Abstract
The Ezh2 protein endows the Polycomb PRC2 and PRC3 complexes with histone lysine methyltransferase (HKMT) activity that is associated with transcriptional repression. We report that Ezh2 expression was developmentally regulated in the myotome compartment of mouse somites and that its down-regulation coincided with activation of muscle gene expression and differentiation of satellite-cell-derived myoblasts. Increased Ezh2 expression inhibited muscle differentiation, and this property was conferred by its SET domain, required for the HKMT activity. In undifferentiated myoblasts, endogenous Ezh2 was associated with the transcriptional regulator YY1. Both Ezh2 and YY1 were detected, with the deacetylase HDAC1, at genomic regions of silent muscle-specific genes. Their presence correlated with methylation of K27 of histone H3. YY1 was required for Ezh2 binding because RNA interference of YY1 abrogated chromatin recruitment of Ezh2 and prevented H3-K27 methylation. Upon gene activation, Ezh2, HDAC1, and YY1 dissociated from muscle loci, H3-K27 became hypomethylated and MyoD and SRF were recruited to the chromatin. These findings suggest the existence of a two-step activation mechanism whereby removal of H3-K27 methylation, conferred by an active Ezh2-containing protein complex, followed by recruitment of positive transcriptional regulators at discrete genomic loci are required to promote muscle gene expression and cell differentiation.
Keywords: Polycomb group, myogenesis, histone methylation, transcription
By preventing inappropriate gene activation, transcriptional repression imposes unique patterns of gene expression and is essential for the specification and maintenance of cell identity (Francis and Kingston 2001). The Polycomb group (PcG) proteins repress transcription of the Drosophila Hox genes and participate in establishing the body anteroposterior axis (Simon et al. 1992). Whereas some PcG genes exert their activities at later stages of development, the PcG Enhancer of zeste, E(z), functions early in development by regulating expression of the gap genes (Pelegri and Lehmann 1994). The PcG coding sequences—and presumably their function—have been conserved throughout evolution. Both in the plant Arabidopsis thaliana and in the nematode Caenorhabditis elegans, the counterparts of Drosophila E(z) regulate homeotic gene expression (Goodrich et al. 1997; Ross and Zarkower 2003; Zhang et al. 2003). E(z) proteins are also involved in initiating X-chromosome inactivation (Plath et al. 2003) and in maintaining the epigenetic patters of pluripotent stem cells (Erhardt et al. 2003). In mammals, two E(z)-related genes have been isolated, Ezh1 and Ezh2 (Laible et al. 1997). Ezh1 expression is prevalent in the adult, whereas Ezh2 is expressed during embryonic development (Laible et al. 1997). Consistent with its expression pattern, Ezh2 is required for early mouse development. Ezh2-null mouse embryos die during the transition from pre- to postimplantation development (O'Carroll et al. 2001).
Among the PcG family, the E(z) proteins are unique in that they are chromatin-modifying enzymes with histone lysine methyltransferase (HKMT) activity (Cao et al. 2002; Czermin et al. 2002; Kuzmichev et al. 2002; Muller et al. 2002). Their catalytic activity resides in the evolutionarily conserved SET domain (Sims et al. 2003). Binding of Drosophila E(z) to a DNA Polycomb response element of the Ultrabithorax (Ubx) gene correlates with H3-K27 methylation and Ubx repression (Cao et al. 2002). Ezh2-mediated methylation of H3-K27 creates a docking site for the subsequent recruitment on the chromatin of the PRC1 (Polycomb repressive complex 1) complex containing additional PcG proteins (Czermin et al. 2002). The interaction of Ezh2 with the histone deacetylase HDAC1 suggests that both histone deacetylation and methylation converge to ensure transcriptional repression (van der Vlag and Otte 1999). The Ezh2 requirement for early mouse development has hampered the study of its role in regulating developmental and postnatal processes. However, a role for Ezh2 in cell cycle progression and cell differentiation has emerged from the analysis of several forms of aggressive tumors. Overexpression of Ezh2 has been reported in hormone-refractory, metastatic prostate cancers (Varambally et al. 2002) and in poorly differentiated and particularly aggressive breast carcinomas (Kleer et al. 2003). Resting cells derived from human lymphomas do not express Ezh2, but Ezh2 is strongly expressed in proliferating lymphoma cells (Visser et al. 2001). Pertinent to its putative role in cell differentiation are the findings that conditional inactivation of Ezh2 results in selectively impaired formation of pre-B and immature B cells but an unaltered development of pro-B cells (Su et al. 2003). Collectively, these and other (Bracken et al. 2003) findings suggest that Ezh2 may regulate cell growth and certain differentiation processes.
Because Ezh2 expression is developmentally regulated in skeletal muscle (Laible et al. 1997), we have tested the hypothesis that Ezh2 may be involved in controlling muscle gene expression and differentiation. Our results indicate that mouse skeletal muscle cells transduced with an Ezh2 retrovirus failed to undergo terminal differentiation and that this differentiation block was mediated by the SET domain, a region responsible for the HKMT activity. Ezh2 interacts with the DNA-binding protein YY1, and both proteins are found—along with the deacetylase HDAC1—on the regulatory regions of transcriptionally inactive muscle specific genes. Their presence correlated with H3-K27 methylation. Upon transcriptional activation, chromatin interaction of Ezh2, HDAC1, and YY1 was lost and replaced by the positive regulators of muscle transcription, SRF and MyoD. This molecular switch was accompanied by H3-K27 hypomethylation and histone hyperacetylation. Thus, our results indicate that the removal of an actively suppressing HKMT protein complex containing Polycomb Ezh2 and the subsequent engagement of positive transcriptional regulators characterize activation of muscle gene expression.
Results
The Polycomb Ezh2 gene is expressed early in the myotomal compartment of developing somites and in proliferating satellite cells and is down-regulated in terminally differentiated muscle cells
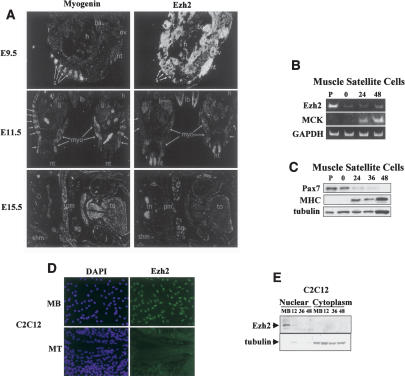
The spatial and temporal expression of Ezh2 was investigated in embryonic day 9.5-15.5 (E9.5-E15.5) mouse embryos by RNA in situ hybridization. At E9.5 (Fig. 1A), Ezh2 was broadly expressed at high levels in most of the anatomical structures of the embryos including the neural tube, otic vesicle, branchial arch, and the myotomes of the developing somites. At this stage of development, the transcripts of myogenin (an early marker of skeletal muscle differentiation) were detected in the myotomes (Fig. 1A, E9.5). At E11.5 (Fig. 1A), Ezh2 expression was significantly decreased but still clearly detectable in the neural tube and, at lower levels, in the liver and limb buds. Ezh2 expression was no longer seen in the developing myotomes, where myogenin continued to be expressed (Fig. 1A, E11.5). By E15.5 (Fig. 1A), Ezh2 mRNAs were only detected in the thymus.
Figure 1.
Ezh2 is expressed early in the myotomal compartment of developing somites and in undifferentiated skeletal myoblasts. (A) Myogenin (left panels) and Ezh2 (right panels) transcripts were analyzed by in situ RNA hybridization on sections from E9.5, E11.5, and E15.5 mouse embryos. In a sagittal section at E9.5, myogenin mRNA was detected in myotomes of the developing somites (s). (nt) Neural tube; (t) tail; (ov) otic vesicle; (ba) branchial arch; (h) heart. Ezh2 mRNA was detected in all structures at a high level. In transverse sections at E11.5, myogenin was detected in developing myotomes (myo; arrows) and at a low level in limb buds (lb). (li) Liver; (h) heart; (nt) neural tube. At E11.5, Ezh2 mRNA levels have decreased significantly from E9.5. Ezh2 was detected in neural tube, liver, limb buds, and paraxial mesoderm. In a transverse section at the level of the jaw (j) at E15.5, myogenin mRNA was detected in tongue (to) muscle, shoulder muscle (shm), pectoral muscle (pm), and all other skeletal muscle. (sg) Salivary gland. At E15.5, Ezh2 mRNAs were only detected in thymus (th). (B) Mouse primary muscle satellite cells were cultured in either growth conditions (P, proliferating) or, once confluent, induced to differentiate for 0, 24, and 48 h. The RNA was isolated and RT-PCR was performed with specific primers for Ezh2, MCK, and GAPDH. (C) Immunoblot of muscle satellite cell extracts with Pax7, MHC, and tubulin antibodies at different differentiation stages. (D) C2C12 proliferating myoblasts (MB) and differentiated myotubes (MT) were immunostained for Ezh2 and nuclei visualized with DAPI. (E) Nuclear and cytoplasmic extracts of proliferating myoblasts (MB) and myoblasts at different stages of differentiation (12, 36, and 48 h in differentiation medium) were fractionated and immunoblotted for Ezh2 and tubulin.
To evaluate the expression of Ezh2 in primary muscle cell cultures, mouse muscle satellite cells were isolated and cultured in conditions favoring either replication or differentiation. Ezh2 mRNA was robustly represented in proliferating satellite cells and progressively diminished as differentiation proceeded (Fig. 1B). Differentiation of satellite cells was accompanied by reduction of Pax7, a specific marker of quiescent and activated satellite cells that is rapidly down-regulated upon myogenic differentiation (Seale et al. 2000), and activation of muscle creatine kinase (MCK) and myosin heavy chain (MHC) gene expression (Fig. 1B,C). Finally, we wished to determine whether C2C12 muscle cells would be amenable to study Ezh2. Ezh2 was found to be expressed in the nucleus of proliferating C2C12 myoblasts and was no longer detectable once myotubes were formed (Fig. 1D,E). Altogether, our results indicate that Ezh2 is expressed early in the myotomal compartment of developing somites and in proliferating myoblasts but becomes undetectable in either more mature developing myotomes or differentiated myotubes.
Ezh2 represses muscle gene expression and contrasts cell differentiation via the SET methyltransferase domain
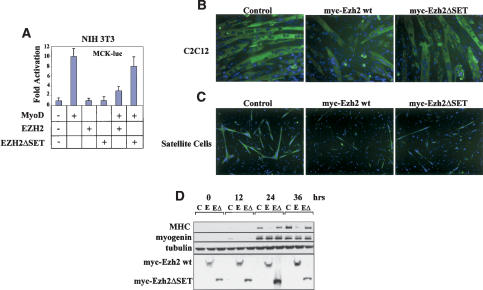
The expression profile of Ezh2 in skeletal muscle suggests the possibility that it may be involved in regulating muscle gene expression. We began to test this hypothesis by transiently transfecting the muscle-specific reporter construct MCK-luc and expression vectors for MyoD and Ezh2 in murine NIH3T3 fibroblasts (Fig. 2A). Although Ezh2 had no effect on the reporter construct alone, MyoD-dependent transactivation of the MCK-luc was repressed by Ezh2. Deletion of the SET domain rendered Ezh2 incapable of repressing MCK-luc expression. To further evaluate the effects of Ezh2 on cell differentiation, we infected either C2C12 myoblasts (Fig. 2B) or primary muscle satellite cells (Fig. 2C) with retroviruses expressing either Ezh2 wild type or Ezh2ΔSET, devoid of HKMT activity (Varambally et al. 2002). Cells expressing Ezh2 were impaired in their differentiation, whereas the Ezh2ΔSET construct had only a marginal effect on differentiation. To quantify the Ezh-2-mediated inhibition of muscle gene expression, we analyzed expression of myogenin and MHC in at different stages of cell differentiation (Fig. 2D). Ezh2 repressed MHC at every time point analyzed, whereas myogenin expression was not affected. Ezh2ΔSET was mostly ineffective in repressing MHC expression (Fig. 2D). In agreement with an essential role of Ezh2 in other critical cellular functions (Cao and Zhang 2004a), attempts of reducing the Ezh2 levels with RNA interference blocked cell growth (Bracken et al. 2003; data not shown). Altogether, these results indicate that Ezh2 overexpression represses muscle gene expression and differentiation through its HKMT activity.
Figure 2.
The SET domain of Ezh2 is required to inhibit muscle gene expression and differentiation. (A) Luciferase assay conducted on NIH 3T3 fibroblasts transfected with the MCK-luc reporter and expression vectors for MyoD, Ezh2, and Ezh2ΔSET mutant and cultured in differentiation medium for 48 h. (B) C2C12 myoblasts were transduced with control, Ezh2 wild-type, or Ezh2ΔSET mutant retroviruses and induced to differentiated for 36 h before immunostaining for MHC. DAPI reveals the nuclei. (C) Mouse primary muscle satellite cells were transduced with control, Ezh2 wild-type, or Ezh2ΔSET mutant retroviruses, induced to differentiate for 36 h, and immunostained for MHC. DAPI reveals the nuclei. (D) Immunoblot of extracts derived from myoblasts transduced with control (C), Ezh2 wild-type (E), or Ezh2ΔSET mutant (EΔ) retroviruses at different stages of differentiation (confluent in growth medium [0]; 12, 24, 36 h in differentiation medium) probed with MHC, myogenin, tubulin, and myc (to detect myc-tagged Ezh2 and Ezh2ΔSET mutant proteins) antibodies. Cells used in B and D belong to the same polyclone and have been cultured within the same experiment.
EZH2 is localized on the chromatin of transcriptionally inactive muscle-specific regulatory regions
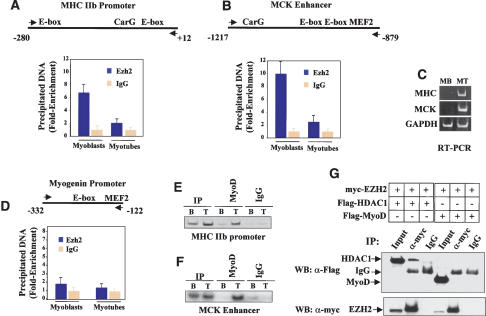
As PcG genes influence gene expression through local chromatin modification (Orlando 2003), we investigated whether Ezh2 may be detected on the chromatin of muscle-specific genes and whether such association may be developmentally regulated. MCK and MHCIIb transcripts were detected in the mouse embryo starting at 13 (Lyons et al. 1991) and 14.5 days postcoitum (dpc) (Lu et al. 1999), respectively, when Ezh2 expression was no longer detected (Fig. 1A). Furthermore, Ezh2 efficiently represses MCK-luc activation and MHC expression (Fig. 2). Therefore, we performed chromatin immunoprecipitation (ChIP) with Ezh2 antibodies and analyzed the immunoprecipitated DNA fragments with real-time PCR using specific primers for the MHCIIb and MCK regulatory regions. Ezh2 was associated with the chromatin regulatory regions of both genes derived from undifferentiated myoblasts but could not be detected on the chromatin of differentiated myotubes (Fig. 3A,B), where MHCIIb and MCK genes are transcribed (Fig. 3C). At variance, the background level of Ezh2 binding to the myogenin promoter did not decrease in differentiated myotubes (Fig. 3D; see below).
Figure 3.
Ezh2 is present at the chromatin regulatory regions of two transcriptionally inactive muscle genes and is no longer detected upon gene activation. (A,B) ChIP with either Ezh2 antibodies or control IgG was performed on chromatin derived from either undifferentiated myoblasts (MB) or differentiated myotubes (MT). The precipitated DNA fragments were amplified with specific oligonucleotides spanning the indicated regulatory regions for the MHCIIb promoter (A), MCK enhancer (B), or myogenin promoter (D) by real-time PCR. (C) Transcripts of MHCIIb, MCK, and GAPDH in either myoblast (MB) or myotubes (MT) were detected by RT-PCR analysis. (E,F) ChIP with either MyoD antibodies or control IgG on chromatin of myoblast (B) or myotubes (T) of MHCIIb promoter (E) and MCK enhancer (F). (G) 293T cells were transfected with expression vectors for Ezh2, HDAC1, and MyoD in different combinations. Cell extracts were prepared and immunoprecipitated with anti-myc antibodies, and the precipitated proteins were revealed with anti-Flag antibodies. Nonspecific IgG was used as the control.
In Drosophila, PcG proteins recognize and exert their activity through specific DNA sequences known as Polycomb Response Elements (PREs) (Lyko and Paro 1999). In mammals, such elements have not been identified yet, even though binding of mammalian PcG proteins to chromatin is well documented (Saurin et al. 1998). Therefore, it is likely that, to be properly positioned on chromatin, PcG may associate with specific DNA-binding proteins. The basic helix-loop-helix (bHLH) MyoD family of transcription factors are pivotal in regulating muscle gene expression (Weintraub et al. 1991). Recently, MyoD has been shown to be functionally linked to gene silencing of muscle-specific transcription through recruitment of the deacetylase HDAC1 onto the regulatory regions of myogenin before this gene is active (Mal and Harter 2003). Because both MHC promoter and MCK enhancer contain MyoD-binding sites and are regulated by myogenic bHLH proteins, we evaluated whether Ezh2 was tethered on the chromatin by MyoD. ChIP performed with specific MyoD antibodies revealed that, at variance with the myogenin promoter (Mal and Harter 2003), neither the MHCIIb promoter nor the MCK enhancer was occupied by MyoD in undifferentiated cells (Fig. 3E,F). This is consistent with earlier in vivo footprinting findings indicating that the E-boxes of the MCK enhancer are unoccupied in undifferentiated myoblasts (Mueller and Wold 1989). Once myotubes were formed, MyoD binding was detected on both regulatory regions (Fig. 3E,F). We were also unable to coimmunoprecipitate MyoD and Ezh2 but were able to confirm the reported (van der Vlag and Otte 1999) interaction of Ezh2 with HDAC1 (Fig. 3G). Interaction of myoblast-derived, endogenous MyoD and Ezh2 was also not observed (data not shown). These results suggest that it is unlikely that Ezh2 is recruited on the chromatin of undifferentiated myoblasts through interaction with MyoD.
Ezh2 associates with YY1, and both proteins are detected on the chromatin of two transcriptionally inactive muscle-specific regulatory regions
YY1 is a specific DNA-binding protein that, depending on cell and promoter context, acts as either a positive or negative regulator of transcription (Shi et al. 1997). YY1 is the mammalian homolog of the Drosophila PcG protein Pho (Brown et al. 1998) and has been shown to interact with the PcG EED protein (Satijn et al. 2001), a component of the PRC2 (Kuzmichev et al. 2002) and PCR3 (Kuzmichev et al. 2004) complexes. Because both the MHC promoter and MCK enhancer contain a DNA-binding site (the CarG-box) recognized by YY1 (Vincent et al. 1993; Galvagni et al. 1998; L'Honore et al. 2003), we evaluated the possibility that Ezh2 may be recruited on these regulatory regions via YY1.
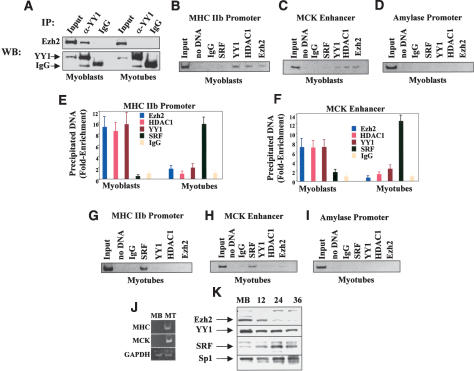
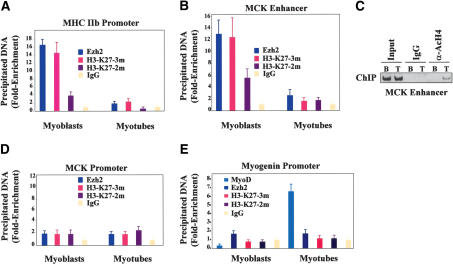
To begin testing this hypothesis, cell extracts derived from either undifferentiated myoblasts or differentiated myotubes were immunoprecipitated with an antibody for YY1, and the precipitated material was immunoblotted with an Ezh2 antibody. As indicated in Figure 4A, Ezh2 coimmunoprecipitated with YY1 from myoblast but not myotube extracts. Next, we investigated whether Ezh2 and YY1 may occupy specific chromatin regions located on the MHC promoter and MCK enhancer. Because Ezh2 interacts with the histone deacetylase HDAC1 (Fig. 3G; van der Vlag and Otte 1999), we also investigated chromatin binding of HDAC1. ChIP experiments were performed with specific antibodies directed against Ezh2, YY1, and HDAC1 while nonspecific IgG served as the negative control. The results of these experiments indicated that Ezh2, YY1, and HDAC1 could be immunoprecipitated from the chromatin of both the MHC promoter and MCK enhancer derived from undifferentiated myoblasts (Fig. 4B,C,E [myoblast], F [myoblast]), in which MHC and MCK are not expressed (Fig. 4J, MB). Amplification of the amylase promoter (Bergstrom et al. 2002) revealed no binding of the above proteins to this genomic region (Fig. 4D). Using electrophoretic mobility shift assay (EMSA), the CarG-box of the MHCIIb promoter was found to bind YY1 from myoblast extracts (Supplementary Fig. 1).
Figure 4.
Ezh2, HDAC1, and YY1 occupy the chromatin of transcriptionally inactive muscle regulatory regions while MyoD and SRF are engaged upon gene activation. (A) Cell extracts from undifferentiated myoblasts or differentiated myotubes were immunoprecipitated with either YY1 antibodies or control IgG and the precipitated material was immunoblotted with Ezh2 antibodies. (B-D) ChIP with antibodies to Ezh2, HDAC1, YY1, and SRF on chromatin obtained from myoblasts. Reactions with no input DNA (no DNA) or ChIP with nonspecific IgG were performed as controls. Precipitated DNA was amplified by PCR with primers for the MHCIIb promoter (B), MCK enhancer (C), or amylase promoter (D). (E,F) ChIP was performed as described in B and C using chromatin derived from either myoblasts or myotubes. The precipitated DNA was amplified by real-time PCR with specific primers for either MHCIIb promoter (E) or MCK enhancer (F). (G-I) ChIP was performed as described in B-D except that chromatin was obtained from differentiated myotubes. (J) Transcripts of MHCIIb, MCK, and GAPDH in either myoblast (MB) or myotubes (MT) were detected by RT-PCR analysis. (K) Immunoblot of extracts derived from myoblasts at different stages of differentiation probed with antibodies against Ezh2, YY1, SRF, and Sp1.
Transcriptional activation of muscle-specific genes is accompanied by loss of Ezh2, HDAC1, and YY1 and chromatin recruitment of MyoD and SRF
In the context of muscle-specific transcription, YY1 is a negative regulator as it competitively prevents binding of the positive regulator SRF to the CarG-box prior to gene activation. At the onset of transcriptional activation, YY1 is replaced by SRF (Gualberto et al. 1992; Lee et al. 1992). Consistent with this result, the CarG-box of the MHCIIb promoter was found to bind SRF from myotube extracts (Supplementary Fig. 1).
We compared the array of transcriptional regulators interacting with the chromatin of the MHCIIb promoter and MCK enhancer in undifferentiated myoblasts (Fig. 4B,C,E [myoblasts], F [myoblasts]) with that of those occupying the same chromatin regions derived from differentiated myotubes. Gene activation of both MHCIIb and MCK (Fig. 4J, MT) was accompanied by chromatin loss of Ezh2, HDAC1, and YY1 and recruitment of SRF (Fig. 4E [myotubes], F-H [myotubes]) and MyoD (Fig. 3E,F) at both the MHCIIb and MCK regulatory regions. Amplification of the amylase promoter served as a negative control (Fig. 4I). Immunoblot analysis indicated that although the levels of Ezh2 and, to a lesser extent, YY1 decreased, those of SRF were increased at later stages of muscle differentiation (Fig. 4K). The levels of the transcription factor Sp1, used as loading control, did not seem to vary.
Histone H3-K27 methylation at inactive versus transcriptional active muscle-specific regulatory regions
The presence of Ezh2 on the transcriptionally inactive MHCIIb promoter and MCK enhancer prompted us to evaluate the status of histone methylation at these regulatory regions. Ezh2 is an in vivo bona fide H3-K27 methyltransferase (Cao and Zhang 2004a). Methylatable lysines in histone H3 and H4 can exist in mono-, di-, and trimethylated states (Paik and Kim 1971). Although the functional significance of this differential methylation remains unclear, the transition from di- to trimethylation of H3-K4 (Santos-Rosa et al. 2002) and H3-K9 (Czermin et al. 2002; Kuzmichev et al. 2002) is biologically relevant. Recently, specific antibodies for mono-, di-, and trimethylated H3-K27 have been used to define the methylation status of pericentric heterochromatin (Peters et al. 2003).
We used these H3-K27 antibodies in ChIP analysis. The histone H3-K27 of the MHCIIb promoter and MCK enhancer—both occupied by Ezh2—were found to be H3-K27 di- and trimethylated in undifferentiated myoblasts, with the trimethylated form being more represented. Loss of Ezh2 binding in differentiated myotubes was associated with hypomethylation of both the di- and trimethylated H3-K27 (Fig. 5A,B) and increased acetylation of H4 histones at the MCK enhancer (Fig. 5C). Interestingly, H3-K27 di- and trimethylation of the MCK proximal promoter regions (Nguyen et al. 2003) were not modified during cell differentiation, and this phenomenon was associated with lack of significant Ezh2 binding to this region (Fig. 5D). The results obtained with the H3-K27 mono-methylated antibody on the MHCIIb promoter and MCK enhancer were not consistently reproduced (data not shown).
Figure 5.
H3-K27 methylation at muscle regulatory regions in undifferentiated myoblasts and differentiated myotubes. (A,B) ChIP with control IgG, Ezh2, H3-K27 α-trimethylated (3m), and α-dimethylated (2m) antibodies on chromatin obtained from myoblasts or myotubes. The precipitated DNA was amplified by real-time PCR using specific primers for the MHCIIb promoter (A) or MCK enhancer (B). (C) ChIP with either control IgG or anti-acetylated H4 antibodies with the chromatin obtained from myoblasts (B) or myotubes (T) on the MCK enhancer. (D) ChIP was performed as described in A and B with specific primers for the MCK promoter—rather than enhancer—were used. (E) ChIP was performed as described in A and B with specific primers for the myogenin promoter.
Results reported throughout this study suggest that myogenin expression was not regulated by Ezh2. In fact, both myogenin and Ezh2 transcripts were coexpressed, albeit transiently, in the myotome (Fig. 1), and Ezh2 overexpression did not interfere with myogenin expression (Fig. 2). Therefore, we evaluated whether Ezh2 was recruited and whether H3-K27 was methylated at the myogenin promoter. At variance with the results from MHCIIb promoter and MCK enhancer experiments, neither Ezh2 nor H3-K27 methylation was detected at the myogenin promoter, whereas MyoD was promptly recruited upon cell differentiation to the same chromatin region (Fig. 5E). This region of the myogenin promoter contains one binding site for MyoD and one for Mef2 but no CarG-box sequences and is sufficient to recapitulate myogenin expression during embryonic development (Cheng et al. 1993; Yee and Rigby 1993).
Presently, we do not know whether di- versus trimethylation of H3-K27 is of physiological significance in the regulation of muscle gene expression. Nonetheless, our experiments have determined that binding of Ezh2 to selected, inactive muscle genes is associated with H3-K27 methylation at discrete chromatin regions.
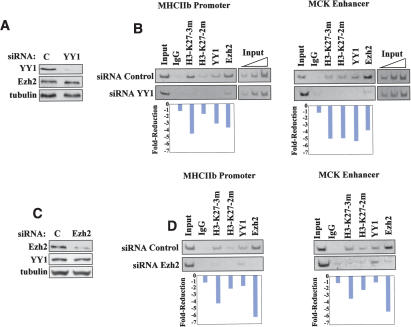
siRNA-mediated reduction of YY1 results in loss of chromatin recruitment of Ezh2 and H3-K27 methylation
Our results suggested that Ezh2 was positioned on discrete chromatin domains through interaction with YY1. If this were true, we would expect that reducing the levels of YY1 would result in decreased Ezh2 chromatin recruitment and, consequently, diminished H3-K27 methylation. To directly test this hypothesis, we depleted YY1 by RNA interference (Fig. 6A). ChIP analysis was performed on the chromatin obtained from skeletal myoblasts transfected with either control or YY1-specific small interfering RNA (siRNA). YY1-specific siRNA abolished YY1 binding, reduced Ezh2 recruitment, and caused loss of H3-K27 methylation at both the MHCIIb promoter and MCK enhancer (Fig. 6B). Importantly, YY1 siRNA did not affect Ezh2 expression (Fig. 6A), indicating that the reduced chromatin recruitment of Ezh2 cannot be ascribed to reduced Ezh2 protein levels but rather to the absence of YY1. Control experiments indicate that, as expected, Ezh2-specific siRNA caused reduced Ezh2 recruitment and loss of H3-K27 methylation at the MHCIIb promoter and MCK enhancer (Fig. 6C,D). Transfection of either siRNA YY1 or siRNA Ezh2 did not anticipate expression of either MHC or MCK in myoblasts and reduced their expression in myotubes (Supplementary Fig. 2). The latter effect is consistent with an antiproliferative effect exerted by either siRNA YY1 or siRNA Ezh2 (Bracken et al. 2003) on myoblasts (data not shown). Because fewer myoblasts were available in the siRNA YY1 and siRNA Ezh2 cultures, compared to control, for the fusion and differentiation process, fewer MHC and MCK transcripts were observed in myotubes. Altogether, the results reported in this paragraph support the hypothesis that YY1 is required to engage Ezh2 on the chromatin of some muscle-specific regulatory regions to promote H3-K27 methylation.
Figure 6.
Efficient chromatin recruitment of Ezh2 and H3-K27 methylation require the presence of YY1. (A) Skeletal myoblasts were transfected with either control (FITC-labeled siRNA; see Materials and Methods) or YY1-specific siRNA, and their extracts were immunoblotted with antibodies to YY1, Ezh2, and tubulin. (B) ChIP assays were performed with control IgG or antibodies to trimethyl, dimethyl H3-K27, YY1, and Ezh2 on the chromatin obtained from cells transfected with either control or YY1 siRNA. The immunoprecipitated chromatin was analyzed by PCR using primers specific for the MHCIIb promoter and MKC enhancer. Three dilutions of the total input are indicated. Quantitation of reduced chromatin binding was performed by scanning the intensity of the individual bands with the NIH Image software and dividing the values obtained for siRNA Control bands for those obtained for the corresponding siRNA YY1 bands. Each value was corrected for the ratio of the two input DNAs (Input DNA siRNA Control/Input siRNA YY1). (C) Skeletal myoblasts were transfected with either control or Ezh2-specific siRNA, and their extracts were immunoblotted with antibodies against Ezh2, YY1, and tubulin. (D) ChIP assays were performed with control IgG and antibodies to trimethyl, dimethyl H3-K27, YY1, and Ezh2 on the chromatin obtained from cells transfected with either control or Ezh2 siRNA. The immunoprecipitated chromatin was analyzed by PCR using primers specific for the MHCIIb promoter and MCK enhancer. Quantitation was performed as described in B. Each value was corrected for the ratio of the two input DNAs (Input DNA siRNA Control/Input siRNA Ezh2).
Discussion
The transcriptional repression exerted by the PcG has been viewed as an epigenetic regulator imposed on chromatin regions fated to be irreversibly silenced. Similarly, histone methylation has been historically considered a stable modification associated with genes whose expression is permanently repressed. This paradigm is gradually changing in view of several observations indicating that histone methylation may be removed (Sims et al. 2003). Moreover, it is becoming increasingly clear that, beside its role in establishing and maintaining constitutive heterochromatic domains at the chromosome level, histone lysine methylation also regulates reversible repression of euchromatic regions (Sims et al. 2003). The findings that the Polycomb Ezh2 protein mediates its repression capabilities through H3-K27 methylation suggest the possibility that such transcriptional repression is also reversible and, therefore, possibly involved in regulating dynamic processes, such as cell specification and differentiation. Molecules that actively suppress transcription may be relevant to prevent expression of genes both in the incorrect cell context and at inappropriate developmental stages.
In skeletal muscle cells, the selection of the muscle gene loci to be activated is directed by the MyoD family of transcription factors, which recognize a specific DNA sequence—the E-box—located in enhancer/promoter regions of muscle-specific genes. At discrete chromatin regions, MyoD recruits ATP-dependent chromatin remodeling complexes and acetyltransferases to redistribute nucleosomes, acetylate histones, and promote transcription (Sartorelli and Puri 2001). MyoD recruitment has also been shown to alter chromatin structure at regulatory regions of muscle genes (Gerber et al. 1997; Bergstrom and Tapscott 2001). Nonetheless, little is known about the mechanisms that actively prevent inappropriate muscle expression.
Because of its expression profile in developing embryos, we postulated that Ezh2 may be involved in targeting certain muscle genes and preventing them from being transcribed in committed, yet undifferentiated, muscle cells. We have found that Ezh2 can be detected in conjunction with methylation of Lys 27 of histone H3 on the chromatin of two regulatory regions of muscle genes when these are transcriptionally silent. Both Ezh2 binding to and H3-K27 methylation of muscle regulatory regions are greatly diminished upon cell differentiation and activation of gene transcription. Although it is still unclear how these methyl groups can be removed, it is possible that histone methylation may be erased through histone replacement (Bannister et al. 2002). Consistent with a regulatory role of the HKMT activity in suppressing muscle gene expression, deletion of the SET domain rendered Ezh2 incapable of blocking cell differentiation and muscle transcription. Because its expression is down-regulated in differentiating mouse primary skeletal myoblasts, Ezh2 is likely to play a role in ensuring replicative capabilities and maintenance of the undifferentiated state to muscle cell precursors.
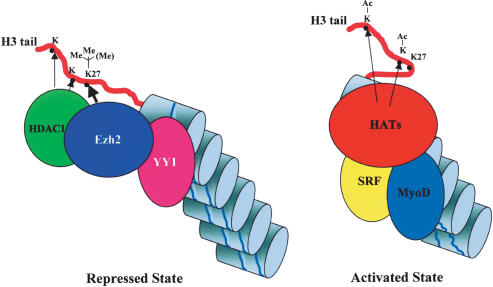
Our results indicate that Ezh2 was recruited at the chromatin of selected muscle regulatory regions by the transcriptional regulator YY1. Both can be coimmunoprecipitated from myoblast and not myotube cell extracts, and the proteins colocalize at the same muscle chromatin regions in a developmentally regulated manner. The interaction of endogenous YY1 and Ezh2 is likely to be mediated by the PcG EED protein because recombinant YY1 and Ezh2 do not directly associate (Satijn et al. 2001). Previous reports have demonstrated a negative role for YY1 in regulating muscle gene expression through interaction with distinct nucleotides within the CarG-box (Gualberto et al. 1992; Lee et al. 1994; MacLellan et al. 1994; Galvagni et al. 1998). Transcriptional activation coincides with replacement of YY1 by the serum response factor (SRF) (Gualberto et al. 1992; Lee et al. 1992), whose interaction with the CarG-box is required for muscle-specific transcription to proceed. Our data suggest a two-step activation model of muscle gene expression (Fig. 7). In the repressed state, YY1 recruits a complex containing both Ezh2 and HDAC1 that silences transcription through histone methylation (H3-K27) and deacetylation. Transcriptional activation entails the initial removal of the YY1-Ezh2-HDAC1 repressive complex and subsequent recruitment of the activators SRF, which replaces YY1, and the MyoD family of transcription factors and associated acetyltransferases. As YY1 binding tolerates a substantial nucleotide heterogeneity in its DNA recognition sites (Hyde-DeRuyscher et al. 1995), muscle and non-muscle-specific CarG-less regulatory regions may be also occupied and regulated in a similar manner. On the other hand, Ezh2 does not appear to promiscuously regulate expression of all muscle-specific genes as indicated by the transient coexpression of Ezh2 and myogenin in the myotome of developing embryos and lack of Ezh2 recruitment and H3-K27 methylation at the myogenin promoter. Distinct histone methyltransferases and deacetylases have been shown to modify histones at the myogenin promoter (Zhang et al. 2002).
Figure 7.
Two-step activation model of muscle gene expression. Regulatory regions of certain muscle-specific genes are occupied by a protein complex containing the DNA-binding protein YY1, the methyltransferase Ezh2, and the deacetylase HDAC1. Deacetylation of lysine residues by HDAC1 and di-/trimethylation of H3-K27 by Ezh2 actively prevent transcription (repressed state). At the onset of transcriptional activation, YY1 is displaced from the chromatin—with consequent loss of Ezh2 and HDAC1—and replaced by SRF. H3-K27 becomes hypomethylated, and loading of the MyoD family of transcription factors allows engagement of histone acetyltransferases (HATs) and permits initiation of transcription (activated state).
Recently, Suz12—a component of the Eed-Ezh2 complexes (Cao et al. 2002; Kuzmichev et al. 2002, 2004)—has been shown to regulate Hox transcription (Cao and Zhang 2004b), and several putative genomic targets of Suz12 have been identified (Kirmizis et al. 2004). H3-K27 methylation of these targets is caused by Suz12-mediated recruitment of Ezh2 (Kirmizis et al. 2004). Interestingly, Suz12 occupies a region of the HoxA9 regulatory regions containing three YY1-binding sites (Cao and Zhang 2004b). A potential role of YY1 in mediating Polycomb-dependent transcriptional repression and histone methylation has been suggested but never experimentally established (Sims et al. 2003; Cao and Zhang 2004b). Our results indicate that Ezh2 is recruited on certain chromatin targets through interaction with YY1 and lend support to the hypothesis that YY1-binding sites may behave as a mammalian equivalent of Drosophila PRE. Consistent with these observations, siRNA-mediated YY1 depletion reduced Ezh2 recruitment and abolished H3-K27 methylation. We note that although RNA interference on YY1 reduced but did not completely abolish Ezh2 recruitment, H3-K27 methylation was erased by YY1 siRNA (Fig. 6B). These findings suggest that additional DNA-binding proteins may recruit Ezh2 on the chromatin and that the residual Ezh2 not associated with YY1 may methylate lysines other than H3-K27. Indeed, biochemical characterization of the Ezh2-containing PRC2 and PRC3 complexes indicates that whereas PRC3 contains nucleosomal H3-K27 HKMT activity, PRC2 preferentially methylates histone H1b (Kuzmichev et al. 2004). Interestingly, the DNA-binding homeoprotein Msx1, a repressor of muscle differentiation (Song et al. 1992), interacts and cooperates with H1b to inhibit myogenesis (Lee et al. 2004). Whether Msx1 interacts with Ezh2 complexes and whether the Ezh2 HKMT activity directed at H1b is involved in mediating the effects of Msx1 on muscle differentiation will have to be addressed in future experiments.
A speculative role for Ezh2 in regulating expression at other muscle genomic loci derives from the molecular characterization of patients with fascioscapulohumeral muscular dystrophy (FSH), a common myopathy. A hallmark of FSH is the deletion of the tandem repeats-containing region D4Z4 located on Chromosome region 4q35. D4Z4 deletion results in lack of binding of a protein complex containing YY1 and consequent inappropriate overexpression of genes located upstream of the D4Z4 region in dystrophic muscles (Gabellini et al. 2002). It should be possible to test if Ezh2 is involved in repressing transcription of genes located around this region to evaluate the hypothesis that Ezh2 regulates muscle gene expression in both physiological and pathological conditions. Finally, because both Ezh2 and YY1 are expressed in several cell types, the mode of transcriptional repression described here may be used to control other tissue-specific transcriptional programs.
Materials and methods
RNA in situ hybridization
The protocol used to fix and embed mouse embryos is described in detail in Lyons et al. (1990). Briefly, embryos were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS), dehydrated, and infiltrated with paraffin. Five- to seven-micron-thick serial sections were mounted on gelatinized slides. One to three sections were mounted per slide, deparaffinized in xylene, and rehydrated. The sections were digested with proteinase K, postfixed, treated with triethanolamine/acetic anhydride, washed, and dehydrated.
The cRNA probes to EZH2 and myogenin were synthesized according to the manufacturer's conditions (Stratagene) and labeled with 35S-UTP (>1000 Ci/mmol; Amersham). cRNA transcripts larger than 100 nt were subjected to alkali hydrolysis to give a mean size of 70 bases for efficient hybridization. Sections were hybridized overnight at 52°C in 50% deionized formamide, 0.3M NaCl, 20 mM Tris-HCl (pH 7.4), 5 mM EDTA, 10 mM NaPO4, 10% dextran sulfate, 1× Denhardt's, 50 μg/mL total yeast RNA, and 50-75,000 cpm/μL 35S-labeled cRNA probe. The tissue was subjected to stringent washing at 65°C in 50% formamide, 2× SSC, 10 mM DTT, and washed in PBS before treatment with 20 μg/mL RNAse A for 30 min at 37°C. Following washes in 2× SSC and 0.1× SSC for 15 min at 37°C, the slides were dehydrated and dipped in Kodak NTB-2 nuclear track emulsion and exposed for 1 wk in light-tight boxes with desiccant at 4°C. Photographic development was carried out in Kodak D-19. Slides were analyzed using both light- and dark-field optics of a Zeiss Axiophot microscope.
Immunofluorescence, immunoprecipitation, and immunoblot
Immunofluorescence was performed as described in Sartorelli et al. (1999) with antibodies against Ezh2 and MHC. 293T cells were cotransfected with plasmids expressing epitope-tagged Ezh2, HDAC-1, and MyoD and lysed as previously described (Puri et al. 2001). Whole cell lysates (1.5 mg) of was incubated either with an anti-myc antibody or normal rabbit IgG. For endogenous YY1/Ezh2 interactions, C2C12 cells were either grown in GM or induced to differentiate in DM for 48 h, and nuclear extracts were prepared as previously described (Schreiber et al. 1989). Five-hundred micrograms of nuclear extract was precipitated with 2 μg either of anti-YY1 antibody or normal mouse IgG. Immunoblots were performed with anti-MHC, Pax7, and tubulin antibodies from the Developmental Studies Hybridoma Bank (University of Iowa), Ezh2 (Zymed), myc (Upstate Biotech), M2 Flag (Sigma) HDAC1, YY1, MyoD, myogenin, SRF, and Sp1 antibodies (Santa Cruz Biotech.)
Satellite cell isolation and culturing
Satellite cells were isolated from SJL/J 4-wk-old mice (Jackson Laboratories) as described (Jackson et al. 1999), plated on Matrigel-coated plates (BD Biosciences), and cultured in DMEM medium supplemented with 20% FBS. Cell differentiation was induced by culturing the cells in DMEM supplemented with 2% horse serum.
Chromatin immunoprecipitation assay and real-time PCR
Chromatin immunoprecipitation was performed as described (Wells et al. 2000) using 2 μg of either normal rabbit IgG or antibodies against Ezh2 (Zymed), HDAC1 (Santa Cruz Biotech., sc-7872), YY1 (sc-7341X), SRF (sc-335X), MyoD (sc-760X), acetyl-histone H4 (Upstate Biotech., 06-866), or with antibodies against di- and trimethylated H3-K27 (Peters et al. 2003). Real-time PCR was performed using the Mx3000P System (Stratagene) with a SyberGreen MasterMix (Applied Biosystems). Real-time PCR was performed at least three times with independent DNA samples. The reported data represent real-time PCR values normalized to input DNAs and to the values obtained with normal rabbit IgG, which were set as one unit in each calculation. Data are presented as fold differences relative to input and values obtained by normal rabbit IgG with the formula 2[(CtIgG - Ct Input) - (CtAb - CtInput)], where Ct is the threshold cycles, IgG is the normal rabbit IgG, Ab is the specific antibody, and Input is the input genomic DNA. To ensure specific PCR amplification, every real-time PCR run was followed by a dissociation phase analysis (Mx3000P software version 1.2). Furthermore, the PCR products were analyzed on agarose gel to ensure that they were of the expected molecular weight. Each of the gel ChIPs was analyzed with at least two (ChIP reported in Fig. 6) but more typically with three to four samples obtained from different experiments with reproducible results.
Cells, retroviral constructs, transduction and transient transfections
293T, C2C12, and NIH3T3 cells were from ATCC. C2C12 cells were cultured in DMEM supplemented with 20% FBS (growth medium, GM) and induced to differentiate with DMEM supplemented with 2% horse serum, 1× insulin, transferrin, and selenium (differentiation medium, DM). 293T and NIH3T3 were cultured in DMEM supplemented with 10% FBS. Myc-tagged Ezh2 and Ezh2ΔSET were cloned into the pBabe retroviral vector and transduced in either C2C12 or satellite cells as described (Costa et al. 2000).
The MCK-luc reporter is described in Puri et al. (1997). Transient transfections were performed with FuGENE6 reagent (Roche) and luciferase assays as described in Puri et al. (1997).
RT-PCR
Total RNA from C2C12 and satellite cells was extracted using TRIzol Reagent (Invitrogen) and purified using the RNAEasy kit (QIAGEN). cDNA was synthesized using the Super-Script double stranded cDNA Synthesis System (Inivitrogen), and 1 μL of cDNA was PCR-amplified.
RNA interference
Skeletal myoblasts were transfected as described (Escobedo and Koh 2003) with duplex siRNA (150 nmol each) for YY1 (Santa Cruz Biotech, sc-36863), Ezh2 (5′-AACACUGUUGCACUGGUUC-3′ and 5′-GAACCAGUGCAACAGUGUU-3′, synthesized by Invitrogen) or control FITC-labeled siRNA (BLOCK-iT Fluorescent Oligo; Invitrogen) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were incubated with the siRNA for 48 h and then harvested, replated, and transfected once again with siRNA. After 48 h, the cells were harvested and used for immunoblot and ChIP assays.
Oligonucleotide sequences
The oligonucleotides used in RT-PCR were GAPDH (GenBank XM_132897): Forward: 5′-AACATCAAATGGGGTGAGGCC-3′, Reverse: 5′-GTTGTCATGGATGACCTTGGC-3′; Ezh2 (GenBank NM_007971): Forward: 5′-CTAATTGGTACTTACTACGATAACTTT-3′, Reverse: 5′-ACTCTAAACTCATACACCTGTCTACAT-3′; MCK (GenBank NM_007710): Forward: 5′-ACTACAAGCCTCAGGAGGAGTA-3′, Reverse: 5′-CTTATCGCGAAGCTTATTGTAG-3′; MHCIIb (GenBank XM_126119): Forward: 5′-TCAATGAGATGGAGATCCAGCTGAAC-3′, Reverse: 5′-GTCCAGGTGCAGCTGTGTGTCCTTC-3′.
The oligonucleotides used in ChIP were MHCIIb Promoter (GenBank M92099): Forward: 5′-CACCCAAGCCGGGAGAAACAGCC-3′, Reverse: 5′-GAGGAAGGACAGGACAGAGGCACC-3′; MCK Enhancer (GenBank M21390): Forward: 5′-GCCACTATGGGTCTAGGCTGC-3′, Reverse: 5′-AAGCCCAGTGCAGGCTGCTCC-3′; Amylase Promoter (GenBank M16540): Forward: 5′-TCAGTTGTAATTCTCCTTGTACGG-3′, Reverse: 5′-CATTCCTTGGCAATATCAACC-3′.
The oligonucleotides used in ChIP real-time PCR were Myogenin Promoter (GenBank X71910): Forward: 5′-CCCTGCCCCACAGGGGCTGTG-3′, Reverse: 5′-ACGCCACAGAAACCTGAGCCC-3′; MCK Promoter (GenBank M21390): Forward: 5′-CGCCAGCTAGACTCAGCACT-3′, Reverse: 5′-CCCTGCGAGCAGATGAGCTT-3′; MCK Enhancer (GenBank M21390): Forward: 5′-AGGGATGAGAGCAGCCACTA-3′, Reverse: 5′-CAGCCACATGTCTGGGTTAAT-3′; MHCIIb Promoter (GenBank M92099): Forward: 5′-CACCCAAGCCGGGAGAAACAGCC-3′, Reverse: 5′-GAGGAAGGACAGGACAGAGGCACC-3′.
Acknowledgments
We thank Marcella Fulco in the Sartorelli Lab for helpful suggestions and stimulating discussion, S. Lnu and A.M Chinnaiyan for providing the Ezh2 constructs, and S. Opravil and T. Jenuwein for the H3-K27 methylation-specific antibodies. The help of S. McCroskery with the isolation and culture of mouse satellite cells is kindly acknowledged.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1241904.
References
- Bannister A.J., Schneider, R., and Kouzarides, T. 2002. Histone methylation: Dynamic or static? Cell 109 801-806. [DOI] [PubMed] [Google Scholar]
- Bergstrom D.A. and Tapscott, S.J. 2001. Molecular distinction between specification and differentiation in the myogenic basic helix-loop-helix transcription factor family. Mol. Cell. Biol. 21 2404-2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstrom D.A., Penn, B.H., Strand, A., Perry, R.L., Rudnicki, M.A., and Tapscott, S.J. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell 9 587-600. [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Pasini, D., Capra, M., Prosperini, E., Colli, E., and Helin, K. 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J. 22 5323-5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.L., Mucci, D., Whiteley, M., Dirksen, M.L., and Kassis, J.A. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol. Cell 1 1057-1064. [DOI] [PubMed] [Google Scholar]
- Cao R. and Zhang, Y. 2004a. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 14 155-164. [DOI] [PubMed] [Google Scholar]
- ____. 2004b. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol. Cell 15 57-67. [DOI] [PubMed] [Google Scholar]
- Cao R., Wang, L., Wang, H., Xia, L., Erdjument-Bromage, H., Tempst, P., Jones, R.S., and Zhang, Y. 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298 1039-1043. [DOI] [PubMed] [Google Scholar]
- Cheng T.C., Wallace, M.C., Merlie, J.P., and Olson, E.N. 1993. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science 261 215-218. [DOI] [PubMed] [Google Scholar]
- Costa G.L., Benson, J.M., Seroogy, C.M., Achacoso, P., Fathman, C.G., and Nolan, G.P. 2000. Targeting rare populations of murine antigen-specific T lymphocytes by retroviral transduction for potential application in gene therapy for autoimmune disease. J. Immunol. 164 3581-3590. [DOI] [PubMed] [Google Scholar]
- Czermin B., Melfi, R., McCabe, D., Seitz, V., Imhof, A., and Pirrotta, V. 2002. Drosophila Enhancer of zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111 185-196. [DOI] [PubMed] [Google Scholar]
- Erhardt S., Su, I.H., Schneider, R., Barton, S., Bannister, A.J., Perez-Burgos, L., Jenuwein, T., Kouzarides, T., Tarakhovsky, A., and Surani, M.A. 2003. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development 130 4235-4248. [DOI] [PubMed] [Google Scholar]
- Escobedo J. and Koh, T.J. 2003. Improved transfection technique for adherent cells using a commercial lipid reagent. Biotechniques 35 936-940. [DOI] [PubMed] [Google Scholar]
- Francis N.J. and Kingston, R.E. 2001. Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell. Biol. 2 409-421. [DOI] [PubMed] [Google Scholar]
- Gabellini D., Green, M.R., and Tupler, R. 2002. Inappropriate gene activation in FSHD: A repressor complex binds a chromosomal repeat deleted in dystrophic muscle. Cell 110 339-348. [DOI] [PubMed] [Google Scholar]
- Galvagni F., Cartocci, E., and Oliviero, S. 1998. The dystrophin promoter is negatively regulated by YY1 in undifferentiated muscle cells. J. Biol. Chem. 273 33708-33713. [DOI] [PubMed] [Google Scholar]
- Gerber A.N., Klesert, T.R., Bergstrom, D.A., and Tapscott, S.J. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: A mechanism for lineage determination in myogenesis. Genes & Dev. 11 436-450. [DOI] [PubMed] [Google Scholar]
- Goodrich J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. 1997. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386 44-51. [DOI] [PubMed] [Google Scholar]
- Gualberto A., LePage, D., Pons, G., Mader, S.L., Park, K., Atchison, M.L., and Walsh, K. 1992. Functional antagonism between YY1 and the serum response factor. Mol. Cell. Biol. 12 4209-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde-DeRuyscher R.P., Jennings, E., and Shenk, T. 1995. DNA binding sites for the transcriptional activator/repressor YY1. Nucleic Acids Res. 23 4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson K.A., Mi, T., and Goodell, M.A. 1999. Hematopoietic potential of stem cells isolated from murine skeletal muscle. Proc. Natl. Acad. Sci. 96 14482-14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirmizis A., Bartley, S.M., Kuzmichev, A., Margueron, R., Reinberg, D., Green, R., and Farnham, P.J. 2004. Silencing of human Polycomb target genes is associated with methylation of histone H3 Lys 27. Genes & Dev. 18 1592-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleer C.G., Cao, Q., Varambally, S., Shen, R., Ota, I., Tomlins, S.A., Ghosh, D., Sewalt, R.G., Otte, A.P., Hayes, D.F., et al. 2003. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc. Natl. Acad. Sci. 100 11606-11611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Nishioka, K., Erdjument-Bromage, H., Tempst, P., and Reinberg, D. 2002. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of zeste protein. Genes & Dev. 16 2893-2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A., Jenuwein, T., Tempst, P., and Reinberg, D. 2004. Different Ezh2-containing complexes target methylation of histone h1 or nucleosomal histone h3. Mol. Cell 14 183-193. [DOI] [PubMed] [Google Scholar]
- L'Honore A., Lamb, N.J., Vandromme, M., Turowski, P., Carnac, G., and Fernandez, A. 2003. MyoD distal regulatory region contains an SRF binding CArG element required for MyoD expression in skeletal myoblasts and during muscle regeneration. Mol. Biol. Cell 14 2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laible G., Wolf, A., Dorn, R., Reuter, G., Nislow, C., Lebersorger, A., Popkin, D., Pillus, L., and Jenuwein, T. 1997. Mammalian homologues of the Polycomb-group gene Enhancer of zeste mediate gene silencing in Drosophila heterochromatin and at S. cerevisiae telomeres. EMBO J. 16 3219-3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.C., Shi, Y., and Schwartz, R.J. 1992. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal α-actin transcription in embryonic myoblasts. Proc. Natl. Acad. Sci. 89 9814-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.C., Zhang, Y., and Schwartz, R.J. 1994. Bifunctional transcriptional properties of YY1 in regulating muscle actin and c-myc gene expression during myogenesis. Oncogene 9 1047-1052. [PubMed] [Google Scholar]
- Lee H., Habas, R., and Abate-Shen, C. 2004. MSX1 cooperates with histone H1b for inhibition of transcription and myogenesis. Science 304 1675-1678. [DOI] [PubMed] [Google Scholar]
- Lu B.D., Allen, D.L., Leinwand, L.A., and Lyons, G.E. 1999. Spatial and temporal changes in myosin heavy chain gene expression in skeletal muscle development. Dev. Biol. 216 312-326. [DOI] [PubMed] [Google Scholar]
- Lyko F. and Paro, R. 1999. Chromosomal elements conferring epigenetic inheritance. Bioessays 21 824-832. [DOI] [PubMed] [Google Scholar]
- Lyons G.E., Ontell, M., Cox, R., Sassoon, D., and Buckingham, M. 1990. The expression of myosin genes in developing skeletal muscle in the mouse embryo. J. Cell Biol. 111 1465-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons G.E., Muhlebach, S., Moser, A., Masood, R., Paterson, B.M., Buckingham, M.E., and Perriard, J.C. 1991. Developmental regulation of creatine kinase gene expression by myogenic factors in embryonic mouse and chick skeletal muscle. Development 113 1017-1029. [DOI] [PubMed] [Google Scholar]
- MacLellan W.R., Lee, T.C., Schwartz, R.J., and Schneider, M.D. 1994. Transforming growth factor-β response elements of the skeletal α-actin gene. Combinatorial action of serum response factor, YY1, and the SV40 enhancer-binding protein, TEF-1. J. Biol. Chem. 269 16754-16760. [PubMed] [Google Scholar]
- Mal A. and Harter, M.L. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. 100 1735-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P.R. and Wold, B. 1989. In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246 780-786. [DOI] [PubMed] [Google Scholar]; Erratum. 1990. 248 802. [DOI] [PubMed] [Google Scholar]
- Muller J., Hart, C.M., Francis, N.J., Vargas, M.L., Sengupta, A., Wild, B., Miller, E.L., O'Connor, M.B., Kingston, R.E., and Simon, J.A. 2002. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111 197-208. [DOI] [PubMed] [Google Scholar]
- Nguyen Q.G., Buskin, J.N., Himeda, C.L., Shield, M.A., and Hauschka, S.D. 2003. Differences in the function of three conserved E-boxes of the muscle creatine kinase gene in cultured myocytes and in transgenic mouse skeletal and cardiac muscle. J. Biol. Chem. 278 46494-46505. [DOI] [PubMed] [Google Scholar]
- O'Carroll D., Erhardt, S., Pagani, M., Barton, S.C., Surani, M.A., and Jenuwein, T. 2001. The Polycomb-group gene Ezh2 is required for early mouse development. Mol. Cell. Biol. 21 4330-4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlando V. 2003. Polycomb, epigenomes, and control of cell identity. Cell 112 599-606. [DOI] [PubMed] [Google Scholar]
- Paik W.K. and Kim, S. 1971. Protein methylation. Science 174 114-119. [DOI] [PubMed] [Google Scholar]
- Pelegri F. and Lehmann, R. 1994. A role of Polycomb group genes in the regulation of gap gene expression in Drosophila. Genetics 136 1341-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H., Kubicek, S., Mechtler, K., O'Sullivan, R.J., Derijck, A.A., Perez-Burgos, L., Kohlmaier, A., Opravil, S., Tachibana, M., Shinkai, Y., et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12 1577-1589. [DOI] [PubMed] [Google Scholar]
- Plath K., Fang, J., Mlynarczyk-Evans, S.K., Cao, R., Worringer, K.A., Wang, H., de la Cruz, C.C., Otte, A.P., Panning, B., and Zhang, Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300 131-135. [DOI] [PubMed] [Google Scholar]
- Puri P.L., Sartorelli, V., Yang, X.J., Hamamori, Y., Ogryzko, V.V., Howard, B.H., Kedes, L., Wang, J.Y., Graessmann, A., Nakatani, Y., et al. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1 35-45. [DOI] [PubMed] [Google Scholar]
- Puri P.L., Iezzi, S., Stiegler, P., Chen, T.T., Schiltz, R.L., Muscat, G.E., Giordano, A., Kedes, L., Wang, J.Y., and Sartorelli, V. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell 8 885-897. [DOI] [PubMed] [Google Scholar]
- Ross J.M. and Zarkower, D. 2003. Polycomb group regulation of Hox gene expression in C. elegans. Dev. Cell 4 891-901. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider, R., Bannister, A.J., Sherriff, J., Bernstein, B.E., Emre, N.C., Schreiber, S.L., Mellor, J., and Kouzarides, T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419 407-411. [DOI] [PubMed] [Google Scholar]
- Sartorelli V. and Puri, P.L. 2001. The link between chromatin structure, protein acetylation and cellular differentiation. Front. Biosci. 6 D1024-D1047. [DOI] [PubMed] [Google Scholar]
- Sartorelli V., Puri, P.L., Hamamori, Y., Ogryzko, V., Chung, G., Nakatani, Y., Wang, J.Y., and Kedes, L. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4 725-734. [DOI] [PubMed] [Google Scholar]
- Satijn D.P., Hamer, K.M., den Blaauwen, J., and Otte, A.P. 2001. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 21 1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin A.J., Shiels, C., Williamson, J., Satijn, D.P., Otte, A.P., Sheer, D., and Freemont, P.S. 1998. The human Polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J. Cell Biol. 142 887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E., Matthias, P., Muller, M.M., and Schaffner, W. 1989. Rapid detection of octamer binding proteins with `mini-extracts,' prepared from a small number of cells. Nucleic Acids Res. 17 6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale P., Sabourin, L.A., Girgis-Gabardo, A., Mansouri, A., Gruss, P., and Rudnicki, M.A. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell 102 777-786. [DOI] [PubMed] [Google Scholar]
- Shi Y., Lee, J.S., and Galvin, K.M. 1997. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta 1332 F49-F66. [DOI] [PubMed] [Google Scholar]
- Simon J., Chiang, A., and Bender, W. 1992. Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114 493-505. [DOI] [PubMed] [Google Scholar]
- Sims R.J., Nishioka, K., and Reinberg, D. 2003. Histone lysine methylation: A signature for chromatin function. Trends Genet. 19 629-639. [DOI] [PubMed] [Google Scholar]
- Song K., Wang, Y., and Sassoon, D. 1992. Expression of Hox-7.1 in myoblasts inhibits terminal differentiation and induces cell transformation. Nature 360 477-481. [DOI] [PubMed] [Google Scholar]
- Su I.H., Basavaraj, A., Krutchinsky, A.N., Hobert, O., Ullrich, A., Chait, B.T., and Tarakhovsky, A. 2003. Ezh2 controls B cell development through histone H3 methylation and Igh rearrangement. Nat. Immunol. 4 124-131. [DOI] [PubMed] [Google Scholar]
- van der Vlag J. and Otte, A.P. 1999. Transcriptional repression mediated by the human Polycomb-group protein EED involves histone deacetylation. Nat. Genet. 23 474-478. [DOI] [PubMed] [Google Scholar]
- Varambally S., Dhanasekaran, S.M., Zhou, M., Barrette, T.R., Kumar-Sinha, C., Sanda, M.G., Ghosh, D., Pienta, K.J., Sewalt, R.G., Otte, A.P., et al. 2002. The Polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419 624-629. [DOI] [PubMed] [Google Scholar]
- Vincent C.K., Gualberto, A., Patel, C.V., and Walsh, K. 1993. Different regulatory sequences control creatine kinase-M gene expression in directly injected skeletal and cardiac muscle. Mol. Cell. Biol. 13 1264-1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser H.P., Gunster, M.J., Kluin-Nelemans, H.C., Manders, E.M., Raaphorst, F.M., Meijer, C.J., Willemze, R., and Otte, A.P. 2001. The Polycomb group protein EZH2 is upregulated in proliferating, cultured human mantle cell lymphoma. Br. J. Haematol. 112 950-958. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Davis, R., Tapscott, S., Thayer, M., Krause, M., Benezra, R., Blackwell, T.K., Turner, D., Rupp, R., Hollenberg, S., et al. 1991. The myoD gene family: Nodal point during specification of the muscle cell lineage. Science 251 761-766. [DOI] [PubMed] [Google Scholar]
- Wells J., Boyd, K.E., Fry, C.J., Bartley, S.M., and Farnham, P.J. 2000. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell. Biol. 20 5797-5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee S.P. and Rigby, P.W. 1993. The regulation of myogenin gene expression during the embryonic development of the mouse. Genes & Dev. 7 1277-1289. [DOI] [PubMed] [Google Scholar]
- Zhang C.L., McKinsey, T.A., and Olson, E.N. 2002. Association of class II histone deacetylases with heterochromatin protein 1: Potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22 7302-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Azevedo, R.B., Lints, R., Doyle, C., Teng, Y., Haber, D., and Emmons, S.W. 2003. Global regulation of Hox gene expression in C. elegans by a SAM domain protein. Dev. Cell 4 903-915. [DOI] [PubMed] [Google Scholar]