Abstract
We found that mammalian Polycomb group (PcG) protein YY1 can bind to Polycomb response elements in Drosophila embryos and can recruit other PcG proteins to DNA. PcG recruitment results in deacetylation and methylation of histone H3. In a CtBP mutant background, recruitment of PcG proteins and concomitant histone modifications do not occur. Surprisingly, YY1 DNA binding in vivo is also ablated. CtBP mutation does not result in YY1 degradation or transport from the nucleus, suggesting a mechanism whereby YY1 DNA binding ability is masked. These results reveal a new role for CtBP in controlling YY1 DNA binding and recruitment of PcG proteins to DNA.
Keywords: YY1; Polycomb, CtBP; transcription; PRE
Polycomb group (PcG) proteins are transcriptional repressors that maintain the spatially restricted expression patterns of hox genes in both flies and vertebrates (Pirrotta 1998). In flies, 15 different PcG proteins are required to repress homeotic genes. These proteins work in concert and absence of any one protein results in derepression of target genes. A number of vertebrate proteins homologous to Drosophila PcG proteins have been identified. These mammalian PcG proteins regulate hox gene expression and are important for skeletal development and hematopoiesis (Jacobs and van Lohuizen 2002). PcG proteins mediate transcription repression by binding to conserved DNA sequence elements known as Polycomb response elements (PREs) (Pirrotta 1997a,b; Francis and Kingston 2001). These sequences have been characterized in Drosophila, but no mammalian counterparts have been identified. PREs contain binding sites for sequence-specific DNA-binding proteins Pleiohomeotic (PHO) and Pleiohomeotic-like (Phol), which are homologs of the ubiquitous mammalian transcription factor, Yin Yang-1 (YY1) (Brown et al. 1998, 2003; Mihaly et al. 1998).
In a transgenic Drosophila system, we previously showed that YY1 repressed transcription in a PcG-dependent manner (Atchison et al. 2003). YY1 also functionally compensated for loss of PHO in pho mutant flies and partially corrected pho mutant phenotypes. These results clearly demonstrated PcG function for YY1 and suggested that YY1 might recruit PcG proteins to DNA.
Results and Discussion
YY1 recruits PcG proteins to DNA
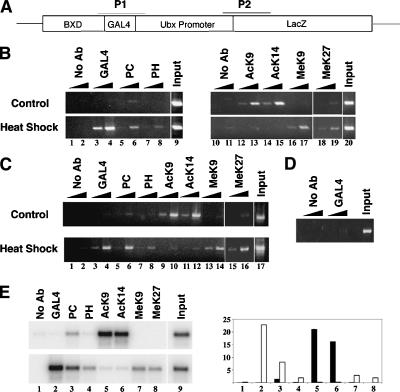
To determine whether YY1 could recruit PcG proteins to DNA, we performed chromatin immunoprecipitation (ChIP) assays in a transgenic Drosophila embryo system consisting of hsp70-driven GALYY1 and a reporter construct containing the LacZ gene under control of the Ultrabithorax (Ubx) BXD enhancer and the Ubx promoter adjacent to GAL4-binding sites (BGUZ) (Fig. 1A; Atchison et al. 2003). The BGUZ reporter is expressed ubiquitously during embryogenesis but is selectively repressed in a PcG-dependent manner by GALYY1 and GALPc (Muller 1995; Atchison et al. 2003). Embryos were either left untreated or heat shocked to induce GALYY1 expression. After immunoprecipitation with various antibodies, the region surrounding the GAL4-binding sites in the BGUZ reporter was detected by PCR. Prior to heat shock, no GALYY1 could be observed at the reporter gene (Fig. 1B, lanes 3,4, top). After heat shock, GALYY1 binding to the reporter gene was easily detected (Fig. 1B, lanes 3,4, bottom). Interestingly, concomitant with GALYY1 binding, there was an increase in binding of the Polycomb (Pc) and Polyhomeiotic (Ph) proteins (Fig. 1B, lanes 5-8). Thus, YY1 DNA binding results in PcG recruitment to DNA.
Figure 1.
YY1 recruits PcG proteins resulting in methylation and deacetylation of histone H3. (A) Schematic representation of the reporter construct in transgenic BGUZ flies. The GAL4 and Ubx-LacZ promoter regions amplified by PCR in ChIP assays are depicted as horizontal lines, P1 and P2, respectively. (B,C) Chip assays showing DNA binding by GALYY1, recruitment of PcG proteins (PC and PH), deacetylation of H3 (at K9 and K14), and methylation of H3 (at K9 and K27). Chromatin was prepared from hsp70GALYY1 BGUZ embryos that were either untreated (top panel), or heat shocked for 45 min at 37°C (bottom panel) to induce GALYY1 expression. The antibody used for immunoprecipitation is shown above each lane. Polymerase chain reaction (PCR) was performed with 0.5 and 5 ng of immunoprecipitated DNA. As controls, mock-precipitated (No Ab) and genomic DNA (Input) samples were amplified. Panel B shows the amplification of region P1, and C depicts region P2. (D) Wild-type GAL4 protein does not bind to the Ubx promoter region. Wild-type GAL4 was induced by heat shock, and binding to the Ubx promoter (P2) was assayed by ChIP and detected by PCR. (E) GALYY1 binds to endogenous PRE sequences and augments PcG recruitment and histone modification. ChIP assays were performed with untreated (top panel) or heat-shocked (bottom panel) hsp70GALYY1 BGUZ embryos. PCR was performed with primers that amplify the ubx PRED region, and samples were subjected to Southern blot analysis using the PRE sequence as probe. Quantitation of the data obtained is shown in the right panel (black bars are uninduced, white bars are heat-shock induced). Numbers on the X-axis in the right panel correspond to lane numbers in the left panel, and numbers on the Y-axis are arbitrary relative units.
Binding of PcG proteins to PRE sequences is known to cause deacetylation of histone H3 (Tie et al. 2003) and methylation on Lys 9 and Lys 27 (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002; Sewalt et al. 2002). Interestingly, induction of GALYY1 binding to the reporter gene resulted in loss of histone H3 acetylation on K9 and K14 (Fig. 1B, lanes 12-15). Simultaneously, there was a gain of methylation on histone H3 Lys 9 and Lys 27 (Fig. 1B, lanes 16-19). Therefore, YY1 binding to the BGUZ reporter results in the recruitment of PcG proteins to DNA and subsequent post-translational modifications of histones characteristic of PcG complexes.
We next determined the presence of PcG proteins and the status of histone H3 modifications at the Ubx promoter region, which is 4 kb downstream of the GALYY1-binding site. To avoid amplification of the endogenous Ubx promoter, immunoprecipitated samples were amplified with primers spanning the Ubx-LacZ boundary (region P2, Fig. 1A). Interestingly, we detected the presence of Pc and Ph at the promoter after GALYY1 induction (Fig. 1C, lanes 5-8). We also detected the presence of GALYY1 at this site (Fig. 1C, lanes 3, 4). The GAL4 protein alone did not bind to the Ubx promoter region, indicating specificity for YY1 sequences (Fig. 1D). The induced GAL4 protein was functional, however, because it efficiently bound to the GAL4-binding site in the BGUZ reporter (see Fig. 3B, below). Binding by GALYY1 could, therefore, be due to either cryptic YY1-binding sites present at the promoter, physical association of GALYY1 with other proteins bound at the promoter, or interactions via looping of DNA between the GAL4-binding sites and the Ubx promoter. Again, induction of GALYY1 resulted in loss of acetylation of H3K9 and H3K14 and simultaneous gain of methylation on H3K9 and H3K27 (Fig. 1C, lanes 9-16). These results are consistent with earlier studies that reported spreading of PcG proteins and histone modifications to flanking DNA (Orlando 2003).
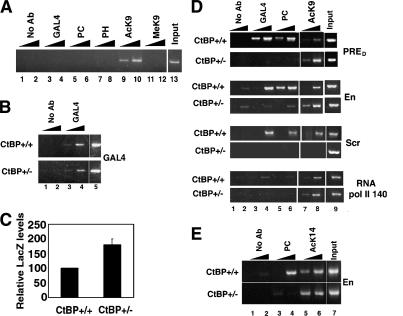
Figure 3.
PcG function of YY1 is controlled by CtBP. (A) Loss of DNA binding by YY1, Pc, and Ph in heterozygous CtBP mutant embryos. ChIP assays were performed with heterozygous CtBP mutant embryos expressing heat shock-induced GALYY1. The antibody used for immunoprecipitation is shown above each lane. PCR was performed with 0.5 and 5 ng of immunoprecipitated DNA. Genomic DNA (Input) served as positive control and mock-immunoprecipitated sample (No Ab) as negative control. (B, lanes 3,4) Chip assays depicting DNA binding by GAL4 protein in wild-type and heterozygous CtBP mutant embryos expressing heat shock-induced GAL4. (C) LacZ expression increases in a CtBP mutant background. Relative differences measured by real-time RT-PCR of LacZ expression in wild-type and ctbp+/- mutant embryos is shown. Actin transcripts served as an internal control. (D) ChIP analysis of protein distribution and histone modification status at Ubx PRED, engrailed (en), and sex combs reduced (scr) PREs and RpII140 promoter regions in wild-type (top panels) and CtBP heterozygous mutant (bottom panels) embryos. (E) Pc binding is reduced in a CtBP mutant background. ChIP assays show loss of Pc DNA binding at the en PRE in a CtBP mutant background.
YY1 recruits PcG proteins to an endogenous PRE
PHO and YY1 bind to the same DNA sequence, and PHO-binding sites have been identified in multiple PREs (Mihaly et al. 1998; Fritsch et al. 1999). Therefore, we reasoned that YY1 would bind to endogenous PREs and perhaps increase recruitment of PcG proteins. For this, we analyzed the major ubx PRE (PRED) that contains multiple PHO-binding sites located in the bxd region (Fritsch et al. 1999). As expected, upon GALYY1 induction, we detected GALYY1 at this endogenous PRE site (Fig. 1E, lane 2). In addition, YY1 binding was accompanied by an increase in Pc and Ph signals when compared with no heat shock controls (Fig. 1E, lanes 3,4) and a loss of H3 K9 and H3 K14 acetylation and gain of H3 K9 and H3 K27 methylation (Fig. 1E, lanes 5-8). Quantitation of changes in PRE occupancy and changes in histone modification are shown in Figure 1E, right panel. Thus, YY1 can bind to an endogenous PRE and can augment PcG recruitment.
E(z) function is crucial for YY1-mediated repression in vivo
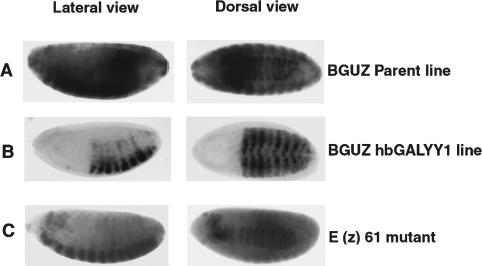
Our results clearly indicated that YY1 DNA binding results in recruitment of PcG proteins, histone deacetylases (HDACs), and histone methyltransferases (HMTases) to DNA. To determine whether the Drosophila E(z) protein (which possesses HMTase activity) was involved, we explored whether YY1 transcriptional repression was lost in an E(z) mutant background. For these studies, we crossed a recombinant chromosome line containing the BGUZ reporter and hunchback-driven GALYY1 (hbGalYY1 BGUZ) (Atchison et al. 2003) into a temperature-sensitive E(z)61 mutant background. The recombinant line gives a pulse of GALYY1 in the anterior ends of developing Drosophila embryos. The parent BGUZ line produces uniform LacZ expression throughout the embryo (Fig. 2A), but in the recombinant chromosome line, LacZ expression is selectively repressed in the anterior ends by hunchback-driven GALYY1 (Fig. 2B). Interestingly, there was a significant derepression of YY1 function in the E(z)61 mutant background when shifted to restricted temperature (Fig. 2C). These results are consistent with the observation that E(z) specifically methylates histone H3 on Lys 27, which creates a binding site for the chromodomain of Pc (Fischle et al. 2003; Min et al. 2003). Thus, the repression we observe with GALYY1 requires function of the E(z) PcG protein.
Figure 2.
Transcriptional repression by YY1 requires E(z) function. Embryos were collected from the BGUZ parent (A), the BGUZ hbGALYY1 recombinant chromosome line (B), and the recombinant chromosome line in an E(z)61 mutant background (C). Embryos were collected at 6 h at 29°C and processed for LacZ staining. Blue staining indicates LacZ expression and the light-colored areas indicate repression of LacZ expression by GALYY1.
CtBP is needed for YY1 DNA binding and PcG recruitment
We previously showed that YY1 interacts with Drosophila CtBP, a well-characterized corepressor molecule (Poortinga et al. 1998; Phippen et al. 2000; Chinnadurai 2002; Atchison et al. 2003). CtBP can also interact with Pc in vivo (Sewalt et al. 1999). These associations led us to propose that CtBP might play a bridging function between YY1 and PcG proteins (Atchison et al. 2003). If true, one would expect loss of PcG recruitment to DNA in a CtBP mutant background. Indeed, ChIP experiments in a CtBP03463/+ background showed greatly reduced Pc and Ph recruitment to the BGUZ reporter (Fig. 3A, lanes 5-8). In addition, histone H3 remained acetylated and unmethylated (Fig. 3A, lanes 9-12). Surprisingly, in a CtBP mutant background, we also observed dramatic loss of GALYY1 DNA binding (Fig. 3A, lanes 3,4). However, full-length GAL4 protein was able to bind to DNA equally well in wild-type and CtBP mutant backgrounds (Fig. 3B, lanes 3,4), indicating that the effect of CtBP mutation was specific for YY1. This was a very unexpected result because CtBP has never been demonstrated to control DNA binding of another protein. The absence of GALYY1 and PcG proteins bound to the BGUZ reporter in the CtBP mutant background suggested that expression of the LacZ gene should be increased. Indeed, LacZ expression was increased in CtBP mutant as compared with wild-type embryos, as revealed by real-time RT-PCR (Fig. 3C). Thus, in a CtBP mutant background, GALYY1 did not bind DNA, PcG proteins were not recruited, histones remained acetylated and unmethylated, and transcription was derepressed.
CtBP mutation affects YY1 and Pc binding to multiple PREs
To be certain that this effect was not peculiar to the BGUZ reporter, we analyzed the effect of CtBP mutation on GALYY1 and PcG binding at endogenous PREs. For this, we chose the Ubx PRED (Fritsch et al. 1999), engrailed (en) PRE (Kassis 1994; Americo et al. 2002), and sex combs reduced (scr) PRE (Gindhart and Kaufman 1995). In addition, we chose RpII140 (the 140-kDa subunit of RNA polymerase II) as a negative control because it previously was shown not to be regulated by PcG proteins (Breiling et al. 2001). Strikingly, GALYY1 and Pc binding to all three PREs was greatly reduced in the CtBP mutant background (Fig. 3D, lanes 3-6). Reduction in GALYY1 and Pc DNA binding correlated with H3 K9 acetylation at the PRED and En PREs. On the other hand, H3 K9 acetylation at the Scr PRE was lost in a CtBP mutant background (Fig. 3D, lanes 7,8). As expected, RpII140 did not bind GALYY1 or PcG proteins and was acetylated on H3 K9 (Fig. 3D). These results clearly indicate an essential role for Drosophila CtBP in PcG recruitment to DNA.
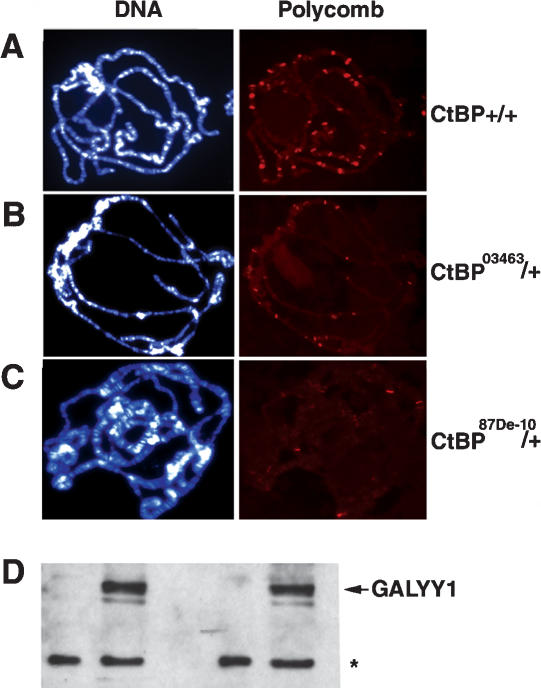
To be certain that reduced PcG binding in the ctbp+/- mutant background was not due to excess GALYY1 causing a shift in cellular equilibrium, we performed ChIP experiments with nontransgenic wild-type and ctbp+/- mutant embryos that do not express GALYY1. As anticipated, Pc binding was greatly reduced at the en PRE in a ctbp+/- background compared with wild type (Fig. 3E, lanes 3,4). Concomitant with loss in PC binding, there was an increase in AcK14 levels in the mutant (Fig. 3E, lanes 5,6). In addition, we explored global Pc binding to polytene chromosomes in nontransgenic wild-type and ctbp+/- mutant backgrounds. As shown in Figure 4A, polytene spreads from wild-type larvae showed ∼60 (SE ± 1.54) specific bands when probed with Pc antisera. On the other hand, polytene spreads from ctbp03463/+ heterozygous larvae (Fig. 4B) showed an average of 15 (SE ± 2.69) specific Pc bands per chromosome spread, indicating a 75% drop in Pc stained bands in the mutant background. To be certain that this effect was not peculiar to the genetic background of the ctbp03463 mutant line, we analyzed a distinct ctbp mutant allele, 87De-10, that fails to complement ctbp03463 (Poortinga et al. 1998). Again, polytene spreads from ctbp87De-10 larvae on average showed a 66% (SE ± 4.8) loss in Pc stained bands (Fig. 4C). Thus, CtBP mutation affected PcG recruitment to a significant subset of PREs.
Figure 4.
(A-C) Pc binding to polytene chromosomes is reduced in CtBP mutant larvae. Salivary gland polytene chromosomes were immunostained with anti-Pc antibodies (red) and DAPI (blue). (A) Polytene spreads from wild-type larvae show ∼60 Pc-binding sites. Pc binding in ctbp03463 (B) and ctbp87De-10 (C) heterozygous mutant larvae showed 75% and 66% (p < 0.0005) reductions in Pc binding on polytene chromosomes, respectively. (D) YY1 levels, mobility, and nuclear localization are unchanged by CtBP mutation. Nuclear and cytoplasmic extracts from wild-type and CtBP mutant embryos were assayed by Western blot with antibodies to GAL4. The position of GALYY1 is indicated. The asterisk denotes a background band that is comparable in all samples.
YY1 stability, mobility, and nuclear localization are unchanged by CtBP mutation
A number of mechanisms might account for the reduced YY1 DNA binding in a CtBP mutant background. YY1 might be degraded, transported from the nucleus to the cytoplasm, post-translationally modified to a form that cannot bind DNA, or sequestered by proteins that inhibit its DNA binding ability. To test these possibilities, we prepared nuclear and cytoplasmic extracts from both wild-type and ctbp03463/+ mutant embryos and assessed GALYY1 by Western blot (Fig. 4D). In both genotypes, the levels of GALYY1 remained indistinguishable, indicating that GALYY1 was not degraded in the mutant background. GALYY1 also remained nuclear, indicating that it was not transported to the cytoplasm. In addition, the electrophoretic mobility of YY1 in both genotypes was the same, indicating an absence of differences in post-translational modification that might alter mobility in SDS-polyacrylamide gels. We also did not detect a difference in ability of GALYY1 extracted from wild-type and mutant embryos to bind to DNA in vitro by electrophoretic mobility shift assay (data not shown). Therefore, GALYY1 is not modified to a form that cannot bind to DNA in vitro. Similarly, we found no evidence of CtBP altering the stability of YY1 bound to DNA in vitro (data not shown). Based on these results, the most likely mechanism is that in a CtBP mutant background, GALYY1 interacts either with proteins that inhibit its ability to bind to DNA or that sequester it to a different subnuclear compartment.
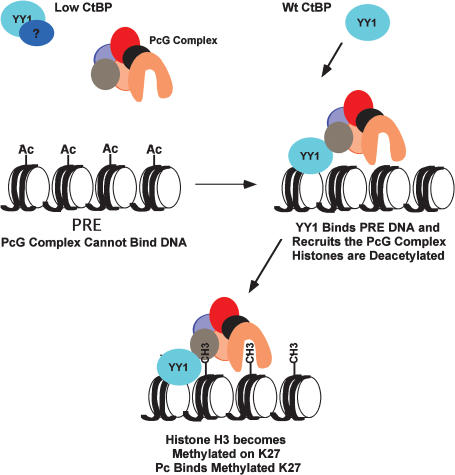
Model of YY1 recruitment of PcG proteins
Collectively, our studies clearly demonstrate PcG recruitment function by the multifunctional transcription factor YY1. This establishes YY1 DNA binding as a key mechanism for targeting PcG proteins to DNA. The loss of YY1 DNA binding and concomitant loss of PcG recruitment to reporters and endogenous PRE sequences in CtBP mutants underscores this mechanism. A model of YY1 and CtBP function is presented in Figure 5. We propose that in a CtBP mutant background, YY1 is sequestered by a protein that inhibits its ability to bind to DNA. In a CtBP wild-type background, YY1 is released from this protein, thus enabling it to bind to DNA. DNA binding by YY1 results in recruitment of PcG complexes that cause deacetylation of histones and methylation of histone H3 at Lys 9 and Lys 27. Deacetylation may also be mediated by HDACs directly recruited by interaction with YY1 (Yao et al. 2001).
Figure 5.
Model of YY1 and CtBP function in PcG recruitment.
The ablation of YY1 DNA binding in a CtBP mutant background was totally unexpected. This represents a new mechanism for controlling YY1 DNA binding and PcG recruitment. The mechanism appears to be exquisitely sensitive to CtBP dose because YY1 DNA binding and PcG recruitment are greatly reduced in heterozygous mutant backgrounds. Heterozygous effects by CtBP on knirps and hairy mutant phenotypes have been observed in other systems (Poortinga et al. 1998; Phippen et al. 2000), suggesting that CtBP levels are limiting in vivo.
The exact role of CtBP in PcG-mediated repression is yet to be elucidated. Our results suggest that CtBP is required for the function of a large subset of PREs that require YY1/PHO for PcG recruitment. Like PcG mutants, CtBP mutants in flies show segmentation defects (Poortinga et al. 1998), but homeotic derepression has not been observed. Heterozygous ctbp mutants can reverse pair-rule phenotypes observed in hairy mutants, and homozygotes show bristle and cuticle defects (Poortinga et al. 1998; Phippen et al. 2000). Furthermore, embryos that are trans-heterozygous for wimp and the ctpb03463 allele die and their cuticle preparations show severe segmentation defects (Poortinga et al. 1998). Similarly, mouse ctbp1 and ctbp2 null mutants show a variety of defects including skeletal abnormalities (Hildebrand and Soriano 2002), but these defects do not precisely match the skeletal posterior transformations seen with mammalian PcG mutants (Akasaka et al. 1996; Schumacher et al. 1996; Bel et al. 1998). Based on the multiple PREs affected by CtBP mutation, it is unclear why a more severe CtBP heterozygous mutant phenotype is not observed. Perhaps a low level of PcG binding to DNA remains that is below detection in immunostains of polytene chromosomes, but which is sufficient to mediate biological effects. In support of this possibility, we occasionally observed polytene spreads that stained with Pc antibodies nearly as well as wild-type spreads. This suggests a possible threshold effect for CtBP involvement in PcG recruitment. ChIP studies on many more PRE sequences will be needed to clarify this issue.
Our results show that modulation of YY1 DNA binding by CtBP is a critical step in the recruitment of PcG proteins to DNA. This mechanism might be differentially used during development to control PcG assembly on PREs. Our demonstration of recruitment of PcG proteins by YY1 should assist in the identification of mammalian PREs since the YY1 recognition sequence is well characterized.
Materials and methods
Drosophila lines, crosses, and antibodies
The BGUZ, hbGALYY1 BGUZ, and hspGALYY1 transgenic fly lines have been described previously (Atchison et al. 2003). The E(z)61 line was provided by Richard Jones (South Methodist University, Dallas, TX) (Carrington and Jones 1996). CtBP mutant lines ctbp03463 (P11590) and ctbp87De-10 (BL1663) were obtained from Bloomington stock center. The hsGAL4 transgenic line was provided by Amita Sehgal (University of Pennsylvania, Philadelphia, PA). Females from the hbGALYY1 BGUZ recombinant line were crossed with E(z)61 males and the resulting males were crossed to virgin females from the E(z)61 mutant stock. Embryos were collected from grape plates after 6 h. Temperature-sensitive E(z) mutant crosses were set up at 18°C and shifted to 29°C 48 h prior to embryo collection. For ChIP assays in a CtBP mutant background, hspGALYY1 BGUZ females were crossed to ctbp03463 balanced over TM3GFP. Rabbit anti-Ph antibodies were raised against a GST-fusion protein containing PH residues 87-431 and affinity purified using the same fusion protein. Antibodies against Pc- and anti-histone H3-di/trimethyl K27 were obtained from V. Pirrotta (University of Geneva, Geneva, Switzerland) and D. Reinberg, (University of Medicine and Dentistry of New Jersey, Piscataway, NJ), respectively. The anti-GAL4 antibody was obtained from Santa Cruz Biotechnology and the rest of histone modification antibodies were from Upstate Cell Signalling Solutions.
ChIP assay
Cross-linked chromatin was prepared from embryos that were either untreated or heat shocked for 45 min at 37°C, and immunoprecipitation was performed as described previously (Orlando et al. 1998). Details about the assay are provided in the Supplemental Material.
LacZ staining and subcellular fractionation of embryos
Staged embryos were stained for LacZ as previously described (Atchison et al. 2003). For subcellular fractionation, embryos were dechorionated with 50% chlorox, washed in PBS plus 0.01% Triton X-100, and homogenized in buffer A (15 mM HEPES at pH 7.6, 10 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 0.5 mM EGTA, 350 mM sucrose, 1 mM DTT, and 0.5 mM PMSF). Samples were centrifuged at 5000g for 10 min to generate the cytoplasmic fraction and the nuclear pellet. Nuclei were lysed in lysis buffer (20 mM HEPES at pH 7.6, 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 25% glycerol, 1 mM DTT, and 0.5 mM PMSF) for 30 min and the extracts were clarified by centrifugation at 12,000g for 10 min. All steps were carried out at 4°C. Equal amounts of cytoplasmic and nuclear extracts were fractionated by SDS-PAGE and then subjected to Western blot procedure with anti-GAL4 (Santa Cruz Biotechnology) antibodies.
Immunostaining of polytene chromosomes
Drosophilapolytene chromosome preparation and immunostaining were performed as previously described (Zink and Paro 1995). Rabbit anti-PC antibody was used at a 1:100 dilution and Cy3 conjugated anti-rabbit IgG (Jackson Immunoresearch) was used at a 1:200 dilution. Chromosomes were mounted in Vectastain (Vector Laboratories) containing DAPI. Spreads were imaged using a Leica fluorescence microscope and the images were processed using Open-lab and Adobe Photoshop software.
RT-PCR
For RT-PCR, RNA was extracted from embryos using Trizol reagent (Invitrogen). Five micrograms of total RNA was used for first-strand cDNA synthesis using Superscript II (Invitrogen) and oligo (dT) primers according to the manufacturer's protocol. Real-time PCR was performed in triplicate using SYBR Green detection and Light Cycler System from Roche Molecular Biochemicals. Quantitative PCR reactions were performed under standardized conditions, and, to compare the relative amount of target in different samples, all values were normalized to actin control. The primers used for quantitative PCR were as follows: actin cDNA primers: CGTCGTTTTACAACGTCGTGAC, CGTTGGTGTAGATGGGCGCATC; and LacZ primers: GGGAATTCACTGGCCGTCGTTTTA, ATTCGCGTCTGGCCTTCCTGTAGC.
Acknowledgments
We thank Richard Jones and Amita Sehgal for fly strains and Vincenzo Pirrotta and Danny Reinberg for plasmids and antibodies. We are grateful to Alexander Mazo and Sheryl Smith for helpful advice on ChIP assays. We thank John Pehrson and Frank Wilkinson for critical reading of the manuscript. This work was supported by NIH grant GM42415 and March of Dimes grant 1-FY02-173 to M.L.A.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1228204.
References
- Akasaka T., Kanno, M., Balling, R., Mieza, M.A., Taniguchi, M., and Koseki, H. 1996. A role for mel-18, a polycomb group-related vertebrate gene, during the anteroposterior specification of the axial skeleton. Development 122: 1513-1522. [DOI] [PubMed] [Google Scholar]
- Americo J., Whiteley, M., Brown, J.L., Fujioka, M., Jaynes, J.B., and Kassis, J.A. 2002. A complex array of DNA-binding proteins required for pairing-sensitive silencing by a polycomb group response element from the Drosophila engrailed gene. Genetics 160: 1561-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchison L., Ghias, A., Wilkinson, F., Bonini, N., and Atchison, M.L. 2003. The YY1 transcription factor functions as a PcG protein in vivo. EMBO J. 22: 1347-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bel S., Core, N., Djaball, M., Kieboo, K., Van der Lugt, N., Alkema, M.J., and Van Lohuizen, M. 1998. Genetic interactions and dosage effects of Polycomb group genes in mice. Development 125: 3543-3551. [DOI] [PubMed] [Google Scholar]
- Breiling A., Turner, B.M., Bianchi, M.E., and Orlando, V. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412: 651-655. [DOI] [PubMed] [Google Scholar]
- Brown J.L., Mucci, D., Whiteley, M., Dirksen, M.-L., and Kassis, J.A. 1998. The Drosophila polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1: 1057-1064. [DOI] [PubMed] [Google Scholar]
- Brown J.L., Fritsch, C., Mueller, J., and Kassis, J.A. 2003. The Drosophila pho-like gene encodes a YY1-related DNA binding protein that is redundant with pleiohomeotic in homeotic gene silencing. Development 130: 285-294. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R.S., and Zhang, Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039-1043. [DOI] [PubMed] [Google Scholar]
- Carrington E.A. and Jones, R.S. 1996. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: Examination of wild-type and mutant protein distribution. Development 122: 4073-4083. [DOI] [PubMed] [Google Scholar]
- Chinnadurai G. 2002. CtBP, an unconventional transcriptional corepressor in development and oncogenesis. Mol. Cell 9: 213-224. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi, R., McCabe,D., Seitz, V., Imhof, A., and Pirrotta, V. 2002. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185-196. [DOI] [PubMed] [Google Scholar]
- Fischle W., Wang, Y., Jacobs, S.A., Kim, Y., Allis, C.D., and Khorasanizadeh, S. 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes & Dev. 17: 1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis N.J. and Kingston, R.E. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2: 409-421. [DOI] [PubMed] [Google Scholar]
- Fritsch C., Brown, J.L., Kassis, J.A., and Muller, J. 1999. The DNA-binding Polycomb group protein pleiohomeotic mediates silencing of a Drosophila homeotic gene. Development 126: 3905-3913. [DOI] [PubMed] [Google Scholar]
- Gindhart J.G.J. and Kaufman, T.C. 1995. Identification of polycomb and trithorax group responsive elements in the regulatory region of the Drosophila homeotic gene sex combs reduced. Genetics 139: 797-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand J.D. and Soriano, P. 2002. Overlapping and unique roles for C-terminal binding protein 1 (CtBP1) and CtBP2 during mouse development. Mol. Cell. Biol. 22: 5296-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.J.L. and van Lohuizen, M. 2002. Polycomb repression: From cellular memory to cellular proliferation and cancer. Biochim. Biophys. Acta 1602: 151-161. [DOI] [PubMed] [Google Scholar]
- Kassis J.A. 1994. Unusual properties of regulatory DNA from the Drosophila engrailed gene: Three “pairing-sensitive” sites within a 1.6 kb region. Genetics 136: 1025-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka, K., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing enhancer of Zeste protein. Genes & Dev. 16: 2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihaly J., Mishra, R.K., and Karch, F. 1998. A conserved sequence motif in Polycomb response elements. Mol. Cell 1: 1065-1066. [DOI] [PubMed] [Google Scholar]
- Min J., Zhang, Y., and Xu, R.M. 2003. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes & Dev. 17: 1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J. 1995. Transcriptional silencing by the polycomb protein in Drosophila embryos. EMBO J. 6: 1209-1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J., Hart, C.M., Francis, N.J., Vargas, M.L., Sengupta, A., Wild, B., Miller, E.L., O'Connor, M.B., Kingston, R.E., and Simon, J.A. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197-208. [DOI] [PubMed] [Google Scholar]
- Orlando V. 2003. Polycomb, epigenomes, and control of cell identity. Cell 112: 599-606. [DOI] [PubMed] [Google Scholar]
- Orlando V., Jane, E.P., Chinwalla, V.B., Harte, P.J., and Paro, R. 1998. Binding of Trithorax and Polycomb proteins to the bithorax complex: Dynamic changes during early Drosophila embryogenesis. EMBO J. 17: 5141-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phippen T.M., Sweigart, A.L., Moniwa, M., Krumm, A., Davie, J.R., and Parkhurst, S.M. 2000. Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both mad and groucho transcriptional repression. J. Biol. Chem. 275: 37628-37637. [DOI] [PubMed] [Google Scholar]
- Pirrotta V. 1997a. Chromatin-silencing mechanisms in Drosophila maintain patterns of gene expression. Trends Genet. 13: 314-318. [DOI] [PubMed] [Google Scholar]
- ____. 1997b. PcG complexes and chromatin silencing. Curr. Opin. Genet. Dev. 7: 249-258. [DOI] [PubMed] [Google Scholar]
- ____. 1998. Polycombing the genome: PcG, trxG, and chromatin silencing. Cell 93: 333-336. [DOI] [PubMed] [Google Scholar]
- Poortinga G., Watanabe, M., and Parkhurst, S.M. 1998. Drosophila CtBP: A Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J. 17: 2067-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A., Faust, C., and Magnuson, T. 1996. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature 383: 250-253. [DOI] [PubMed] [Google Scholar]
- Sewalt R.G.A.B., Gunster, M.J., van der Vlag, J., Satijn, D.P.E., and Otte, A.P. 1999. C-terminal binding protein is a transcriptional repressor that interacts with a specific class of vertebrate Polycomb proteins. Mol. Cell. Biol. 19: 777-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sewalt R.G.A.B., Lachner, M., Vargas, M.L., Hamer, K.M., den Blaauwen, J.L., Hendrix, T., Mlecher, M., Schwiezer, D., Jenuwein, T., and Otte, A.P. 2002. Selective interactions between vertebrate Polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol. Cell. Biol. 22: 5539-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F., Prasad-Sinha, J., Birve, A., Rasmuson-Lestander, A., and Harte, P.J. 2003. A 1-megadalton ESC/E(Z) complex from Drosophila that contains Polycomblike and RPD3. Mol. Cell. Biol. 23: 3352-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Y.-L., Yang, W.-M., and Seto, E. 2001. Regulation of transcription factor YY1 by acetylation and deacetylation. Mol. Cell. Biol. 21: 5979-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D. and Paro, R. 1995. Drosophila Polycomb-group regulated chromatin inhibits the accessibility of a trans-activator to its target DNA. EMBO J. 14: 5660-5671. [DOI] [PMC free article] [PubMed] [Google Scholar]