Abstract
Nuclear export of mRNA is a central step in gene expression that shows extensive coupling to transcription and transcript processing. However, little is known about the fate of mRNA and its export under conditions that damage the DNA template and RNA itself. Here we report the discovery of four new factors required for mRNA export through a screen of all annotated nonessential Saccharomyces cerevisiae genes. Two of these factors, mRNA surveillance factor Rrp6 and DNA repair protein Lrp1, are nuclear exosome components that physically interact with one another. We find that Lrp1 mediates specific mRNA degradation upon DNA-damaging UV irradiation as well as general mRNA degradation. Lrp1 requires Rrp6 for genomic localization to genes encoding its mRNA targets, and Rrp6 genomic localization in turn correlates with transcription. Further, Rrp6 and Lrp1 are both required for repair of UV-induced DNA damage. These results demonstrate coupling of mRNA surveillance to mRNA export and suggest specificity of the RNA surveillance machinery for different transcript populations. Broadly, these findings link DNA and RNA surveillance to mRNA export.
Keywords: mRNA export, mRNA surveillance, DNA repair, nuclear exosome, Lrp1, Rrp6
Nuclear export of mRNA is central to eukaryotic gene expression. mRNA export proteins bind mRNAs and facilitate their translocation through the nuclear pores in the form of messenger ribonucleoprotein (mRNP) complexes (for review, see Vinciguerra and Stutz 2004). mRNA export factors extensively couple mRNA export to other processes in gene expression, including transcription, splicing, and 3′ end processing (for review, see Reed 2003; Vinciguerra and Stutz 2004). This coupling is often mediated by factors that act in an upstream process and in mRNA export itself. Such export factors commonly function directly in the upstream process and then subsequently bind or recruit downstream export factors, presumably to facilitate export-competent mRNP formation (Strasser and Hurt 2001; Libri et al. 2002; Strasser et al. 2002). For example, mRNA export is coupled to transcriptional elongation by the TREX (transcription/export) complex (Strasser et al. 2002), which is required for both processes and interacts with the transcriptional and export machinery (Chang et al. 1999; Libri et al. 2002; Strasser et al. 2002). Splicing is thought to be coupled to mRNA export through the splicing factor Sub2/UAP56 (Reed 2003). In this case, Sub2 is required for both splicing and mRNA export (Libri et al. 2002; Strasser and Hurt 2002) and interacts with the mRNA export factor Yra1 (Strasser and Hurt 2001; Zenklusen et al. 2002).
Prior to export, transcripts undergo surveillance mediated by the nuclear exosome (Burkard and Butler 2000; Hilleren et al. 2001; Butler 2002; Torchet et al. 2002). The nuclear exosome is a conserved complex of 3′ → 5′ exoribonucleases that processes snRNAs, snoRNAs, and rRNAs and degrades aberrant mRNAs (Allmang et al. 1999; van Hoof et al. 2000; Butler 2002; Peng et al. 2003). The exosomal proteins Rrp6 (Briggs et al. 1998; Allmang et al. 1999) and Lrp1 (Erdemir et al. 2002; Mitchell et al. 2003; Peng et al. 2003) are the only exclusively nuclear components of the exosome (Burkard and Butler 2000; Kumar et al. 2002), whereas the other exosomal components participate in both nuclear and cytoplasmic exosomal activities.
Lrp1 and Rrp6 have overlapping functions in RNA processing and degradation (Butler 2002; Mitchell et al. 2003; Peng et al. 2003). Both participate in nuclear rRNA, snoRNA, and snRNA processing (Allmang et al. 1999; Butler 2002; Mitchell et al. 2003). Rrp6 also mediates nuclear mRNA surveillance, while the role of Lrp1 in this process is largely undefined. Rrp6 is required for the degradation of aberrantly spliced and 3′-end processed transcripts generated in processing mutants (Burkard and Butler 2000; Butler 2002; Torchet et al. 2002). Further, Rrp6 degrades mRNAs trapped in the nucleus upon mutation of the nucleoporin Nup116 (Das et al. 2003). Lrp1 is not required for degradation of aberrantly polyadenylated mRNAs in the case of two tested transcripts (Mitchell et al. 2003); however, its role in surveillance of other mRNA species has not been determined.
Rrp6 is thought to mediate mRNA surveillance through retention and degradation of mRNAs at transcriptional sites. Drosophila exosomal components, including Rrp6, are cotranscriptionally recruited and interact with elongation factors Spt5 and Spt6 (Andrulis et al. 2002). In splicing and 3′-end processing Saccharomyces cerevisiae mutants, Rrp6 retains transcripts at sites of transcription where they are presumed to be degraded (Jensen et al. 2001; Hilleren et al. 2001; Butler 2002; Libri et al. 2002; Zenklusen et al. 2002; Thomsen et al. 2003). Rrp6 also mediates retention of transcripts at transcriptional foci in TREX mutants (Libri et al. 2002; Zenklusen et al. 2002); these transcripts may represent mRNAs that are not correctly or completely packaged into mRNPs. These findings have lead to the view that Rrp6 cotranscriptionally monitors mRNP state and degrades mRNAs that are aberrant or otherwise export incompetent.
However, the fate of mRNAs in the absence of Rrp6-mediated mRNP quality control is poorly understood (Hilleren et al. 2001; Thomsen et al. 2003; Galy et al. 2004). While mRNAs move to the nucleolus and the nuclear periphery in export factor mutants lacking Rrp6 (Thomsen et al. 2003), the localization of transcripts in cells lacking Rrp6 alone has never been established. It is therefore unknown how cells deal with loss of mRNP quality control under conditions where inherent and environmentally induced errors may produce aberrant transcripts.
Such aberrant transcripts can be produced by DNA- and RNA-damaging radiation. Radiation may produce aberrant transcripts through RNA damage and increased transcriptional stalling at DNA damage sites (Svejstrup 2002); yet, it is not known if mRNA surveillance mechanisms exist to repair or degrade mRNA under such conditions. Interestingly, Lrp1 and the exosome have been previously linked to DNA repair and cancer as well as to RNA processing. Lrp1 was originally characterized as a DNA repair factor homologous to C1D (Erdemir et al. 2002), a DNA/RNA-binding protein that increases the RNA binding of the recombination-localized mRNA transport protein translin (Aoki et al. 1995; Chennathukuzhi et al. 2001). Lrp1 is required for normal levels of homologous recombination and nonhomologous end-joining (Erdemir et al. 2002). Other exosome components have been found as autoantigens in chronic myelogenous leukemia (Yang et al. 2002). Further, scleroderma diseases that exhibit exosome auotantigenicity have been linked to increased cancer incidence (Hill et al. 2003). Lrp1 and the exosome therefore have various ties to DNA damage and repair, but the connection between their roles in RNA metabolism and DNA damage is unclear. More generally, RNA surveillance under DNA damaging conditions is poorly understood.
Here we address questions about mRNA surveillance under wild type and RNA- and DNA-damaging conditions and about the fate of mRNAs in the absence of such surveillance. We identify four new mRNA export factors, including the exosomal factors Lrp1 and Rrp6, in a screen of all annotated nonessential S. cerevisiae genes. Whole-genome analysis of Lrp1 and Rrp6 genomic localization reveals that wild-type Lrp1 localization depends on Rrp6, and that Rrp6 localization in turn correlates with transcription. We find that Lrp1 and Rrp6 act in a novel RNA degradation response mediated by Lrp1 upon DNA- and RNA-damaging UV irradiation, in addition to mRNA degradation under nonirradiated conditions. Further, Rrp6 and Lrp1 are required for full DNA repair upon UV irradiation. Together, Lrp1 and Rrp6 serve as a checkpoint that prevents mRNA export in the absence of mRNA quality control. Broadly, Lrp1 and Rrp6 link DNA and RNA surveillance to mRNA export.
Results
Screen of all nonessential S. cerevisiae genes identifies four new mRNA export factors
To identify all annotated nonessential genes required for mRNA export in S. cerevisiae, we screened a comprehensive deletion strain collection (Winzeler et al. 1999) for nuclear poly(A)+ RNA accumulation by fluorescence in situ hybridization (FISH). We identified four novel factors required for poly(A)+ RNA export: Slx9, Apq12, Lrp1/Rrp47, and Rrp6 (Fig. 1A). Subsequently, Apq12 was independently shown to be required for nuclear poly(A)+ RNA export (Baker et al. 2004). Known mRNA export factors were also identified (data not shown). SLX9 deletion results in subnuclear accumulation of poly(A)+ RNA, and APQ12 deletion produces an elongated cellular phenotype with subnuclear poly(A)+ RNA accumulation in 20%-50% of cells (Fig. 1A). In contrast, deletion of RRP6 and LRP1 each results in poly(A)+ RNA accumulation throughout the nucleus in all cells (Fig. 1A).
Figure 1.
A screen of all annotated nonessential S. cerevisiae genes reveals four novel mRNA export factors, Rrp6, Lrp1, Apq12, and Slx9, which interact genetically. (A) A screen of a comprehensive deletion strain collection identified four new strains with poly(A)+ RNA nuclear accumulation, shown here by Cy3-dT50 fluorescence in situ hybridization (FISH) with DAPI costaining. (B) Δslx9, Δapq1, Δlrp1, and Δrrp6 strains show nuclear accumulation of YOR095C mRNA as determined by FISH and DAPI staining. (C) Lrp1 and Rrp6 genetically interact with each other and other mRNA export factors. The Δlrp1 Δslx9, Δrrp6 Δapq1, and Δlrp1 Δrrp6 double mutants have synthetic slow growth phenotypes. Dilution series grown for 2 d are shown. All mutants are in the BY4741/2 background except for Δrrp6 (BP0-12F) crossed with Δapq1.
The specific requirement for Slx9, Apq12, Lrp1, and Rrp6 in poly(A)+ RNA export was verified in several ways. The deletion marker consistently segregated with each mRNA export defect. Reintroduction of the deleted protein by plasmid expression abolished the export defect in all cases (Supplementary Fig. S1). We also verified that aberrant RNA processing and degradation resulting from general inhibition of exosome function in rrp4-1, rrp43-1, and ski6-2 mutants does not, in and of itself, result in nuclear poly(A)+ RNA accumulation (data not shown).
The nuclear accumulation of poly(A)+ RNA in Slx9, Apq12, Lrp1, and Rrp6 mutants reflects their requirement in mRNA export. Because polyadenylation of snoRNA and snRNA precursors (van Hoof et al. 2000) and rRNA (Kuai et al. 2004) could account for the observed nuclear poly(A)+ RNA accumulation, we tested whether these factors were required for export of mRNA by single transcript FISH. Cells lacking these factors show nuclear accumulation of YOR095C mRNA, a transcript identified as a target of these export factors (Fig. 1B; see below). Thus, Slx9, Apq12, Lrp1, and Rrp6 act as mRNA export factors.
The novel mRNA export factors exhibit a number of specific genetic and transcriptional connections to one another. LRP1 and RRP6 interact genetically, as indicated by the slight synthetic slow growth phenotype of Δlrp1 Δrrp6 strains at 37°C (Fig. 1C). Codeletion of LRP1 and SLX9 also resulted in synthetic sickness at 25°C and 37°C, as did deletion of RRP6 and APQ12 (Fig. 1C). In addition, the expression of these genes shows correlated changes under various conditions (Supplementary Fig. S2). LRP1 and RRP6 expression is correlated during sporulation and over a variety of environmental and DNA damage conditions (Pearson correlation coefficient, r ≥ 0.5). LRP1 and SLX9 show correlated expression over these environmental conditions and sporulation (r ≥ 0.8). RRP6 and APQ12 levels are correlated during sporulation (r ≥ 0.5). In contrast, none of the novel factors show correlated gene expression with the nucleoporin NUP116; mRNA export factors THP1, NPL3, and YRA1; or the negative control glucose-6-phosphate dehydrogenase gene ZWF1.
Genome-wide effects and genomic localization of the novel mRNA export factors Lrp1 and Rrp6
Of the novel mRNA export factors, the conserved factors Lrp1 and Rrp6 interact (Mitchell et al. 2003; Peng et al. 2003) and act in exosome-mediated RNA metabolism (Butler 2002; Mitchell et al. 2003; Peng et al. 2003). To further investigate the functional similarities of Lrp1 and Rrp6, we carried out mRNA expression profiling of Δlrp1 and Δrrp6 deletion strains relative to a wild-type strain by using whole-genome cDNA microarrays (Supplementary Data 1). Consistent with the functional overlap between Lrp1 and Rrp6, deletion of these proteins results in highly correlated changes in mRNA expression levels across the whole genome (r = 0.88). They also show similar functional biases (Supplementary Table S1), with both Δlrp1 and Δrrp6 cells showing upregulation of transcriptional machinery (p = 0.01), ribo-some biogenesis factors (p = 0.001), and proteasome components (p < 0.001). The increased expression of ribosomal and transcriptional machinery is consistent with compensation for decreased rRNA processing and mRNA export. Lrp1 and Rrp6 therefore have common cellular effects as well as common functions.
In addition to this genome-wide functional correlation, Lrp1 and Rrp6 show functional interdependence in their genomic localization. After verifying the nuclear localization and physical interaction of tagged Lrp1 and Rrp6 (data not shown), we determined their genomic localization by microarray analysis of coimmunoprecipitating chromatin. In these experiments, immunoprecipitated and total chromatin was competitively hybridized to whole-genome cDNA microarrays in order to establish the relative binding levels of Lrp1 and Rrp6 to individual genes across the whole genome (Fig. 2A, black and gray lines; Supplementary Data 2). Rrp6 binding levels are distributed fairly close to the mean binding level, with most genes showing relatively similar binding levels (Fig. 2A, black line). In contrast, Lrp1 binding levels range more widely (Fig. 2A, gray line). These trends are consistent with Rrp6 having a general role at chromatin and Lrp1 having a more targeted function.
Figure 2.
Chromatin immunoprecipitation profiling reveals Lrp1 and Rrp6 genomic localization. (A) Rrp6 binding levels are relatively uniform across the genome, while Lrp1 binding levels vary more widely. The distributions of Lrp1 (gray line) and Rrp6 (black line) binding levels in wild-type cells are shown as the log of coimmunoprecipitated chromatin to input chromatin ratio. Binding level distributions for genes bound by Lrp1 (gray) or Rrp6 (black) above the mean binding level with P < 0.01 are shown by the color-filled region. (B) Lrp1 and Rrp6 show code-pendent genome-wide DNA localization, with Lrp1 localization exhibiting particularly strong dependence on Rrp6. Lrp1 and Rrp6 ChIP profiles were clustered by principal component analysis (PCA). Their similarity scores are represented chromatically for all enriched genes (left) and genes exhibiting more than twofold enrichment and de-enrichment (right). (C) Lrp1 binding levels become more uniform in the absence of Rrp6, while the Rrp6 binding level distribution is largely unchanged by Lrp1 deletion. The distributions of Lrp1 (gray line) and Rrp6 (black line) binding levels in deletion cells lacking the other factor are shown as the log of coimmunoprecipitated chromatin to input chromatin ratio. Distributions of binding levels for genes bound by Lrp1 in Δrrp6 cells (gray) or Rrp6 in Δlrp1 cells (black) above the mean binding level for P < 0.01 are indicated by the color-filled region. (D) Rrp6 binding levels correlate with transcriptional frequency. The histogram of median transcriptional frequency as a function of binding level is shown.
The localization patterns of Lrp1 and Rrp6 across the genome are similar but not identical. The overlap of genes bound by both proteins is highly significant but not absolute, as indicated by the shared number of genes and its statistical significance (P = 10-104) (Table 1). We also determined the similarity of Lrp1 and Rrp6 binding across all genes by principal component analysis (PCA), data reduction-based clustering approach that establishes the similarity of multiple microarray data sets. This similarity is represented here chromatically, such that the similarity of any two microarray profiles is indicated by the similarity of the two colors representing them. Lrp1 and Rrp6 chromatin binding profiles, light green and blue, respectively, are therefore similar but not identical (Fig. 2B, left, light green and blue). When only highly bound or unbound genes are analyzed by PCA, Lrp1 and Rrp6 chromatin binding profiles are even more similar, as indicated here by their almost identical PCA color representation (Fig. 2B, right, dark blue). Interestingly, PCA clustering of the genomic localization of Lrp1, Rrp6, and several other mRNA export factors (Yu et al. 2004) reveals that Lrp1 and Rrp6 localization are most similar to that of Tho2, Yra1, and Npl3, all of which have ties to the nuclear exosome (Supplementary Fig. S3). Lrp1 and Rrp6 therefore have genomic localizations similar to that of functionally related mRNA export factors and of each other.
Table 1.
Overlap between genes affected and bound by Lrp1 and Rrp6 and genes encoding transcripts degraded by Lrp1
| Lrp1 bound in WT
|
Lrp1 bound in Δrrp6
|
Rrp6 bound in WT
|
Rrp6 bound in Δlrp1
|
up in Δlrp1
|
down in Δlrp1
|
up in Δrrp6
|
down in Δrrp6
|
||
|---|---|---|---|---|---|---|---|---|---|
| Microarray experiment | self | −log P (#) | −log P (#) | −log P (#) | −log P (#) | −log P (#) | −log P (#) | −log P (#) | −log P (#) |
| Bound by Lrp1 (WT) | (1233) | ||||||||
| Bound by Lrp1 (Δrrp6) | (643) | 10 (203) | |||||||
| Bound by Rrp6 (WT) | (524) | 104 (334) | 4 (82) | ||||||
| Bound by Rrp6 (Δlrp1) | (387) | 57 (223) | 7 (75) | 2 (147) | |||||
| Up in Δlrp1 | (305) | 4 (91) | 0 (28) | 0 (49) | 2 (30) | ||||
| Down in Δlrp1 | (234) | 2 (31) | 3 (41) | 0 (10) | 0 (14) | n/a | |||
| Up in Δrrp6 | (847) | 19 (228) | 0 (90) | 5 (109) | 1 (66) | 02 (203) | 0 (0) | ||
| Down in Δrrp6 | (972) | 5 (151) | 3 (73) | 4 (53) | 4 (37) | 0 (6) | 65 (152) | n/a | |
| Stabilized in Δlrp1 (relative to WT) | (1383) | 56 (91) | 13 (75) | 2 (146) | 0 (30) | 3 (46) | 0 (55) | 0 (191) | 5 (146) |
The statistical significance of overlap between microarray data categories is given as the negative log of the P-value. The absolute number of common genes between microarray data categories in the overlap is given in parentheses. (WT) Wild type.
We then examined the genomic localization of Lrp1 and Rrp6 in the presence and absence of the other to determine the interdependence of their localization. The distribution of Rrp6 binding levels is largely unaffected by deletion of Lrp1 (Fig. 2C, black line). In contrast, Lrp1 binding levels distribute more closely around the mean in the absence of Rrp6 (Fig. 2C, gray line). PCA also shows that Lrp1 localization changes significantly when Rrp6 is absent, as indicated by the difference in the PCA colors of the Lrp1 chromatin binding profile in wild-type and Δrrp6 cells (blue and magenta; Fig. 2B); Rrp6 localization is largely unaffected by the absence of Lrp1, shown by the similarity of PCA colors of Rrp6 chromatin binding in wild-type and Δlrp1 cells (green; Fig. 2B). Further, the absolute number of genes that Lrp1 binds above the mean binding level decreases by half in the absence of Rrp6 relative to wild type, while the number of genes that Rrp6 binds decreases by only one-fourth in the absence of Lrp1 (Table 1). Thus, while both Lrp1 and Rrp6 genomic localization show some codependence, proper Lrp1 localization strongly requires Rrp6.
Lrp1 and Rrp6 localization is also linked to transcription. All strains were grown under galactose induction for genomic location profiling, and correspondingly, the galactose-induced genes are enriched in all localization profiles (P < 0.025). This enrichment indicates that yeast Lrp1 and Rrp6 are recruited to transcriptionally induced genes, as are Drosophila exosome components (Andrulis et al. 2002). In addition, Rrp6 binding level correlates with the transcriptional frequency of the bound genes (Fig. 2D). Transcription therefore correlates with Rrp6 localization, and Rrp6 in turn plays a role in Lrp1 genomic localization.
Lrp1 and Rrp6 play a role in genome-wide mRNA degradation under normal and DNA-damaging conditions
Since Rrp6 and Lrp1 functionally interact at chromatin and have known roles in mRNA degradation and DNA repair, respectively, we hypothesized that Lrp1 and Rrp6 couple mRNA export to mRNA and DNA surveillance. To address this hypothesis, we first established whether Lrp1 functions in mRNA surveillance. We determined the role of Lrp1 in mRNA degradation by quantifying the relative changes in transcript abundance of all yeast mRNAs over 45 min in wild-type and Δlrp1 cells treated with the transcriptional inhibitor thiolutin (Das et al. 2003). The abundance changes during this time, as assayed by whole-genome microarray, directly reflect degradation and hence relative degradation rates (Supplementary Data 3). We found that more than one-fifth of all transcripts (22%) become stabilized in Δlrp1 cells relative to wild type (Table 2), while Δrrp6 cells did not show significant changes in mRNA stability as previously observed (Anderson and Parker 1998). Almost no mRNAs (0.4%) are destabilized in Δlrp1 cells (Table 2). The median log stability ratio is 23% higher in Δlrp1 cells than wild-type cells. Moreover, Lrp1-bound genes significantly overlap with genes encoding transcripts degraded by Lrp1 (P = 10-56) (Table 1). Thus, Lrp1 plays a role in degradation of mRNAs encoded by genes it binds.
Table 2.
Comparison of transcript stability in wild-type, Δlrp1, and Δrrp6 cells with and without UV irradiation
| Cell types compared
|
Transcripts stabilized
|
Transcripts destabilized
|
Transcripts unchanged
|
|
|---|---|---|---|---|
| type 1 | type 2 | # (%) | # (%) | # (%) |
| Δlrp1 | wt | 1383 (22) | 22 (0) | 4816 (77) |
| Δrrp6 | wt | 217 (3) | 22 (0) | 5982 (96) |
| Δlrp1 + UV | wt + UV | 1698 (27) | 6 (0) | 4517 (73) |
| Δrrp6 + UV | wt + UV | 39 (1) | 20 (0) | 6162 (99) |
| Δlrp1 + UV | Δlrp1 | 1205 (19) | 214 (3) | 4802 (77) |
| Δrrp6 + UV | Δrrp6 | 1399 (23) | 817 (13) | 4005 (64) |
| wt + UV | wt | 1617 (26) | 726 (12) | 3878 (62) |
The number and percentage of transcripts stabilized, destabilized, or unchanged in stability in cell type 1 relative to cell type 2 is listed. (wt) Wild type.
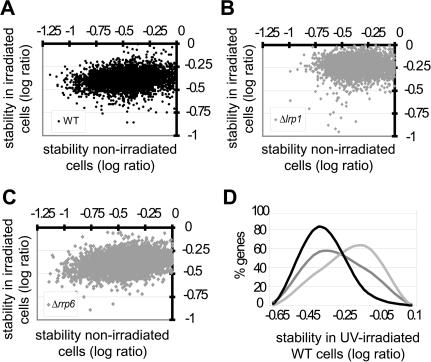
Next, we asked whether Lrp1 and Rrp6 function in mRNA degradation upon DNA-damaging irradiation. Wild-type cells exhibit an overall narrowing in their range of mRNA stabilities upon UV irradiation (Fig. 3A). Notably, they show destabilization of the most stable transcript subpopulation, as indicated by a gap in the population of mRNAs with the highest log stability ratio. This results in a 66% decrease in the range of log stability ratios in wild-type cells upon UV irradiation. While Δrrp6 cells also exhibit this phenotype (Fig. 3C), Δlrp1 cells do not show UV-induced destabilization of the most stable mRNA subpopulation (Fig. 3B). Only 3% of mRNAs are destabilized in Δlrp1 cells upon UV irradiation in contrast to 12% in wild-type cells (Table 2). The median mRNA stability in Δlrp1 cells increases 30% upon irradiation. Moreover, the stability frequency distribution of transcripts degraded in an Lrp1-dependent manner upon irradiation (Fig. 3D, dark gray line) is similar to that of transcripts destabilized irradiated wild-type cells (black line). These results suggest that Lrp1 is required for mRNA degradation upon UV damage beyond its role under nonirradiated conditions.
Figure 3.
Lrp1 is required for UV-induced RNA degradation. (A) Upon UV-irradiation, wild-type cells show destabilization of the most stable mRNA subpopulation (black). The stability of each transcript in nonirradiated cells is plotted against its stability after UV irradiation, as the log ratio of abundance after 45-min degradation to original abundance. (B) Transcripts in Δlrp1 cells (gray) do not exhibit the UV-induced degradation pattern of wild-type cells. (C) Transcripts in Δrrp6 cells show the same UV-induced degradation phenotype as do wild-type cells. (D) The distribution of mRNAs over the mRNA stability range of UV-irradiated wild-type cells is shown for mRNAs that exhibit Lrp1-dependent degradation in UV-irradiated cells (black) and increased (dark gray) and unaltered (light gray) degradation in UV-irradiated wild-type cells.
To establish the role of Rrp6 in this process, we examined the requirement for Rrp6 in Lrp1 localization to genes encoding Lrp1 degradation targets. Lrp1 genomic localization overlaps less significantly with genes encoding its target transcripts in the absence of Rrp6 (P = 10-13) than in the presence of Rrp6 (P = 10-56), indicating that Rrp6 is partially required for Lrp1 localization to chromatin encoding Lrp1 targets under nonirradiated conditions (Table 1). Lrp1 genomic localization under nonirradiated conditions also correlates to some degree with mRNA stability in irradiated Δlrp1 cells (r = 0.4). This correlation disappears when Lrp1 genomic localization in the absence of Rrp6 is considered. Rrp6 is therefore involved in localizing Lrp1 to genes encoding mRNAs Lrp1 degrades under normal and DNA damaging conditions.
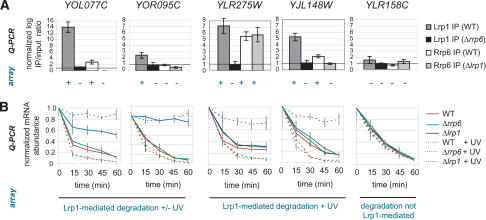
The microarray-based localization and degradation results were validated by real-time quantitative PCR of five genes and their transcripts. Quantitative PCR confirmed the microarray-determined enrichment or under-enrichment of Rrp6 and Lrp1 at the five genes upon immunoprecipitation (Fig. 4A). In addition, mRNA degradation time courses established the role of Lrp1 and Rrp6 in degradation of the corresponding transcripts with and without UV irradiation (Fig. 4B). Two transcripts (YOL077C, YOR095C) showed Lrp1-mediated degradation under normal conditions and additional degradation upon UV irradiation by both microarray- and quantitative PCR-based analysis. Two others exhibited only UV-dependent degradation by Lrp1 (YLR275W, YJL148W), and another showed no Lrp1-dependent stability effects (YLR158C) by both analysis approaches. All five transcripts therefore showed the Lrp1 and Rrp6 binding patterns and the Lrp1-mediated degradation trends observed by microarray.
Figure 4.
Quantitative PCR validates location and degradation profiling. (A) Quantitative PCR (Q-PCR) confirms the microarray analysis of Lrp1 and Rrp6 genomic localization at five genes, YOL077C, YOR095C, YLR275W, YJL148W, and YLR158C. Lrp1 binding levels in the presence (dark gray) and absence (black) of Rrp6 and Rrp6 binding levels in the presence (white) and absence (light gray) of Lrp1 to these genes are shown as the normalized log ratio of IP to input chromatin. The log IP/input chromatin ratios were normalized to that of a control intergenic region. Error bars represent the standard deviation of three experiments. Microarray-determined binding above or below the mean binding level are indicated by plus and minus signs below each column. (B) Degradation time courses of YOL077C, YOR095C, YLR275W, YJL148W, and YLR158C mRNA in wild-type (red), Δlrp1 (blue), and Δrrp6 (green) cells with (solid line) and without (dashed line) UV irradiation over 60 min validate the role of Lrp1 in general and UV-mediated mRNA degradation. mRNA abundance was normalized to starting abundance and to the abundance of a spiked-in control Arabidopsis mRNA. Error bars represent the standard deviation of three experiments.
Lrp1 and Rrp6 are required for normal DNA repair
We next sought to determine the role of Lrp1 and Rrp6 in DNA repair upon UV irradiation. Repair of UV-induced cyclobutane pyrimidine dimers (CPDs) was assayed in Δlrp1 and Δrrp6 strains by quantitative PCR of RPB2 after T4 endonuclease V-mediated CPD cleavage (Sweder and Hanawalt 1992). This assay is not strand-specific and therefore does not distinguish between transcription-coupled repair (TCR, Svejstrup 2002) and nontranscriptional global genomic repair (GGR); however, GGR occurs at lower rates than TCR and would contribute less significantly to repair over the assayed time period. Both Δrrp6 and Δlrp1 strains show significant decreases in CPD repair relative to wild type (Fig. 5A). The observed repair defect is comparable to the TCR defect seen in Δrad7 mutants and roughly half that seen in Δrad26 mutants (Sweder and Hanawalt 1992; Gonzalez-Barrera et al. 2002). Lrp1 and Rrp6 are therefore required for wild-type repair of UV-induced DNA damage.
Figure 5.
Rrp6 and Lrp1 are involved in DNA repair. (A) Lrp1 and Rrp6 are required for repair of UV-induced cyclobutane pyrimidine dimers (CPDs). The repair of UV-induced CPDs at the RPB2 gene in wild-type (black), Δlrp1 (light gray), and Δrrp6 (dark gray) strains over 60 min is shown at 15-min intervals, as the percent of wild-type DNA repair after 120 min. The error bars represent the standard deviation of three replicates. (B) Deletion of RAD26 results in a synthetic growth defect with deletion of RRP6. Dilution series grown for 2 d are shown.
RRP6 deletion also results in synthetic sickness with deletion of RAD26 (Fig. 5B), a yeast homolog of the Cockayne's syndrome protein CSB that acts in transcriptional elongation and TCR (Svejstrup 2003). Since the exosome interacts with the transcription elongation machinery (Andrulis et al. 2002), the Δrrp6 Δrad26 synthetic phenotype might reflect a shared involvement of Rad26 and Rrp6 in transcriptional elongation. However, codeletion of Rrp6 and elongation factor Thp1 does not result in synthetic slow growth (data not shown). Deletion of Rad26 with transcription elongation factor Spt4 also does not give rise to synthetic defects and instead abolishes the requirement for Rad26 in TCR (Jansen et al. 2000; Svejstrup 2002). The genetic interaction of Rrp6 and Rad26 is therefore likely to indicate a common role of these proteins in DNA repair.
Discussion
Nuclear export of mRNA is highly coupled to other processes in gene expression, including transcription and transcript processing (for review, see Reed 2003; Vinciguerra and Stutz 2004). Moreover, mRNA export factor recruitment occurs cotranscriptionally, as do processes central to transcriptional fidelity such as DNA repair and mRNA surveillance (Dimaano and Ullman 2004; Vinciguerra and Stutz 2004). However, little is known about mRNA and its export in the absence of these fidelity mechanisms (Hilleren et al. 2001; Thomsen et al. 2003; Galy et al. 2004).
We set out to identify all nonessential S. cerevisiae mRNA export factors in order to gain a comprehensive picture of the mRNA export factor population. Since mRNA export factors are typically defined by nuclear accumulation of poly(A)+ RNA upon abrogation of their function (Amberg et al. 1992; Strasser and Hurt 2001; Strasser et al. 2002; Rodriguez-Navarro et al. 2004), we screened a complete deletion strain collection for nuclear poly(A)+ RNA accumulation to achieve this end. The novel mRNA export factors identified here represent almost one-fifth of known mRNA export factors, excluding factors required nonspecifically for general nuclear transport.
Of the new export factors, Slx9 and Apq12 have no previously known functions outside of mRNA export and polyadenylation (Baker et al. 2004). However, synthetic phenotypes link them to recombination and transcription. Slx9 deletion causes synthetic sickness with deletion of DNA repair proteins Sgs1 and Rad27 (Ooi et al. 2003). Apq12 deletion is synthetically lethal with deletion of the transcription factor Cdc73 (Tong et al. 2004). Such connections to transcription and recombination are prevalent among mRNA export factors (Jimeno et al. 2002; Vinciguerra and Stutz 2004).
In contrast, Lrp1 and Rrp6 have previously been linked to RNA metabolism. The broader nuclear mRNA retention in Rrp6 single mutants is initially surprising, given that Rrp6 deletion results in loss of mRNA retention at the site of transcription in other mutant backgrounds. This retention is, however, consistent with previously observed nuclear accumulation of a control reporter transcript in Δrrp6 cells (Galy et al. 2004). Nuclear mRNA accumulation in Δrrp6 and Δlrp1 cells could potentially reflect an mRNA degradation defect that increases mRNA levels and consequently saturates the export machinery, rather than a nuclear export defect. In the case of Δrrp6 cells, such an explanation is inconsistent with their lack of significant mRNA stabilization. This explanation is unlikely even for strains with decreased mRNA degradation, since increased mRNA levels due to increased mRNA synthesis or transcription factor mutation do not result in visible nuclear mRNA accumulation. Thus, Lrp1 and Rrp6 are required for mRNA export. This requirement may serve as a second level of quality control that prevents nuclear export of transcripts that escape exosome-mediated surveillance.
Lrp1 mediates novel mRNA surveillance under both normal and DNA- and RNA-damaging conditions. Since UV irradiation may induce both RNA damage and aborted transcription at DNA damage sites, the transcripts degraded by Lrp1 upon irradiation are likely to represent damaged transcripts and/or aberrant transcripts generated by aborted transcription (Svejstrup 2002). DNA damage commonly results in stalled transcription, and without subsequent lesion resolution, it results in RNA polymerase II (RNAPII) degradation and aborted transcription (Svejstrup 2002). Although TFIIS can facilitate RNA cleavage at stalled RNAPII to release short 3′ mRNA stretches (Kettenberger et al. 2003), the fate of mRNA upon aborted transcription is less clear. Since transcription is often aborted under both normal and DNA damaging conditions, the role of Lrp1 in both general and UV-induced mRNA degradation may represent mRNA surveillance of aborted transcription. Additional selectivity beyond transcriptional coupling is indicated by the high stability of the UV-induced Lrp1 target population. In the absence of Lrp1-mediated surveillance and degradation, this population would persist upon damaging irradiation.
Rrp6 is involved in Lrp1 localization to genes encoding its mRNA targets and thereby plays a role in Lrp1-mediated mRNA surveillance. Lrp1 and Rrp6 genomic location analysis provides the first genome-wide view of the exosome's chromatin localization. This analysis revealed that Rrp6 has a more uniform, transcription-correlated distribution of binding levels across the genome than Lrp1. Rrp6 in turn plays a role in the targeted genomic localization of Lrp1, consistent with the substoichiometric coimmunoprecipitation of Lrp1 by Rrp6 (Peng et al. 2003). While Rrp6 is not directly required for detectible mRNA degradation under normal or DNA damage conditions, redundant mechanisms for Lrp1 localization to its target transcripts are likely to exist. These may include post-transcriptional processes. Lrp1 and Rrp6 therefore act in a novel RNA degradation response under normal conditions and further upon DNA- and RNA-damaging UV irradiation.
Lrp1 and Rrp6 are also required for full DNA surveillance and repair upon UV irradiation. Rrp6 and Lrp1 may function in DNA repair by helping resolve stalled RNAPII complexes and increasing the accessibility of DNA lesions to DNA repair machinery, as does Rad26 (Svejstrup 2003); such resolution could occur through Lrp1 localization to sites of stalled transcription and subsequent mRNA clearing at these sites by Lrp1-mediated mRNA degradation. Alternatively, lack of Lrp1-mediated mRNA degradation could lead to transcriptional defects and consequent TCR inhibition. For example, the transcriptional elongation and export factor Hpr1 prevents formation of DNA:RNA hybrids that otherwise impair transcription (Huertas and Aguilera 2003) and is further required for TCR (Gonzalez-Barrera et al. 2002). However, cells lacking Hpr1 exhibit hyper-recombination rather than the decreased recombination seen in the Lrp1 knockout mutant (Chavez and Aguilera 1997); this suggests that the mechanistic role of Hpr1 in DNA repair differs from that of Lrp1 and Rrp6. Broadly, the role of Lrp1 and Rrp6 in mRNA surveillance upon UV irradiation may explain their requirement in DNA repair.
Lrp1 and Rrp6 link mRNA export to DNA and RNA surveillance. In particular, Lrp1 and Rrp6 couple surveillance to mRNA export through their dual functions in these processes. Functional coupling of mRNA export to other processes has been previously concluded from the concurrent activity of factors in both export and other events, as observed for Lrp1 and Rrp6 (Vinciguerra and Stutz 2004). Such coupling suggests that surveillance is required for export competence of mRNAs. Cells therefore degrade aberrant mRNAs through exosome-mediated surveillance (Butler 2002) and, second, prevent export of transcripts that escape this surveillance. Thus, the exosome serves in two linked mRNA quality control checkpoints.
Specificity of post-transcriptional mRNP factors for different mRNA subpopulations is an emerging theme in mRNA export and general mRNP biology (Keene and Tenenbaum 2002; Hieronymus and Silver 2003; Rehwinkel et al. 2004). Specificity may arise from differential coupling among post-transcriptional processes (Dimaano and Ullman 2004) and differential combinatorial interactions of proteins that mediate these processes (Keene and Tenenbaum 2002). Our findings suggest that such specificity extends to nuclear mRNA surveillance and degradation. Lrp1 is differentially required for degradation of mRNA subpopulations under normal and DNA damaging conditions. Moreover, Rrp6 and Lrp1 are involved in surveillance of different types of aberrant mRNAs. While Rrp6 is broadly required for mRNA surveillance of many aberrant mRNA forms, Lrp1 is required for UV-induced mRNA degradation but not for degradation of aberrantly polyadenylated transcripts (Mitchell et al. 2003). Last, Rrp6 and Lrp1 are differentially localized across the genome. This indicates that different exosomal factors preferentially bind different genes or genomic areas and represents the first time the chromatin binding specificities of the exosome has been established on a large scale. Thus, although specificity is widely observed in cytoplasmic mRNA degradation (Yang et al. 2003), this work reveals specificity of the nuclear mRNA surveillance and export machinery.
In sum, these results show that mRNA surveillance is coupled to mRNA export by the nuclear exosome. Moreover, they suggest that exosomal components link mRNA export to DNA and mRNA surveillance. This work also provides the first global view of exosomal targets and indicates that post-transcriptional mRNP specificity extends to exosome-mediated nuclear mRNA surveillance. Broadly, this work expands the current view of mRNA export coupling to include surveillance and its specificity.
Materials and methods
Yeast strains and plasmids
All strains and plasmids used here are as listed (Supplementary Tables S2, S3). Lrp1 was tagged with 13 tandem copies of the myc epitope by C-terminal integration of myc13::KAN; myc-Lrp1 is therefore integrated and expressed under its own promoter. The GST-tagged Rrp6 used in this work (Burkard and Butler 2000) was expressed under galactose induction over Rrp6 deletion (Δrrp6).
Screen for mRNA export mutants
A complete haploid deletion strain collection (Research Genetics; Winzeler et al. 1999) was screened for mutants showing nuclear accumulation of poly(A)+ RNA using FISH as described (Hieronymus and Silver 2003). The poly(A)+ and mRNA export defects of the novel mutants (PSY3013, PSY3014, PSY3015, PSY3214) were verified more than three times. The identity of the deleted ORF in each strain was verified by PCR as described (Giaever et al. 2002). Segregation of the kanamycin deletion marker with the poly(A)+ RNA export defect was examined in the progeny of >20 tetrads of each strain. Poly(A)+ RNA localization was examined in deletion strains transformed with Rrp6 (PPS2884), GST-Lrp1 (PPS2954), Slx9, and GST-Apq12 (PPS2953) and in exosome mutants rrp4-1, rrp43-1, and ski6-2. Single transcript FISH was carried out as described (Hieronymus and Silver 2003) except with probes against YOR095C (Supplementary Table S4).
Testing for genetic interactions
Haploid double mutants were generated from all pair-wise matings of Δrrp6, Δlrp1, Δslx9, Δapq12, Δrad7, Δrad16, and Δrad26 strains. Double mutants were tested for synthetic sickness by growing 104, 103, 102, and 10 cells on YEPD plates at 25°C and 37°C for 2 d.
Genomic location profiling
Genomic location profiling was performed in triplicate as described (Casolari et al. 2004) with strains expressing (1) myc-Lrp1 and GST-Rrp6 (PSY3251), (2) myc-Lrp1 in a Δrrp6 background (PSY3230), and (3) GST-Rrp6 in a Δlrp1 background (PSY3252). All strains were grown in 1% raffinose, 0.1% galactose at 30°C to 0.5-0.75 OD600. Immunoprecipitation was performed using anti-rabbit IgG beads (Dynal) with rabbit anti-myc 9E11 antibody or anti-GST antibody (Santa Cruz Biotechnology). Binding was considered significantly enriched at genes with an immunoprecipitated DNA to input DNA ratio >1 (P < 0.01).
Expression profiling
Expression profiling was carried out in triplicate for Δrrp6 (PSY3013) and Δlrp1(PSY3014) strains relative to wild type (PSY1930). Cells were grown in YEPD at 30°C to 1-2 × 107 cells/mL. In vitro transcribed Arabidopsis chlorophyll synthase mRNA (PPS2865) was spiked into each pelleted cell sample (47 ng/108 cells). RNA preparation, cDNA preparation, and hybridization were carried out as described (Casolari et al. 2004). cDNA from each knockout strain was compared to wild-type cDNA by competitive hybridization. RNAs with nonzero log ratios (ChIP/input DNA) at P < 0.01 were considered up- or down-regulated.
mRNA degradation profiling
mRNA degradation profiling was carried out in triplicate for wild-type (PSY1930), Δrrp6 (PSY3013), and Δlrp1(PSY3014) strains with and without UV irradiation. The strains were grown to 1-2 × 107 cells/mL in YEPD at 30°C. Equal numbers of cells were irradiated with 0 or 30 J/m2 256-nm UV light. The cells were then grown in YEPD with 4 ug/mL thiolutin (National Cancer Institute) in the dark for 0 or 45 min at 30°C. Arabidopsis control RNA was added to pelleted cell samples (47 ng/108 cells for 0-min samples, 94 ng/108 cells for 45-min samples). RNA preparation, cDNA preparation, and hybridization were carried out as described for expression profiling. Labeled cDNA from cells incubated for 0 and 45 min were compared by competitive hybridization. The resulting data was normalized to Arabidopsis control spot intensities. The stability of a transcript was considered to differ between cell types if the log ratios of the quantity after 45 min degradation relative to its starting quantity differed by >2 standard deviations.
Further data analysis
Coexpression between the novel mRNA export factor genes was analyzed as previously implemented (Saccharomyces Genome Database). PCA was carried out with 90% data reduction (Rosetta Resolver). ChIP profiling data was compared to transcriptional frequency as described (Casolari et al. 2004). Functional enrichment analysis was performed with Funcassociate (Berriz et al. 2003). The significance of overlap between gene populations was determined by Fisher's exact test.
Microarray validation by quantitative real-time PCR
Quantitative real-time PCR was used to determine the amount of YOL077C, YOR095C, YLR275W, YJL148W, and YLR158C coimmunoprecipitating with myc-Lrp1 and GSTRrp6 (PSY3251) in wild-type cells; myc-Lrp1 in Δrrp6 cells (PSY3230); and GST-Rrp6 in Δlrp1 cells (PSY3252) as well as in the IP input. It was also used to determine the amount of the corresponding transcripts in wild-type, Δrrp6, and Δlrp1 strains over a 1-h degradation time course. For the time courses, cells were treated with ultraviolet (UV) irradiation and thiolutin as described above, and samples were collected at 15-min intervals. RNA was then isolated and reverse transcribed, and the resulting cDNA was analyzed by real-time PCR. The PCR reaction was carried out with 1× Sybr green master mix (Applied Biosystems), 50 mM forward and reverse primers each, and ∼50 pg DNA in 15 μL. Primer sequences are listed in the Supplemental Material (Supplementary Table S4). The resulting product was quantified in real-time by Sybr green fluorescence (Applied Biosystems Prism 7700 Sequence Detector). The log linear fit of the reaction and initial input amount of the gene was determined as described (TAQ software). The ChIP amounts were normalized to the amount of chromatin in the input and then to the IP/input ratio of a nontranscribed intergenic region. The mRNA levels were normalized to the amount of the spiked-in Arabidopsis mRNA and then to the initial abundance of the transcript. The average level and standard deviation was calculated from three replicates.
DNA repair assay
CPD repair was assayed by using a T4 endonuclease V (TEV)-based quantitative PCR approach modified from existing TCR assays. Wild-type (PSY1930), Δrrp6 (PSY3013), and Δlrp1(PSY3014) strains were grown to 1-2 × 107 cells/mL at 30°C, irradiated with 30 J/m2 256-nm light, and grown in the dark for 0, 20, 40, 60, 90, and 120 min as described (Sweder and Hanawalt 1992). DNA was isolated from the cells, precipitated, and treated with TEV to cleave at CPD sites also as described (Sweder and Hanawalt 1992). DNA repair of a 1.5-kb region of RPB2 was then measured by quantitative PCR (Amplitaq gold, Applied Biosystems) over five twofold dilutions with the following temperature cycles: 2 min at 50°C; 10 min at 95°C; 26 cycles of 95°C for 20 sec, 57°C for 30 sec, and 70°C for 2 min 15 sec; and 7 min at 70°C. A 100-bp control region was similarly amplified. Primer sequences are given in the Supplemental Material (Supplementary Table S4). The PCR products were quantified on gels stained with Sybr green. The resulting signal of the 1.5-kb region was normalized to that of the 100-bp control region and to wild-type repair after 120 min.
Acknowledgments
We thank K. Burchett for help with genetic analysis. We also thank S. Komili, J. Casolari, and F. Bachand for comments on the manuscript. This work was supported by a Howard Hughes Medical Institute fellowship (to H.H.), an NIH postdoctoral fellowship (to M.C.Y.), and NIH grants (to P.A.S.).
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1241204.
References
- Allmang C., Petfalski, E., Podtelejnikov, A., Mann, M., Tollervey, D., and Mitchell, P. 1999. The yeast exosome and human PM-Scl are related complexes of 3′ → 5′ exonucleases. Genes Dev. 13: 2148-2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amberg D.C., Goldstein, A.L., and Cole, C.N. 1992. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 6: 1173-1189. [DOI] [PubMed] [Google Scholar]
- Anderson J.S. and Parker, R.P. 1998. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 17: 1497-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrulis E.D., Werner, J., Nazarian, A., Erdjument-Bromage, H., Tempst, P., and Lis, J.T. 2002. The RNA processing exosome is linked to elongating RNA polymerase II in Drosophila. Nature 420: 837-841. [DOI] [PubMed] [Google Scholar]
- Aoki K., Suzuki, K., Sugano, T., Tasaka, T., Nakahara, K., Kuge, O., Omori, A., and Kasai, M. 1995. A novel gene, Translin, encodes a recombination hotspot binding protein associated with chromosomal translocations. Nat. Genet. 10: 167-174. [DOI] [PubMed] [Google Scholar]
- Baker K.E., Coller, J., and Parker, R. 2004. The yeast Apq12 protein affects nucleocytoplasmic mRNA transport. RNA. 10: 1352-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriz G.F., King, O.D., Bryant, B., Sander, C., and Roth, F.P. 2003. Characterizing gene sets with FuncAssociate. Bioinformatics 19: 2502-2504. [DOI] [PubMed] [Google Scholar]
- Briggs M.W., Burkard, K.T., and Butler, J.S. 1998. Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem. 273: 13255-13263. [DOI] [PubMed] [Google Scholar]
- Burkard K.T. and Butler, J.S. 2000. A nuclear 3′-5′ exonuclease involved in mRNA degradation interacts with Poly(A) polymerase and the hnRNA protein Npl3p. Mol. Cell. Biol. 20: 604-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J.S. 2002. The yin and yang of the exosome. Trends Cell. Biol. 12: 90-96. [DOI] [PubMed] [Google Scholar]
- Casolari J.M., Brown, C.R., Komili, S., West, J., Hieronymus, H., and Silver, P.A. 2004. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117: 427-439. [DOI] [PubMed] [Google Scholar]
- Chang M., French-Cornay, D., Fan, H.Y., Klein, H., Denis, C.L., and Jaehning, J.A. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 19: 1056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez S. and Aguilera, A. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes & Dev. 11: 3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennathukuzhi V.M., Kurihara, Y., Bray, J.D., and Hecht, N.B. 2001. Trax (translin-associated factor X), a primarily cytoplasmic protein, inhibits the binding of TB-RBP (translin) to RNA. J. Biol. Chem. 276: 13256-13263. [DOI] [PubMed] [Google Scholar]
- Das B., Butler, J.S., and Sherman, F. 2003. Degradation of normal mRNA in the nucleus of Saccharomyces cerevisiae. Mol. Cell. Biol. 23: 5502-5515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano C. and Ullman, K.S. 2004. Nucleocytoplasmic transport: Integrating mRNA production and turnover with export through the nuclear pore. Mol. Cell. Biol. 24: 3069-3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdemir T., Bilican, B., Cagatay, T., Goding, C.R., and Yavuzer, U. 2002. Saccharomyces cerevisiae C1D is implicated in both non-homologous DNA end joining and homologous recombination. Mol. Microbiol. 46: 947-957. [DOI] [PubMed] [Google Scholar]
- Galy V., Gadal, O., Fromont-Racine, M., Romano, A., Jacquier, A., and Nehrbass, U. 2004. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63-73. [DOI] [PubMed] [Google Scholar]
- Giaever G., Chu, A.M., Ni, L., Connelly, C., Riles, L., Veronneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418: 387-391. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Barrera S., Prado, F., Verhage, R., Brouwer, J., and Aguilera, A. 2002. Defective nucleotide excision repair in yeast hpr1 and tho2 mutants. Nucleic Acids Res. 30: 2193-2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector R.E., Nykamp, K.R., Dheur, S., Anderson, J.T., Non, P.J., Urbinati, C.R., Wilson, S.M., Minvielle-Sebastia, L., and Swanson, M.S. 2002. Dual requirement for yeast hnRNP Nab2p in mRNA poly(A) tail length control and nuclear export. EMBO J. 21: 1800-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H. and Silver, P.A. 2003. Genome-wide analysis of RNA-protein interactions illustrates specificity of the mRNA export machinery. Nat. Genet. 33: 155-161. [DOI] [PubMed] [Google Scholar]
- Hill C.L., Nguyen, A.M., Roder, D., and Roberts-Thomson, P. 2003. Risk of cancer in patients with scleroderma: A population based cohort study. Ann. Rheum. Dis. 62: 728-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilleren P., McCarthy, T., Rosbash, M., Parker, R., and Jensen, T.H. 2001. Quality control of mRNA 3′-end processing is linked to the nuclear exosome. Nature 413: 538-542. [DOI] [PubMed] [Google Scholar]
- Huertas P. and Aguilera, A. 2003. Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol. Cell 12: 711-721. [DOI] [PubMed] [Google Scholar]
- Jansen L.E., den Dulk, H., Brouns, R.M., de Ruijter, M., Brandsma, J.A., and Brouwer, J. 2000. Spt4 modulates Rad26 requirement in transcription-coupled nucleotide excision repair. EMBO J. 19: 6498-6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen T.H., Patricio, K., McCarthy, T., and Rosbash, M. 2001. A block to mRNA nuclear export in S. cerevisiae leads to hyperadenylation of transcripts that accumulate at the site of transcription. Mol. Cell 7: 887-898. [DOI] [PubMed] [Google Scholar]
- Jimeno S., Rondon, A.G., Luna, R., and Aguilera, A. 2002. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 21: 3526-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. and Tenenbaum, S.A. 2002. Eukaryotic mRNPs may represent post-transcriptional operons. Mol Cell 9: 1161-1167. [DOI] [PubMed] [Google Scholar]
- Kettenberger H., Armache, K.J., and Cramer, P. 2003. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell 114: 347-357. [DOI] [PubMed] [Google Scholar]
- Kleiman F.E. and Manley, J.L. 1999. Functional interaction of BRCA1-associated BARD1 with polyadenylation factor CstF-50. Science 285: 1576-1579. [DOI] [PubMed] [Google Scholar]
- Kuai L., Fang, F., Butler, J.S., and Sherman, F. 2004. Polyadenylation of rRNA in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. 101: 8581-8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Cheung, K.H., Tosches, N., Masiar, P., Liu, Y., Miller, P., and Snyder, M. 2002. The TRIPLES database: A community resource for yeast molecular biology. Nucleic Acids Res. 30: 73-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D., Dower, K., Boulay, J., Thomsen, R., Rosbash, M., and Jensen, T.H. 2002. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 22: 8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P., Petfalski, E., Houalla, R., Podtelejnikov, A., Mann, M., and Tollervey, D. 2003. Rrp47p is an exosome-associated protein required for the 3′ processing of stable RNAs. Mol. Cell. Biol. 23: 6982-6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi S.L., Shoemaker, D.D., and Boeke, J.D. 2003. DNA helicase gene interaction network defined using synthetic lethality analyzed by microarray. Nat. Genet. 35: 277-286. [DOI] [PubMed] [Google Scholar]
- Peng W.T., Robinson, M.D., Mnaimneh, S., Krogan, N.J., Cagney, G., Morris, Q., Davierwala, A.P., Grigull, J., Yang, X., Zhang, W., et al. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113: 919-933. [DOI] [PubMed] [Google Scholar]
- Reed R. 2003. Coupling transcription, splicing and mRNA export. Curr. Opin. Cell Biol. 15: 326-331. [DOI] [PubMed] [Google Scholar]
- Rehwinkel J., Herold, A., Gari, K., Kocher, T., Rode, M., Ciccarelli, F.L., Wilm, M., and Izaurralde, E. 2004. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat. Struct. Mol. Biol. 11: 558-566. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S., Fischer, T., Luo, M.J., Antunez, O., Brettschneider, S., Lechner, J., Perez-Ortin, J.E., Reed, R., and Hurt, E. 2004. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75-86. [DOI] [PubMed] [Google Scholar]
- Strasser K. and Hurt, E. 2001. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature 413: 648-652. [DOI] [PubMed] [Google Scholar]
- Strasser K., Masuda, S., Mason, P., Pfannstiel, J., Oppizzi, M., Rodriguez-Navarro, S., Rondon, A.G., Aguilera, A., Struhl, K., Reed, R., et al. 2002. TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304-308. [DOI] [PubMed] [Google Scholar]
- Svejstrup J.Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell. Biol. 3: 21-29. [DOI] [PubMed] [Google Scholar]
- ____. 2003. Rescue of arrested RNA polymerase II complexes. J. Cell Sci. 116: 447-451. [DOI] [PubMed] [Google Scholar]
- Sweder K.S. and Hanawalt, P.C. 1992. Preferential repair of cyclobutane pyrimidine dimers in the transcribed strand of a gene in yeast chromosomes and plasmids is dependent on transcription. Proc. Natl. Acad. Sci. 89: 10696-10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen R., Libri, D., Boulay, J., Rosbash, M., and Jensen, T.H. 2003. Localization of nuclear retained mRNAs in Saccharomyces cerevisiae. RNA 9: 1049-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Lesage, G., Bader, G.D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G.F., Brost, R.L., Chang, M., et al. 2004. Global mapping of the yeast genetic interaction network. Science 303: 808-813. [DOI] [PubMed] [Google Scholar]
- Torchet C., Bousquet-Antonelli, C., Milligan, L., Thompson, E., Kufel, J., and Tollervey, D. 2002. Processing of 3′-extended read-through transcripts by the exosome can generate functional mRNAs. Mol. Cell 9: 1285-1296. [DOI] [PubMed] [Google Scholar]
- van Hoof A., Lennertz, P., and Parker, R. 2000. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell Biol. 20: 441-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinciguerra P. and Stutz, F. 2004. mRNA export: An assembly line from genes to nuclear pores. Curr. Opin. Cell. Biol. 16: 285-292. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Shoemaker, D.D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J.D., Bussey, H., et al. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901-906. [DOI] [PubMed] [Google Scholar]
- Yang X.F., Wu, C.J., Chen, L., Alyea, E.P., Canning, C., Kantoff, P., Soiffer, R.J., Dranoff, G., and Ritz, J. 2002. CML28 is a broadly immunogenic antigen, which is overexpressed in tumor cells. Cancer Res. 62: 5517-5522. [PMC free article] [PubMed] [Google Scholar]
- Yang E., van Nimwegen, E., Zavolan, M., Rajewsky, N., Schroeder, M., Magnasco, M., and Darnell Jr., J.E. 2003. Decay rates of human mRNAs: Correlation with functional characteristics and sequence attributes. Genome Res. 13: 1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M.C., Bachand, F., McBride, A.E., Komili, S., Casolari, J.M., and Silver, P.A. Arginine methyltransferase affects interaction and recruitment of mRNA processing and export factors. 18: 2024-2035. [DOI] [PMC free article] [PubMed]
- Zenklusen D., Vinciguerra, P., Wyss, J.C., and Stutz, F. 2002. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol. Cell Biol. 22: 8241-8253. [DOI] [PMC free article] [PubMed] [Google Scholar]