Table 1.
Results from simulation scenario 1 (1000 data sets, n = 200)
| Parameter | Method | Bias | Coverage | ESD |
|---|---|---|---|---|
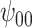 : Intercept : Intercept |
REG | 0.01 | 0.93 | 0.72 |
| IPTW | - 0.05 | 0.93 | 1.31 | |
| IPTWtr | - 0.14 | 0.93 | 1.06 | |
| IPTWaug | 0.02 | 0.95 | 1.05 | |
| TMLE | - 0.05 | 0.90 | 1.08 | |
| TMLEboot | - 0.05 | 0.94 | 1.08 | |
| BNP | - 0.07 | 0.97 | 1.00 | |
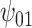 : V : V
|
REG | - 0.03 | 0.94 | 1.05 |
| IPTW | - 0.07 | 0.92 | 2.25 | |
| IPTWtr | - 0.13 | 0.93 | 1.67 | |
| IPTWaug | - 0.01 | 0.96 | 1.75 | |
| TMLE | 0.14 | 0.86 | 1.93 | |
| TMLEboot | 0.14 | 0.94 | 1.93 | |
| BNP | 0.13 | 0.96 | 1.27 | |
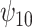 : A : A
|
REG | - 0.04 | 0.95 | 1.16 |
| IPTW | 0.34 | 0.91 | 2.05 | |
| IPTWtr | 0.74 | 0.91 | 1.52 | |
| IPTWaug | - 0.05 | 0.95 | 1.86 | |
| TMLE | 0.42 | 0.70 | 2.22 | |
| TMLEboot | 0.42 | 0.93 | 2.22 | |
| BNP | 0.16 | 0.96 | 1.24 | |
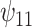 : A × V : A × V
|
REG | 0.04 | 0.95 | 1.53 |
| IPTW | - 0.06 | 0.91 | 3.11 | |
| IPTWtr | - 0.17 | 0.94 | 2.31 | |
| IPTWaug | 0.03 | 0.95 | 2.58 | |
| TMLE | - 0.39 | 0.71 | 3.28 | |
| TMLEboot | - 0.39 | 0.94 | 3.28 | |
| BNP | - 0.37 | 0.96 | 1.60 |
The true values were: 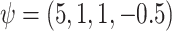 . REG is the correctly specified regression model. IPTW and IPTWtr use a correctly specified propensity score. IPTWaug uses a correctly specified outcome and propensity score model. TMLE uses a correctly specified propensity score and Super Learner for the outcome model. TMLEboot uses bootstrap confidence intervals, rather than asymptotic intervals. BNP is the proposed method. Bias is the absolute bias and ESD is the empirical standard deviation.
. REG is the correctly specified regression model. IPTW and IPTWtr use a correctly specified propensity score. IPTWaug uses a correctly specified outcome and propensity score model. TMLE uses a correctly specified propensity score and Super Learner for the outcome model. TMLEboot uses bootstrap confidence intervals, rather than asymptotic intervals. BNP is the proposed method. Bias is the absolute bias and ESD is the empirical standard deviation.
