFAM molecules, endogenous TFAM, and exogenous TFAM. The amount of mtDNA was measured by quantitative PCR. Error bars indicate ±1 standard deviation from the mean of three independent experiments.
Both mouse TFAM-HA and human TFAM-ΔC-HA can maintain the amount of mtDNA.
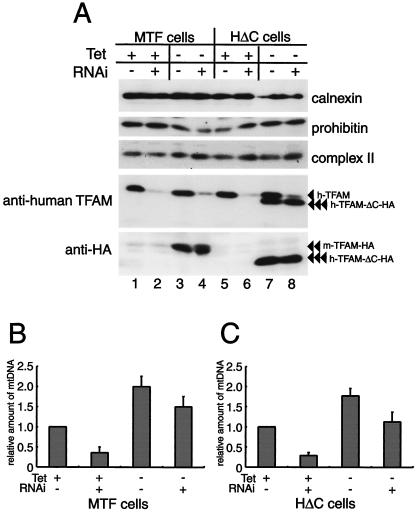
mtDNA was increased by the overexpression of C-tail-deleted human TFAM, which is considered not to activate transcription (6), raising the possibility that the increase in mtDNA does not require the upregulation of transcription. In order to clarify this point, we performed RNAi in the TFAM-overexpressing cells. To avoid suppression of recombinant human TFAM by the RNAi treatment, silent mutations were introduced into a cDNA of human TFAM (see Methods). In medium with doxycycline, MTF cells and HΔC cells expressed little or no mouse TFAM-HA (Fig. 5A, double arrowheads; lanes 1 and 2) and human TFAM-ΔC-HA (Fig. 5A, triple arrowheads; lanes 5 and 6), respectively. However, In medium without doxycycline, the exogenous TFAM proteins were overexpressed (Fig. 5A, bottom panel, lanes 3, 4, 7, and 8). The RNAi treatment (i.e., 3 days after RNAi) suppressed the endogenous TFAM (Fig. 5A, fourth panel, lanes 2, 4, 6, and 8), but exogenous TFAMs were not affected (Fig. 5A, bottom panel, lanes 4 and 8; double and triple arrowheads), showing selective downregulation of endogenous human TFAM.
FIG. 5.
(A) MTF and HΔC cells were cultured with doxycycline [tet (+)] or without doxycycline [tet (−)]. Three days after RNAi treatment (RNAi +) or without RNAi treatment (RNAi −), the cells were collected and analyzed. The expression of proteins in MTF and HΔC cells was analyzed by Western blotting. Endogenous human TFAM (arrowhead), exogenous mouse TFAM-HA (double arrowheads), and exogenous human TFAM-ΔC-HA (triple arrowheads) are indicated. (B and C) The amount of mtDNA was measured in cells with and without doxycycline and RNAi treatments, as indicated. Error bars indicate ± 1 standard deviation from the mean of three independent experiments.
In MTF cells in which mouse TFAM is not yet expressed but endogenous TFAM is repressed by RNAi (Fig. 5B), mtDNA was reduced compared to that in the MTF cells in which endogenous TFAM is not repressed (Fig. 5B). In MTF cells in which mouse TFAM is expressed but endogenous TFAM is not repressed (Fig. 5B), mtDNA was almost doubled with the twofold increase in total TFAM. mtDNA was reduced to the control level by the RNAi treatment in the mouse TFAM-expressing cells (Fig. 5B). The reduced amount of mtDNA by RNAi in medium without doxycycline roughly equaled that in medium with doxycycline (compare the difference between RNAi with and without doxycycline) (Fig. 5B). A similar pattern was observed in HΔC cells (Fig. 5C). These results suggest that both exogenous TFAMs are as competent in the maintenance of mtDNA as endogenous human TFAM under conditions in which endogenous human TFAM remains at about 15% of the control level.
In vitro and in organello transcription.
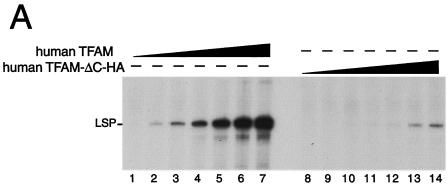
The C-tail of TFAM is considered essential for initiation of transcription from LSP (6). To confirm that the HA tag does not function like the C-tail, we performed an in vitro transcription assay for human LSP. Human TFAM-ΔC without an HA tag was about 100-fold less active than full-length human TFAM (unpublished data). As expected, a transcript from LSP in the presence of His-human TFAM-ΔC-HA was about 100-fold less than that in the presence of His-human TFAM (Fig. 6A), indicating that LSP-dependent transcription is not increased by adding the HA tag to human TFAM-ΔC.
FIG. 6.
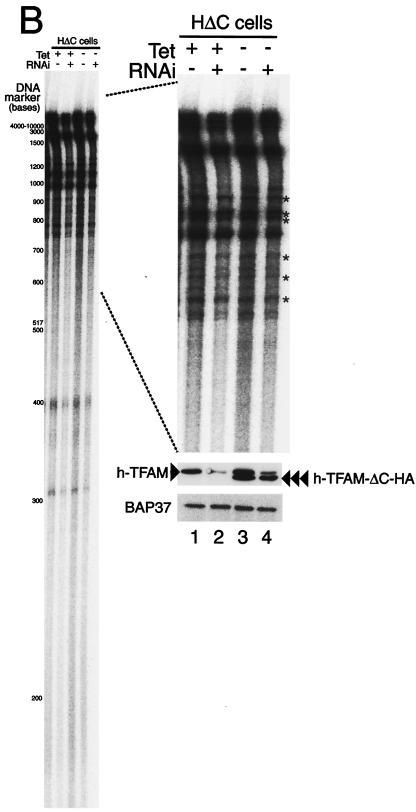
In vitro and in organello transcription assays. (A) TFAM lacking its C-terminal tail only supports low levels of transcription from LSP. A template containing the light-strand promoter (LSP, 85 fmol) of human mtDNA was used for in vitro runoff transcription assays. The reactions were performed with the following pure recombinant proteins: human mitochondrial RNA polymerase (250 fmol), human TFB2 M (500 fmol), and wild-type human TFAM with an N-terminal His tag or human ΔC-tail TFAM with an N-terminal His tag and C-terminal HA tag (human TFAM-ΔC-HA) in increasing amounts (0.02, 0.05, 0.2, 0.5, 1.6, 5, and 15 pmol). (B) A representative image of five independent experiments is shown. The HΔC cells were cultured with and without doxycycline and treated or not by RNAi, and mitochondria were isolated from these cells 3 days after RNAi treatment. The newly synthesized transcripts were labeled with [α-32P]UTP for 30 min in mitochondria and analyzed by urea-polyacrylamide gel electrophoresis. The approximate size of nucleotides is shown based on the [γ-32P]ATP-labeled DNA size markers (left panel). The part showing bands larger than 600 bases was enlarged (right upper panel). The asterisks indicate measured bands used for estimation. The isolated mitochondria (5 μg of protein) were analyzed by Western blotting to check the protein amount (anti-BAP37 antibody, right lower panel) and the efficiency of RNAi (anti-human TFAM antibody, right middle panel).
With the isolated mitochondria from HΔC cells cultured with and without doxycycline and with and without RNAi treatment, we measured transcription in organello by measuring newly synthesized transcripts. The newly synthesized transcripts were labeled with [32P]UTP for 30 min in mitochondria and analyzed by urea-polyacrylamide gel electrophoresis (Fig. 6B). The transcripts were not detected in mitochondria from Rho0 206 cells (data not shown), excluding any background contribution from genomic transcription. Between the 3,000- and 700-base size markers, the signals for transcription were seen as bands, although the bands were not identified (right panel). In addition to these bands, a smearing background was seen. When human TFAM-ΔC-HA was overexpressed, this smearing background was about 1.6-fold higher (left panel, lane 3) than in the control (lane 1). This nonspecific background was reduced by RNAi treatment (lane 2; by about 60%) and returned to the control level with expression of human TFAM-ΔC-HA (lane 4). The level of the nonspecific smear appeared to be correlated with the amount of mtDNA. The background might reflect nonspecific LSP/HSP-independent transcription or highly heterogeneous RNA processing of LSP/HSP-specific transcription. Another possibility may be RNA primers remaining in nascent mtDNA strands that initiated at diverse sites according to the strand-coupled replication model.
Because processed transcripts of mtDNA are distributed mainly between bases 2000 and 700 according to their expected lengths, we hypothesized that the bands found between bases 2000 and 700 are promoter-dependent transcripts (bands marked by asterisks, right panel). The signal intensities were corrected by subtracting the background. When human TFAM-ΔC-HA was overexpressed (lane 3), the specific signals were not changed or somewhat decreased (61 to 113%; average, 77%) in spite of the twofold increase in the amount of mtDNA. This inhibition may be caused by competition between TFAM-ΔC-HA and endogenous TFAM for the promoters. When wild-type human TFAM was reduced by RNAi, the specific signals were reduced to about 46% (41 to 53%) and 49% (44 to 74%) of the control in control and human TFAM-ΔC-HA-overexpressing cells, respectively (lanes 2 and 4), while the amount of mtDNA was about 0.2 and 1 times that of the control cells, respectively (Fig. 5C). Compensatory upregulation of the transcription seems to occur in the former situation. The decrease in the latter may also be due to the competition. Thus, the amount of mtDNA was not correlated with the transcription level.
Both mouse TFAM-HA and TFAM-ΔC-HA are components of the mitochondrial nucleoid.
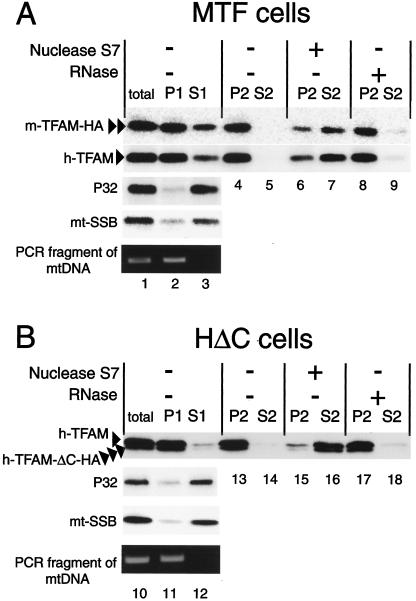
We have previously demonstrated that mtDNA and human TFAM were mostly included in the NP-40-insoluble fraction but that other mitochondrial matrix proteins, P32 and mtSSB, were mostly recovered from the soluble fraction, while part of mtSSB was bound to mtDNA as a component of the mitochondrial nucleoid (1). To examine whether overexpressed TFAM is bound to mtDNA, we prepared mitochondria from MTF and HΔC cells cultured without doxycycline and then separated them into NP-40-insoluble (P1) and -soluble (S1) fractions (Fig. 7, lanes 2 to 3 and 11 to 12). As reported previously (1), most of the endogenous human TFAM (arrowhead) was recovered from the P1 fraction (Fig. 7, lanes 2 and 11), whereas most of the p32 and mtSSB were recovered from the S1 fraction (Fig. 7, lanes 3 and 12). Mitochondrial DNA was detected only in the P1 fraction by PCR (lanes 2 and 11). Both mouse TFAM-HA (double arrowheads) and human TFAM-ΔC-HA (triple arrowheads) were also mostly recovered from the P1 fraction (Fig. 7, compare lane 2 with 3 and lane 11 with 12).
FIG. 7.
Mitochondria were prepared from MTF (A) and HΔC (B) cells cultured without doxycycline. The mitochondria were solubilized with 0.5% NP-40 (total) and separated into pellet (P1) and supernatant (S1). The P1 fractions were treated with nuclease S7 or DNase-free RNase A and then separated again into pellet (P2) and supernatant (S2). Each sample was analyzed by Western blotting with anti-human TFAM, anti-P32, anti-mtSSB, and anti-HA antibodies. mtDNA in total, P1, and S1 were detected by PCR. Human TFAM (arrowhead), mouse TFAM-HA (double arrowheads), and human TFAM-ΔC-HA (triple arrowheads) are indicated.
The P1 fractions were treated with nuclease S7 or DNase-free RNase A and then separated into insoluble (P2) and soluble (S2) fractions. After treatment with nuclease S7, most of the endogenous human TFAM, exogenous mouse TFAM-HA, and human TFAM-ΔC-HA were recovered from the soluble fractions (S2) (lanes 7 and 16). However, after the RNase A treatment, the TFAMs were recovered from insoluble fractions (P2) (lanes 8 and 17). These findings suggest that both endogenous and exogenous TFAM molecules are bound to mtDNA as mitochondrial nucleoid components.
DISCUSSION
We showed that the amounts of TFAM and mtDNA were reduced in parallel (Fig. 2B). There are already two reports on suppression of TFAM in vertebrate cells: heterozygous targeted disruption of the Tfam gene in mice (19) and in chicken DT40 cells (22). In both cases, both TFAM and mtDNA were decreased by about 50%. These reports, however, showed only steady-state values a long time after the 50% decrease in TFAM, whereas we measured the daily change in the amount of TFAM and mtDNA after the start of RNAi treatment. In the present study, we first showed that the amount of mtDNA was strongly correlated to that of TFAM on a time scale that appears to be less than 1 day. Taking into account that the changes in TFAM and mtDNA were nearly the same at each day, mtDNA levels may reach the corresponding levels of TFAM within hours.
The replication of mammalian mtDNA is coupled with transcription according to the strand displacement model (34). Thus, one possible mechanism by which mtDNA decreases or increases with the decrease or increase in TFAM is that replication of mtDNA is down- or upregulated due to the down- or upregulation of TFAM-activated transcription. However, we found that the transcription level was not correlated with the amount of mtDNA (Fig. 5C and 6B). On the other hand, we noticed a striking correlation between the levels of TFAM and mtDNA. TFAM is a transcription factor (10, 11, 23, 30), but it is also an HMG protein having DNA-binding properties regardless of DNA sequence (11, 30). TFAM molecules are abundant enough to cover mtDNA entirely, and indeed most of them bind mtDNA, suggesting that mtDNA is packaged with TFAM (1, 36) and TFAM is in functional excess (33).
Such a nucleoid structure may also be required for the stability of mtDNA. In agreement with this notion, mouse TFAM and human TFAM-ΔC were as active in the maintenance of mtDNA as wild-type TFAM, at least under conditions in which promoter-specific transcription was maintained at a certain level (Fig. 5C and 6B). About 85% of full-length TFAM could be replaced without reduction in mtDNA by TFAM-ΔC-HA, which has only 1% of the LSP-dependent transcription activity of full-length TFAM (Fig. 5A and C). If the majority of TFAM molecules maintained mtDNA through transcription-coupled replication, the amount of mtDNA would decrease. Therefore, this result raises the possibility that the majority of TFAM molecules participate architecturally in maintenance.
The overexpressed recombinant TFAM was mostly recovered from the insoluble fraction together with mtDNA (Fig. 7, lanes 2 and 11). Nuclease S7 treatment but not RNase A treatment released recombinant TFAM as well as endogenous TFAM to the soluble fraction (Fig. 7, lanes 6 to 9 and 15 to 18), supporting the idea of TFAM-mtDNA binding. Taken together with the fact that the level of mtDNA rapidly and finely corresponds to the level of TFAM (Fig. 2), one likely explanation is that the vast majority of TFAM molecules are bound to mtDNA and that only mtDNA covered with TFAM can be maintained. According to this model, the mass but not the copy number of mtDNA would be titrated by TFAM, consistent with a finding by Tang et al. that the total mass but not the copy number of mtDNA is constant among cells harboring wild-type, deleted, and partially duplicated mtDNAs (37). Conversely, when mtDNA was depleted with ethidium bromide in HeLa cells, TFAM was reduced to the same extent as mtDNA (33). TFAM and mtDNA may thus stabilize each other when bound together.
We presume that TFAM exists abundantly and that the majority of TFAM molecules are bound to sites other than the promoter regions for packaging mtDNA. Goto et al. reported that TFAM levels can be substantially reduced by RNAi without significant inhibition of transcription per mtDNA in insect cells (15). Here we also showed that transcription per mtDNA was not decreased but rather upregulated by RNAi-induced suppression of TFAM (see Fig. 5C and Fig. 6B, lane 2). These observations suggest that the amount of TFAM can be reduced without lowering the transcription level per mtDNA if the amount of TFAM is manipulated within a cell. In contrast, Garstka et al. reported that import of TFAM into isolated mitochondria significantly enhances transcription in organello (14).
The reason for these apparently contradictory observations is currently unknown. However, the differences may reflect differences in the experimental systems. Garstka et al. manipulated the amount of TFAM with isolated mitochondria, while we in the present study and Goto et al. did so in living cells. A possible but hypothetical explanation is to assume that TFAM preferentially binds to promoters with a higher affinity but can also be preferentially displaced from the promoters by an unknown mechanism. In a cell, continuous input of TFAM would compensate for this selective displacement. However, in isolated mitochondria, there is no input of TFAM unless TFAM is imported artificially, as done by Garstka et al. When the promoter regions are selectively vacant at a certain level, a small amount of TFAM could enhance transcription because TFAM has a higher affinity for the promoters. A human Lon protease homologue is one candidate for such selective displacement, because the homologue was recently reported to bind preferentially to the GT-rich sequence overlapping the LSP of human mtDNA (21).
TFAM is essential for transcription of mtDNA, and TFAM-enabled transcription may be involved in the replication of mtDNA. However, we propose that TFAM may have a dual role in the maintenance of mtDNA, transcription and nucleoid formation. It is probable that the majority of TFAM molecules are involved in architecturally maintaining the higher structure of mtDNA, because at any time most TFAM molecules should be bound to nonpromoter regions. The higher structure is called nucleoid in general. However, it might be more appropriately described as a mitochondrial chromosome or mitochromosome when based on the whole structure of mtDNA.
Acknowledgments
This work was supported in part by the Uehara Memorial Foundation, the Naito Foundation, and Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Technology, Sports and Culture of Japan.
We extend special thanks to Eric A. Schon (Department of Genetics and Development and Neurology, Columbia University) for critical discussion. We are grateful to Keiji Hisaeda (Department of Medical Biochemistry, Kyushu University) for useful technical advice.
REFERENCES
- 1.Alam, T. I., T. Kanki, T. Muta, K. Ukaji, Y. Abe, H. Nakayama, K. Takio, N. Hamasaki, and D. Kang. 2003. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 31:1640-1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albring, M., J. Griffith, and G. Attardi. 1977. Association of a protein structure of probable membrane derivation with HeLa cell mitochondrial DNA near its origin of replication. Proc. Natl. Acad. Sci. USA 74:1348-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barat, M., D. Rickwood, C. Dufresne, and J. C. Mounolou. 1985. Characterization of DNA-protein complexes from the mitochondria of Xenopus laevis oocytes. Exp. Cell Res. 157:207-217. [DOI] [PubMed] [Google Scholar]
- 4.Bowmaker, M., M. Y. Yang, T. Yasukawa, A. Reyes, H. T. Jacobs, J. A. Huberman, and I. J. Holt. 2003. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 278:50961-50969. [DOI] [PubMed] [Google Scholar]
- 5.Caron, F., C. Jacq, and J. Rouviere-Yaniv. 1979. Characterization of a histone-like protein extracted from yeast mitochondria. Proc. Natl. Acad. Sci. USA 76:4265-4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dairaghi, D. J., G. S. Shadel, and D. A. Clayton. 1995. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J. Mol. Biol. 249:11-28. [DOI] [PubMed] [Google Scholar]
- 7.DeFrancesco, L., and G. Attardi. 1981. In situ photochemical crosslinking of HeLa cell mitochondrial DNA by a psoralen derivative reveals a protected region near the origin of replication. Nucleic Acids Res. 9:6017-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diffley, J. F., and B. Stillman. 1992. DNA binding properties of an HMG1-related protein from yeast mitochondria. J. Biol. Chem. 267:3368-3374. [PubMed] [Google Scholar]
- 9.Enriquez, J. A., A. Perez-Martos, M. J. Lopez-Perez, and J. Montoya. 1996. In organello RNA synthesis system from mammalian liver and brain. Methods Enzymol. 264:50-57. [DOI] [PubMed] [Google Scholar]
- 10.Falkenberg, M., M. Gaspari, A. Rantanen, A. Trifunovic, N. G. Larsson, and C. M. Gustafsson. 2002. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 31:289-294. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, R. P., and D. A. Clayton. 1988. Purification and characterization of human mitochondrial transcription factor 1. Mol. Cell. Biol. 8:3496-3509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fisher, R. P., T. Lisowsky, M. A. Parisi, and D. A. Clayton. 1992. DNA wrapping and bending by a mitochondrial high mobility group-like transcriptional activator protein. J. Biol. Chem. 267:3358-3367. [PubMed] [Google Scholar]
- 13.Garrido, N., L. Griparic, E. Jokitalo, J. Wartiovaara, A. M. van der Bliek, and J. N. Spelbrink. 2003. Composition and dynamics of human mitochondrial nucleoids. Mol. Biol. Cell 14:1583-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garstka, H. L., W. E. Schmitt, J. Schultz, B. Sogl, B. Silakowski, A. Perez-Martos, J. Montoya, and R. J. Wiesner. 2003. Import of mitochondrial transcription factor A (TFAM) into rat liver mitochondria stimulates transcription of mitochondrial DNA. Nucleic Acids Res. 31:5039-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goto, A., Y. Matsushima, T. Kadowaki, and Y. Kitagawa. 2001. Drosophila mitochondrial transcription factor A (d-TFAM) is dispensable for the transcription of mitochondrial DNA in Kc167 cells. Biochem. J. 354:243-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holt, I. J., H. E. Lorimer, and H. T. Jacobs. 2000. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100:515-524. [DOI] [PubMed] [Google Scholar]
- 17.Kaufman, B. A., S. M. Newman, R. L. Hallberg, C. A. Slaughter, P. S. Perlman, and R. A. Butow. 2000. In organello formaldehyde crosslinking of proteins to mtDNA: identification of bifunctional proteins. Proc. Natl. Acad. Sci. USA 97:7772-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, M. 1989. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol. Cell. Biol. 9:5073-5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Larsson, N. G., J. Wang, H. Wilhelmsson, A. Oldfors, P. Rustin, M. Lewandoski, G. S. Barsh, and D. A. Clayton. 1998. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat. Genet. 18:231-236. [DOI] [PubMed] [Google Scholar]
- 20.Lee, D. Y., and D. A. Clayton. 1998. Initiation of mitochondrial DNA replication by transcription and R-loop processing. J. Biol. Chem. 273:30614-30621. [DOI] [PubMed] [Google Scholar]
- 21.Liu, T., B. Lu, I. Lee, G. Ondrovicova, E. Kutejova, and C. K. Suzuki. 2004. DNA and RNA binding by the mitochondrial lon protease is regulated by nucleotide and protein substrate. J. Biol. Chem. 279:13902-13910. [DOI] [PubMed] [Google Scholar]
- 22.Matsushima, Y., K. Matsumura, S. Ishii, H. Inagaki, T. Suzuki, Y. Matsuda, K. Beck, and Y. Kitagawa. 2003. Functional domains of chicken mitochondrial transcription factor A for the maintenance of mitochondrial DNA copy number in lymphoma cell line DT40. J. Biol. Chem. 278:31149-31158. [DOI] [PubMed] [Google Scholar]
- 23.McCulloch, V., B. L. Seidel-Rogol, and G. S. Shadel. 2002. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol. Cell. Biol. 22:1116-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCulloch, V., and G. S. Shadel. 2003. Hum. mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol. Cell. Biol. 23:5816-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Megraw, T. L., and C. B. Chae. 1993. Functional complementarity between the HMG1-like yeast mitochondrial histone HM and the bacterial histone-like protein HU. J. Biol. Chem. 268:12758-12763. [PubMed] [Google Scholar]
- 26.Miyakawa, I., N. Sando, S. Kawano, S. Nakamura, and T. Kuroiwa. 1987. Isolation of morphologically intact mitochondrial nucleoids from the yeast, Saccharomyces cerevisiae. J. Cell Sci. 88:431-439. [DOI] [PubMed] [Google Scholar]
- 27.Muta, T., D. Kang, S. Kitajima, T. Fujiwara, and N. Hamasaki. 1997. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 272:24363-24370. [DOI] [PubMed] [Google Scholar]
- 28.Ohno, T., S. Umeda, N. Hamasaki, and D. Kang. 2000. Binding of human mitochondrial transcription factor A, an HMG box protein, to a four-way DNA junction. Biochem. Biophys. Res. Commun. 271:492-498. [DOI] [PubMed] [Google Scholar]
- 29.Pardue, M. L., J. M. Fostel, and T. R. Cech. 1984. DNA-protein interactions in the Drosophila virilis mitochondrial chromosome. Nucleic Acids Res. 12:1991-1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parisi, M. A., B. Xu, and D. A. Clayton. 1993. A human mitochondrial transcriptional activator can functionally replace a yeast mitochondrial HMG-box protein both in vivo and in vitro. Mol. Cell. Biol. 13:1951-1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Potter, D. A., J. M. Fostel, M. Berninger, M. L. Pardue, and T. R. Cech. 1980. DNA-protein interactions in the Drosophila melanogaster mitochondrial genome as deduced from trimethylpsoralen crosslinking patterns. Proc. Natl. Acad. Sci. USA 77:4118-4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sasaki, N., H. Kuroiwa, C. Nishitani, H. Takano, T. Higashiyama, T. Kobayashi, Y. Shirai, A. Sakai, S. Kawano, K. Murakami-Murofushi, and T. Kuroiwa. 2003. Glom is a novel mitochondrial DNA packaging protein in Physarum polycephalum and causes intense chromatin condensation without suppressing DNA functions. Mol. Biol. Cell 14:4758-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seidel-Rogol, B. L., and G. S. Shadel. 2002. Modulation of mitochondrial transcription in response to mtDNA depletion and repletion in HeLa cells. Nucleic Acids Res. 30:1929-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shadel, G. S., and D. A. Clayton. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66:409-435. [DOI] [PubMed] [Google Scholar]
- 35.Spelbrink, J. N., F. Y. Li, V. Tiranti, K. Nikali, Q. P. Yuan, M. Tariq, S. Wanrooij, N. Garrido, G. Comi, L. Morandi, L. Santoro, A. Toscano, G. M. Fabrizi, H. Somer, R. Croxen, D. Beeson, J. Poulton, A. Suomalainen, H. T. Jacobs, M. Zeviani, and C. Larsson. 2001. Hum. mitochondrial DNA deletions associated with mutations in the gene encoding Twinkle, a phage T7 gene 4-like protein localized in mitochondria. Nat. Genet. 28:223-231. [DOI] [PubMed] [Google Scholar]
- 36.Takamatsu, C., S. Umeda, T. Ohsato, T. Ohno, Y. Abe, A. Fukuoh, H. Shinagawa, N. Hamasaki, and D. Kang. 2002. Regulation of mitochondrial D-loops by transcription factor A and single-stranded DNA-binding protein. EMBO Rep. 3:451-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang, Y., E. A. Schon, E. Wilichowski, M. E. Vazquez-Memije, E. Davidson, and M. P. King. 2000. Rearrangements of human mitochondrial DNA (mtDNA): new insights into the regulation of mtDNA copy number and gene expression. Mol. Biol. Cell 11:1471-1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang, M. Y., M. Bowmaker, A. Reyes, L. Vergani, P. Angeli, E. Gringeri, H. T. Jacobs, and I. J. Holt. 2002. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111:495-505. [DOI] [PubMed] [Google Scholar]
- 39.Zelenaya-Troitskaya, O., S. M. Newman, K. Okamoto, P. S. Perlman, and R. A. Butow. 1998. Functions of the high mobility group protein, Abf2p, in mitochondrial DNA segregation, recombination and copy number in Saccharomyces cerevisiae. Genetics 148:1763-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]