Abstract
It is generally assumed that there are only two ways to maintain the ends of chromosomes in yeast and mammalian nuclei: telomerase and recombination. Without telomerase and recombination, cells enter senescence, a state of permanent growth arrest. We found that the decisive role in preventing senescent budding yeast cells from dividing is played by the Exo1 nuclease. In the absence of Exo1, telomerase- and recombination-defective yeast can resume cell cycle progression, despite degradation of telomeric regions from many chromosomes. As degradation progresses toward internal chromosomal regions, a progressive decrease in viability would be expected, caused by loss of essential genes. However, this was not the case. We demonstrate that extensive degradation and loss of essential genes can be efficiently prevented through a little-studied mechanism of DNA double-strand-break repair, in which short DNA palindromes induce formation of large DNA palindromes. For the first time, we show that large palindromes form as a natural consequence of postsenescence growth and that they become essential for immortalization in the absence of telomerase activity.
Keywords: PAL-mechanism, telomere, Exo1, palindrome
The stability of eukaryotic chromosomes depends on the structural and functional integrity of their ends, called telomeres. It is generally accepted that telomeres have different properties from internal double-strand breaks, because functional telomeres do not alert DNA-damage surveillance mechanisms or repair pathways. Several hypotheses have been proposed to explain what distinguishes telomeres from internal double-strand breaks (Cervantes and Lundblad 2002; Lydall 2003; Ferreira et al. 2004; Harrington 2004). One involves a particular telomeric structure called the t-loop, where a 3′ single-stranded DNA extension loops back and intercalates into the double-stranded DNA (Griffith et al. 1999). Another hypothesis attributes to telomere-associated proteins the major role in telomere protection, although many (Ku, Sir, MRX) are also associated with double-strand breaks. Recent evidence suggests that telomerase itself plays a role in telomere protection (Masutomi et al. 2003).
Another critical function of telomeres in dividing cells is to solve the end-replication problem. The end-replication problem, originally recognized by Olovnikov and Watson, is caused by the incapacity of conventional DNA replication to fully replicate the end of linear DNA molecules (Watson 1972; Olovnikov 1973). All forms of life with linear DNA molecules, from bacteriophage to mammals, have developed mechanisms to solve the end- replication problem. One such mechanism is provided by telomerase. Telomerase, found in most eukaryotic organisms, is a reverse transcriptase-based enzyme able to synthesize DNA de novo, using species-specific RNA templates.
In many multicellular organisms, telomerase expression appears to be limited, except in stem cells, germ cells, and cancer cells. Somatic cells can divide in the absence of telomerase, but their telomeres shorten with each division. After several divisions, telomeres of many somatic cells are sufficiently short to lose their “capping” properties and activate checkpoint proteins and repair pathways, as DNA double-strand breaks do. Telomere dysfunction is thought to be the major cause of replicative senescence, a state of continued cell viability without cell division. It has been shown that inactivation of checkpoint pathways (Rb and p53) permits human cells to bypass replicative senescence. However, such checkpoint-defective postsenescent cells enter crisis and lose viability after several divisions (Shay and Wright 1989; Hara et al. 1991). Conversely, telomerase expression is thought to allow many tumor cells to divide indefinitely.
Budding yeast cells express telomerase and divide indefinitely. Yeast cells from which telomerase has been removed behave like mammalian somatic cells, dividing for a limited number of generations before cell division is inhibited by short or defective telomeres (Lundblad and Szostak 1989). After a period of senescence, rare telomerase-negative survivors escape senescence using recombination-dependent mechanisms to amplify telomeric and subtelomeric repeats (Lundblad and Blackburn 1993). Similarly, some immortalized human cells are able to maintain telomeres without expressing telomerase. These cells use a mechanism that has been termed ALT (Alternative Lengthening of Telomeres) (Henson et al. 2002). The ALT mechanism is also thought to be recombination-dependent (Dunham et al. 2000; Varley et al. 2002).
Here we report that telomerase- and recombination-defective yeast can escape senescence in the absence of a telomere-active nuclease, Exo1. These cells proliferate indefinitely with linear, but abnormally sized chromosomes that have lost, in most cases, telomeric and subtelomeric sequences. In these strains, essential genes are maintained by formation of palindromes at the ends of chromosomes.
Results
Exo1-defective cells proliferate with chromosomes lacking telomeres
To date, there are only two ways known to maintain linear chromosomes in budding yeast and mammalian nuclei: telomerase and recombination. To facilitate identification of other ways to maintain linear chromosomes, we eliminated telomerase and recombination by deleting the TLC1 and RAD52 genes in budding yeast. TLC1 encodes the RNA component of telomerase (Singer and Gottschling 1994), whereas RAD52 is required for virtually all homologous recombination events (Paques and Haber 1999). Normally, telomeraseand recombination-defective cells rapidly enter senescence and do not recover to generate survivors (Lundblad and Blackburn 1993).
We recently showed that the Exo1 nuclease was important for cell cycle arrest (Maringele and Lydall 2002) and death (L. Maringele, unpubl.) of telomere-defective mutants and contributed to senescence of telomerase-deficient cells (Maringele and Lydall 2004). Therefore, we reasoned that deletion of Exo1 might rescue growth of telomerase- and recombination-defective cells.
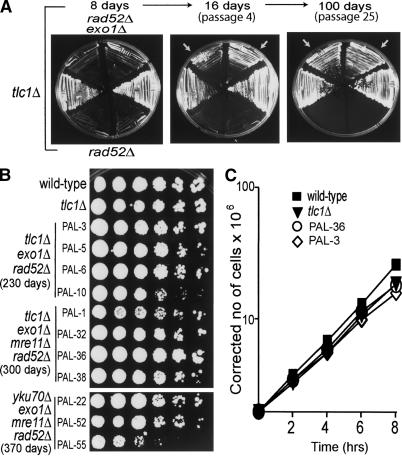
To test the role of Exo1, several independent haploid tlc1Δ rad52Δ exo1Δ strains were generated, along with controls, and ∼10 million cells from each strain were serially propagated on solid plates (Fig. 1A). As previously found, RAD52+ tlc1Δ control strains escaped senescence within a few days, whereas none of the 100 million tested tlc1Δ rad52Δ cells (from 10 independent strains) was able to generate survivors (Fig. 1A; Table 1). In contrast, 10 out of 22 independent tlc1Δ rad52Δ exo1Δ strains generated survivors (indicated by arrows in Fig. 1A; Table 1) after a period of senescence. Thus, it is clear that EXO1 opposes telomerase- and recombination-independent survival.
Figure 1.
Exo1 inhibits growth of tlc1Δ rad52Δ strains. (A) Two independent strains of each genotype were passaged on plates every 4 d and photographed 4, 16, and 100 d after being taken from germination plates. Top segments are tlc1Δ rad52Δ exo1Δ strains, equatorial segments are tlc1Δ strains, and bottom segments are tlc1Δ rad52Δ strains. (B) Twelve independent strains, propagated for the indicated periods, were grown overnight in liquid, and fivefold dilution series were prepared before small aliquots were spotted onto agar plates. The top 10 strains were photographed after 4 d, the bottom three strains, which grew more slowly, after 6 d. The wild-type strain was DLY641, and the tlc1Δ strain was DLY2146, propagated for >60 d. (C) Four strains, described in B, were grown in liquid culture at 25°C. Cells were diluted in fresh medium and counted, by hemocytometer, every 2 h.
Table 1.
Exo1 opposes escape from senescence and immortalization
| Genotype | Fraction escaped senescence | Fraction immortalized | Total time in culture (days) | Strain numbers |
|---|---|---|---|---|
| tlc1Δ rad52Δ | 0/10 | |||
| tlc1Δ exo1Δ rad52Δ | 10/22 | 5/5 | >310 | PAL-2, PAL-3, PAL-5, PAL-6, PAL-10 |
| tlc1Δ mre11Δ rad52Δ | 0/10 | |||
| tlc1Δ exo1Δ mre11Δ rad52Δ | 10/10 | 6/6 | >390 | PAL-1, PAL-11, PAL-30, PAL-32, PAL-36, PAL-38 |
| yku70Δ mre11Δ rad52Δ | 0/5 | |||
| yku70Δ mre11Δ exo1Δ rad52Δ | 5/5 | 4/5 | >460 | PAL-15, PAL-22, PAL-52, PAL-55 |
Column 1 shows the genotypes of strains generated from germinated spores and propagated as described in Materials and Methods. Column 2 indicates the fraction of strains that escaped senescence and that were passaged for at least 60 d. Column 3 shows the fractions of strains that were passaged for longer. Only one of 16 propagated strains lost viability, after ∼70 d. Column 4 shows the current length of time in culture. Column 5 shows strain numbers.
Previous experiments showed overlapping roles for Exo1 and another nuclease, Mre11, in DNA repair (Tsubouchi and Ogawa 2000; Moreau et al. 2001; Lee et al. 2002; Lewis et al. 2002; Tran et al. 2002). Therefore, we tested the role of Mre11 in tlc1Δ rad52Δ cells. We found that in the presence of EXO1, none of the 100 million tlc1Δ rad52Δ mre11Δ cells, from 10 independent strains, escaped senescence (Table 1). Thus, deletion of MRE11 is insufficient to permit tlc1Δ rad52Δ cells to escape senescence.
Interestingly, we found that deletion of MRE11 increased by hundreds of fold the frequency of telomeraseand recombination-independent survivors in an exo1Δ background. All tlc1Δ rad52Δ exo1Δ mre11Δ strains escaped senescence at a frequency higher than 1 × 10-5 cells. This frequency was deduced from the lowest number of tlc1Δ rad52Δ exo1Δ mre11Δ cells in 3/3 liquid cultures shown to generate survivors (Maringele and Lydall 2004). Taken together, our data indicate that Exo1 strongly opposes telomerase- and recombination-independent survival, whereas Mre11 plays an obvious role only in the absence of Exo1, when it decreases the number of telomerase- and recombination-defective cells that escape senescence.
It is possible to induce senescence without removing telomerase. For example, we and others have shown that cells defective in two telomere capping proteins, Yku70 and Mre11, rapidly enter senescence, even though telomerase is expressed (DuBois et al. 2002; Maringele and Lydall 2002, 2004). We wanted to know if removal of Exo1 also permitted growth in such situations. We found that all tested yku70Δ mre11Δ rad52Δ exo1Δ strains escaped senescence, but none of the yku70Δ mre11Δ rad52Δ strains escaped (Table 1). Together with data in the tlc1Δ background, this indicates that deletion of EXO1 permits proliferation of a variety of telomere-induced senescent cells. This experiment also shows that Yku70 is not essential for growth in the absence of both recombination and efficient telomerase activity.
To simplify description, we call the telomerase-, recombination-, and Exo1-defective strains PAL-survivors, based on the mechanism that permits their immortalization, as described later. PAL-survivors grew slowly during passages 4-10, then growth improved (Fig. 1A, cf. growth and colony size at passages 4 and 25).
To compare the growth of PAL-survivors with strains using telomerase or recombination to maintain telomeres, we plated serial dilutions on agar. Figure 1B shows that although some strains grew slower than others, the viability of PAL-survivors was high. Furthermore, the best growing PAL-survivors (PAL-3, PAL-36) had similar growth rates to a tlc1Δ strain (maintaining telomeres using recombination). We found that PAL-survivors lacking YKU70 grew more slowly, perhaps because of more pronounced DNA repair defects.
PAL-survivors not only grow well, but are probably immortalized, because 15/16 were still growing 300-460 d after they were generated. These time periods correspond to ∼3000-4600 generations for wild-type cells grown under the same conditions. In summary, PAL-survivors grow well and indefinitely, yet they lack the classical mechanisms to maintain chromosome ends. How is this possible?
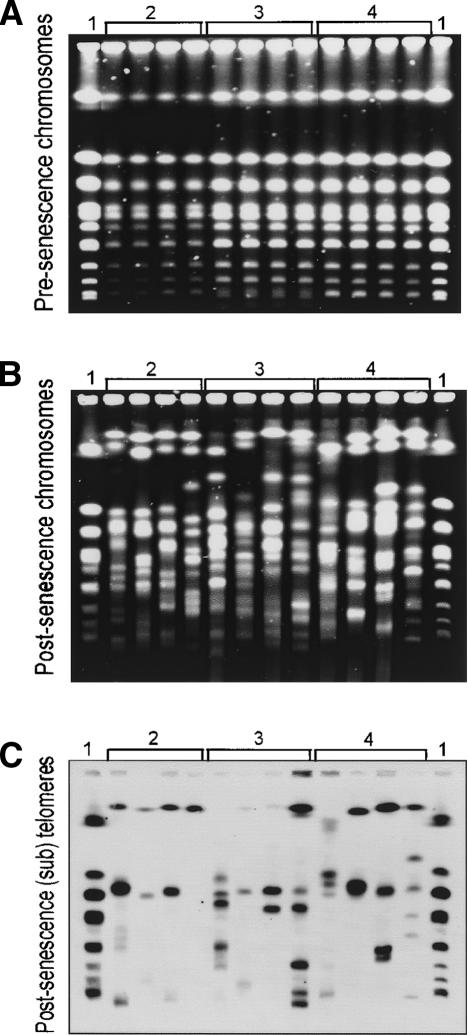
One possibility to explain growth of yeast cells without telomerase and recombination would be that they circularized their chromosomes, like telomerase-defective fission yeast (Nakamura et al. 1998). To test this hypothesis, we performed pulsed field gel electrophoresis (PFGE) on pre- and postsenescent strains, because only linear chromosomes should enter pulsed field gels. Figure 2A and B shows that both pre- and postsenescent strains contained linear chromosomes, recognizable as tight bands of similar intensity as the wild-type chromosome bands. This PFGE analysis also revealed that all “mature” (older than 200 d) PAL-survivors had extensive chromosome size abnormalities. Moreover, each PAL-survivor had a different pattern of extensive chromosomal changes.
Figure 2.
PAL survivors contain linear, abnormally sized chromosomes. (A) PFGE of strains unable to maintain telomeres using telomerase, at early passage. Twelve independent freshly germinated spores were grown on agar plates for 5 d, then overnight in liquid culture, before chromosomes were analyzed. Lane 1 is the wild type strain DLY641; lanes bracketed by 2 are yku70Δ mre11Δ exo1Δ rad52Δ strains; lanes bracketed by 3 are tlc1Δ exo1Δ rad52Δ strains; and lanes bracketed by 4 are tlc1Δ mre11Δ exo1Δ rad52Δ strains. (B) As in A, except that PAL-survivors had been in culture for 200-340 d before chromosomal analysis. The genotypes bracketed by the numbers are as in A. From left to right, lane 1 is wild type; lanes bracketed by 2 are PAL-15, PAL-22, PAL-52, PAL-55 (340 d); lanes bracketed by 3 are PAL-2, PAL-5, PAL-6, PAL-10 (200 d); and lanes bracketed by 4 are PAL-1, PAL-30, PAL-36, PAL-38 (270 d). (C) The gel shown in B was hybridized to a telomeric and subtelomeric probe, previously described (Maringele and Lydall 2004).
To test if PAL-survivors contained telomeric DNA, we hybridized the chromosomes to a probe that detected telomeric and subtelomeric repeats. Figure 2C shows that all 12 PAL-survivors gave few telomeric signals, ranging from one to six bands, whereas wild-type strains had eight hybridization bands. In conclusion, it is clear that PAL-survivors escaped senescence and proliferated with linear, but abnormally sized chromosomes, that have lost, in most cases, telomeric and subtelomeric DNA.
PAL-survivors progressively lose single gene loci
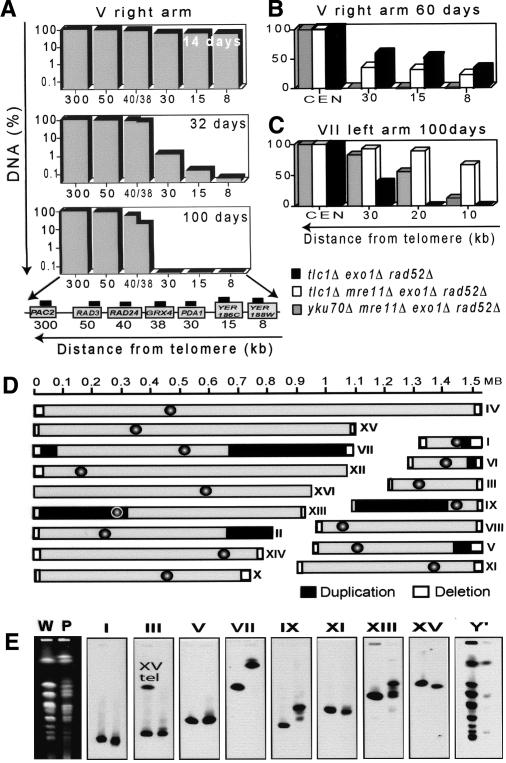
To address the question of whether chromosomes of PAL-survivors had lost more DNA than just telomeric and subtelomeric regions, we analyzed, by quantitative PCR, single gene loci on chromosomes V and VII. The amount of DNA at all loci was measured in three independent PAL-survivors, and the mean amount of DNA is shown in Figure 3A-C. We found that DNA levels at internal single copy loci were getting progressively less with time. For example, after 32 d, the right arm of chromosome V had lost 30 kb of DNA in ∼99% of cells (Fig. 3A). The extent of degradation exceeded the DNA loss expected from the end-replication problem, which causes <10 bp loss per generation in budding yeast (Lundblad and Szostak 1989). If survivors divided 10 times per day for 32 d, they should lose a maximum of 3.2 kb of DNA because of the end-replication problem. After 100 d in culture, the right arm of chromosome V had lost 38 kb in >50% of cells, and degradation largely stopped before the RAD24 gene at 40 kb (Fig. 3A). DNA loss showed a similar pattern on different chromosomes and in different genetic backgrounds: it started at telomeres and progressed toward centromeres (Fig. 2B,C). This pattern and the speed of degradation are consistent with the activity of nucleases degrading the ends of chromosomes V and VII, in addition to the end-replication problem.
Figure 3.
DNA loss and amplification in PAL-survivors. (A-C) Each column shows the average amount of DNA relative to wild type, measured by quantitative PCR in three independent strains with the same genotype, indicated by the key below C. (A) DNA levels were measured at seven loci along the right arm of chromosome V after 4, 32, and 100 d in culture. The X-axis indicates the distance from the right end of chromosome V. (Bottom) A cartoon indicates genes on the right arm of chromosome V (gray boxes) and the relative position of the probes (black boxes). (B) Loss of DNA along the right arm of chromosome V, after 60 d in culture, measured using a subset of the probes described in A. (C) Loss of DNA along the left arm of chromosome VII, after 100 d in culture, measured using probes at 10-kb intervals. (D) Genomic analysis of PAL-22 at 240 d. Wild-type and PAL-22 DNA were separately labeled and hybridized to Affymetrix S98 microarrrays, and the PAL/wild-type ratio was calculated for each gene. Yeast chromosomes sizes and centromere positions (circles) are according to the Saccharomyces Genome Database. Deletions detected by microarrays are marked in white, duplications are marked in dark gray, and normal levels of DNA are light gray. We classified DNA in PAL-22 as deleted if the ratio to wild type was 0-0.5 and as duplicated if the ratio was >1.4. We ignored single amplified genes. See Supplementary Figure 1 for primary data. (E) The first column shows PFGE of wild-type (W) and PAL-22 strain (P) stained with ethidium bromide. The following eight columns are Southern blots of the gel shown in the first column. The chromosomes detected are indicated above each blot. Probes hybridized to regions of duplicated DNA (except for chromosome XIII). Primers used to make probes are listed in Supplementary List 1. In the case of chromosome III, the probe also bound to a region close to the left end of the wild-type chromosome XV, and this region was deleted in PAL-22. The last column shows chromosomes hybridized with the telomere probe used in Figure 2C.
We further asked if Mre11 or Yku70, both involved in telomere protection, influenced chromosomal degradation in PAL-survivors. When we examined DNA loss on the right arm of chromosome V, yku70Δ mre11Δ exo1Δ rad52Δ strains showed more degradation than other PAL-survivors (Fig. 3B). However, on the left arm of chromosome VII, we found that YKU70+ MRE11+ (tlc1Δ exo1Δ rad52Δ) strains had more degradation (Fig. 3C). Therefore, we conclude that neither Mre11 nor Yku70 plays a critical role in protecting chromosome ends from degradation in PAL-survivors. Degradation appears to be a stochastic process. The apparently different degradation rates on different chromosomes (Fig. 3, cf. B and C) might be partially caused by unannotated sequences near telomeres, some as large as 30 kb (E. Louis, pers. comm.). Alternatively, different chromosome ends may have different susceptibilities to degradation.
To better understand the extent of degradation on all chromosome ends, we used genomic microarray analysis. Figure 3D is a schematic representation of all chromosomes from one PAL-survivor (PAL-22, yku70Δ mre11Δ exo1Δ rad52Δ, 240 d) and shows that 28/32 chromosome ends have lost DNA, some up to 45 kb. Primary data are presented in Supplementary Figure 1. Importantly, microarray analysis confirmed that 38 kb was lost from the right arm of chromosome V, as found by quantitative PCR as shown in Figure 3A.
Interestingly, 8/32 chromosome ends of PAL-22 showed 50-400-kb regions of duplicated DNA (Fig. 3D; Supplementary Fig. 1). Duplications detected by microarrays were larger (on chromosomes VII, IX, XIII), similarly sized (chromosome V), or smaller (chromosome I) than deletions on the same chromosome (Fig. 3D). We found a corresponding variation in chromosome size by PFGE (Fig. 3E), suggesting that duplications were not translocated to other chromosomes. Consistent with duplications not being translocated is the fact that the probes used in Southern blots were directed to the duplicated DNA (except for XIII, which was a dicentric chromosome), and they detected mainly single bands.
Unprotected chromosome ends might fuse to other chromosome ends. However, Figure 3E shows this was not the case for PAL-22, because chromosomes III, XI, and XV were shorter or similarly sized to the corresponding wild-type chromosomes (Fig. 3E). Degradation of telomeres in this strain was confirmed by hybridization with a telomeric probe: only three weak signals were detected (Fig. 3E, last column, labeled Y′). We also used microarrays to analyze whole genomes of six independent tlc1Δ exo1Δ rad52Δ and tlc1Δ mre11Δ exo1Δ rad52Δ strains and found a similar picture of absent distal genes and large duplicated chromosome regions (Supplementary Fig. 2; data not shown).
Palindromes limit loss of DNA and rescue essential genes
Duplications detected by microarray were frequently adjacent to regions of degradation (Fig. 3D; Supplementary Figs. 1, 2). This pattern suggested that cells with uncapped chromosomes limit progressive DNA loss by duplicating large chromosomal regions.
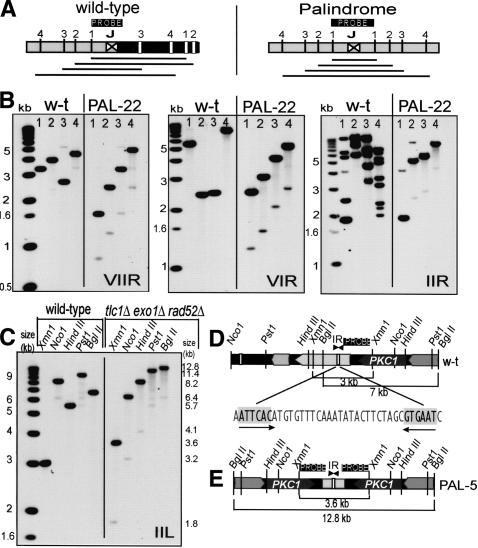
Microarray analysis allowed us to map (within 15 kb) the loci where degradation stopped and duplication began. Duplications of DNA on a single chromosome could be, in principle, caused by direct or inverted repeats (palindromes). To differentiate between these possibilities by Southern blot, we used a succession of restriction enzymes (1, 2, 3, 4) chosen to cut progressively further from the junction between two repeats, marked with J (Fig. 4A). At the wild-type locus or in the case of several-kilobase-large direct repeats, different fragments would most likely give a random pattern. In contrast, if a palindrome formed, the cut sites would be symmetric around the junction, and increasing fragment sizes would appear as a ladder (Fig. 4A). Figure 4B shows that ladders were clearly detected on the right arms of chromosomes VII, VI, and II in PAL-22, indicating that duplications were palindromes. It is also clear that a weaker ladder of bands, half the size of the palindrome ladder, accompanied each palindrome ladder. The origin of these half-sized bands is discussed below. Palindromes also formed in other PAL-survivors with different genotypes (Figs. 4C, 5, 6).
Figure 4.
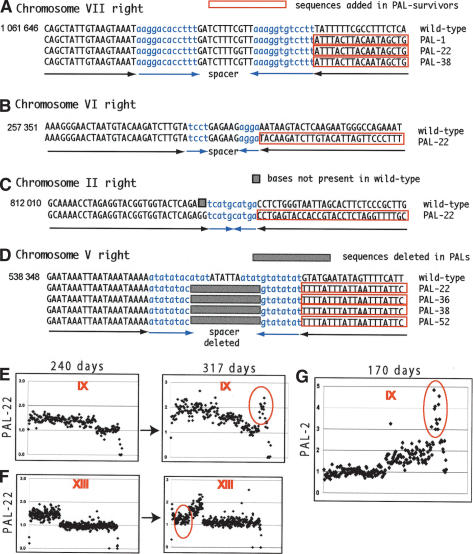
Palindromes form on chromosome arms. (A) Cartoons demonstrating how palindrome symmetry can be identified by appropriate choice of restriction enzymes and probe location. (Left cartoon) A succession of restriction enzymes (1, 2, 3, 4) will give a random pattern of bands when cutting wild-type DNA. (Right cartoon) If a palindrome has formed at this locus (by deleting the right half of wild-type DNA and duplicating the left half), enzymes will cut symmetrically around the junction between the palindrome arms, and increasing fragment sizes will generate a ladder. (B) Palindromes detected on the right arm of chromosomes VII, VI, and II in PAL-22. In each case, the first lane from the left shows molecular weight markers, the next four lanes are wild-type DNA, and the four right lanes are PAL-22 DNA (yku70Δ mre11Δ exo1Δ rad52Δ at 240 d). The enzymes used to cut DNA are indicated with numbers above each lane as follows: for chromosome VII, Stu1 (1), Bst11 (2), EcoR1 (3), and Bgl2 (4); for chromosome VI, Bst11 (1), Kpn1 (2), Pst1 (3), and Sac2 (4); and for chromosome II, Nsi1 (1), Afl2 (2), EcoR5 (3), and Nsp1 (4). The junction loci are shown in Figure 6. In the case of chromosome IIR, the probe hybridized close to the end of chromosome II, but also to several other wild-type chromosome ends, which were lost in this PAL-survivor by 240 d. Primers used to make probes are listed in Supplementary List 1. (C) A palindrome formed near PKC1 on the left arm of chromosome II. The first lane from the left shows molecular weight markers, the next five lanes are wild-type DNA, the five right lanes are DNA from PAL-5 (tlc1Δ exo1Δ rad52Δ strain at 100 d). The enzymes used to cut DNA and their positions are shown in D and E. (D) A map of the wild-type locus examined in C. Restriction enzymes sites, probe location, open reading frames, and an inverted repeat are shown. (E) A map of the palindrome detected in C.
Figure 5.
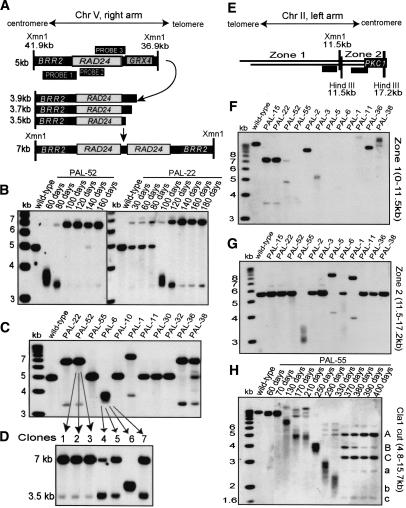
Palindrome formation in PAL-survivors. (A-D) Examination of the right end of chromosome V. (E-H) Examination of the left end of chromosome II. The strain genotypes are listed in Table 1. (A) A cartoon explaining the nature of the DNA fragments detected in B, C, and D. In B-D, an Xmn1 restriction enzyme was used to cut DNA; the distance between the telomere and the relevant cut sites is shown. Also indicated are the positions of the probes used in B-D. The primers used to generate probes are in Supplementary List 1. (B) Southern blot analysis of DNA extracted from PAL-52 and PAL-22 at intervals indicated above each lane. (C) Southern blot analysis of 12 independent PAL survivors: PAL-22, PAL-52, PAL-55 at 180 d; PAL-6, PAL-10 at 110 d; PAL-1, PAL-11, PAL-30, PAL-32, PAL-36, PAL-38 at 140 d. (D) Southern blot analysis of clones purified from two strains analyzed in C. (Lanes 1-3) DNA from PAL-52 clones. (Lanes 4-7) DNA purified from PAL-6 clones. All clones were grown for 7 d after the time point shown blot in C, when single clones were selected. (E) A map of the PKC1 locus near the left telomere of chromosome II. HindIII and Xmn1 restriction enzyme sites are shown. Probes used to examine zones 1 and 2 by Southern blot are shown as black boxes. (F) Twelve independent PAL-survivors were examined by Southern blot after Xmn1 digestion, using the probe directed to zone 1: PAL-15, PAL-22, PAL-52 at 390 d; PAL-55 at 340 d; PAL-2, PAL-3, PAL-5, PAL-6 at 250 d; and PAL-1, PAL-11, PAL-36, PAL-38 at 320 d. (G) The same DNA preparation examined in F were examined by Southern blot after HindIII digestion, using the probe directed to zone 2. (H) Southern blot analysis of DNA purified from PAL-55 at the indicated time points and cut with Cla1 (sites 4.8 and 15.7 kb from the telomere). The probe was as in G.
Figure 6.
Palindrome junctions contain unduplicated wild-type inverted repeats. (A-D) The sequences at the junction of nine palindromes from four independent strains are shown. The coordinates indicated on the right are according to the Saccharomyces Genome Database. Blue lowercase letters indicate the arms of wild-type inverted repeats, and red boxes indicate palindrome duplications. Black and blue arrows indicate complementary arms of inverted repeats and palindromes. PAL-22 and PAL-52 had been in culture for 317 d and PAL-1, PAL-36, and PAL-38 for 240 d. To facilitate PCR amplification and sequencing of palindrome junctions, DNA was treated with sodium meta-bisulfite to convert cytosine to uracil. The sequences shown have been deconverted, meaning some Ts were manually changed back to Cs, to facilitate reading of the palindrome sequences. Supplementary Figure 4 shows primary data. (E) Microarray analysis of chromosome IX of PAL-22 at 240 and 317 d. (F) Microarray analysis of chromosome XIII of PAL-22 at 240 and 317 d. (G) Microarray analysis of chromosome IX of PAL-2 at 170 d.
In PAL-survivors, there is strong selective pressure to maintain essential genes that could be degraded at chromosome ends. One hypothesis to explain palindrome formation in PAL-survivors is that palindromes always form close to essential genes under selective pressure. Because all genes essential for cell viability have been mapped in budding yeast, it was possible to test if palindromes only formed close to essential genes. We found this was not always the case. For example, on the right arm of chromosome II of PAL-22 (Fig. 4B), a palindrome formed in a subtelomeric repetitive region, 72 kb away from TSC10, the closest essential gene to the right telomere. However, we cannot exclude the possibility of selective pressure caused by synthetic lethal interactions (specific subtelomeric genes may become essential when other genes are deleted).
On other chromosomes and in other strains, it is very likely that palindromes rescued essential genes from degradation, and therefore, palindromes were critical for immortalization of these strains. Consistent with this, on the left arm of chromosome II (IIL) in PAL-5 (tlc1Δ exo1Δ rad52Δ; Fig. 4C-E), palindromes originated 2 kb from PKC1, the most telomere-proximal essential gene. Interestingly, we found a 6-bp inverted repeat (short palindrome) at the origin of palindrome formation on IIL (Fig. 4D).
Another situation in which palindromes rescued essential genes occurred on chromosome V. On the right arm of chromosome V, the most telomere-proximal essential gene is BRR2, 41 kb from the natural telomere. We used the restriction mapping approach described in Figure 4, in this case using nine different restrictions enzymes, and found palindromes on the right arm of chromosome V in five independent PAL-survivors (data are only shown for Xmn1 in Fig. 5C, and EcoRV in Supplementary Fig. 3). 4 PAL-survivors formed palindromes ∼2 kb from BRR2, and restriction mapping showed an AT-rich short inverted repeat (IR) at the junction between palindrome arms (Supplementary Fig. 3).
Weak half-sized bands were always detected in addition to strong palindrome bands (Figs. 4, 5). These might indicate sister chromatids joined together through noncovalent bonds (e.g., through base pairing between single-stranded inverted repeats) and able to separate under certain circumstances (Supplementary Fig. 3C). However, we found that the bond between palindrome arms was covalent, because it did not break under heat (Supplementary Fig. 3D) or alkali (Supplementary Fig. 3E). Also, separation of palindrome arms did not appear to be cell cycle phase-specific (Supplementary Fig. 3G).
We think it is more likely that half-sized bands are related to a fraction of palindromes that form a cruciform-like structure, caused by intrastrand base pairing around the junction (Supplementary Fig. 3H). Because cruciform structures are similar to recombination intermediates, Holliday junctions, it is possible that the enzyme required to resolve Holliday junctions, resolvase, cleaves around the junction of cruciforms (cut site labeled with 1 in Supplementary Fig. 3H). Alternatively, the half-sized bands might be generated in vitro by cutting large cruciforms (extending over several kilobases) with restriction enzymes that recognize sites on the vertical arms (a virtual cut site is labeled with 2 in Supplementary Fig. 3H). Irrespective of the origin of half-sized bands, we consider these bands a signature of large palindromes on chromosome arms.
Mechanism of palindrome formation
To understand the sequence of events that leads to palindrome formation, we analyzed the BRR2 locus on chromosome V by Southern blot. A simple model explaining the series of events that might lead to palindrome formation is shown in Figure 5A. To analyze the sequence of events in vivo, we cut genomic DNA and monitored the BRR2 locus by Southern blot, in two independent PAL-survivors, at regular periods of time, as indicated in Figure 5B. Consistent with the high specificity of our probes, we obtained a single 5-kb band in wild-type DNA (Fig. 5B). We had shown by real-time PCR that 30-38 kb from the right telomere of chromosome V had been lost in these two PAL-survivors (Fig. 3A), indicating that degradation was approaching the BRR2 locus. In PAL-52 at 60 d, we detected a diffuse band, 3.5-3.9 kb in size (Fig. 5B). This diffuse hybridization pattern, stretching over 400 bp, shows that DNA cut with Xmn1 and hybridized to the probe is heterogeneous in size. This heterogeneity reflects the range of chromosome ends in a large population of cells. At 80 d, the band became smaller (∼3.5 kb), but also tighter, suggesting that degradation had slowed in many cells. After 100 d, a strong 7-kb band, caused by a palindrome, appeared and persisted for the rest of the experiment (Fig. 5B). Another independent strain, PAL-22, appeared to be comprised of two populations of cells. A minor population formed a 7-kb palindrome early during the time course, because a weak 7-kb band can be seen by 30 d. The major population contained the wild-type 5-kb band for 80 d, then showed a diffuse band at 100 d before, finally, a palindrome formed and persisted for the remainder of the time course.
To see if palindromes formed at the BRR2 locus in other strains, we analyzed 11 independent PAL-survivors (Fig. 5C). Four appeared to have formed a palindrome at the same location (PAL-22, PAL-52, PAL-36, PAL-38). This suggests that palindromes may be specifically formed when degradation reaches a hot spot, presumably the short inverted repeat at this locus (Supplementary Fig. 3C). One strain, PAL-1, showed a larger 7.6-kb band, indicating that in this case, a palindrome initiated 300 bp before degradation reached the major hot spot on chromosome V. In this case also, a short inverted repeat mapped to the origin of the large palindrome (data not shown). Other PAL-survivors analyzed in Figure 5C had diffuse (PAL-6) or wild-type-like bands (PAL-55, PAL-10, PAL-11, PAL-30, PAL-32) at this time point and, according to the model in Figure 5A, either had not formed palindromes yet, or formed palindromes closer to the original telomere.
The sequence of events presented in Figure 5A could occur in many cells from the same strain, rather than in a single clone that overtook the culture. For example, when we analyzed four clones purified from cells with diffuse bands (PAL-6, Fig. 5C) we found that within 7 d, three of these clones had started to generate 7-kb bands from 3.5-kb bands (Fig. 5D, lanes 4,5,7), whereas one clone still had a diffuse band (Fig. 5D, lane 6).
Microarray analysis showed duplications (palindromes) at only a fraction of chromosome ends. Moreover, the same chromosome end appeared to have formed a palindrome in some PAL-survivors, but not in others (Supplementary Figs. 1, 2; data not shown). Therefore, we questioned whether the “palindrome-free” ends might have another form of end-protection, or if they formed palindromes at later time points. To systematically address this question, we visualized the left end of chromosome II in 12 independent PAL-survivors that had been in culture for 250-390 d. We chose chromosome II because the first essential gene, PKC1, is only 15 kb from the chromosome end. Importantly, all 12 strains had changed the left end of chromosome II, and four of 12 strains (PAL-15, PAL-22, PAL-5, PAL-1) showed clear evidence for palindrome formation, based on the presence of a novel band accompanied by a half-sized band (Fig. 5F,G).
Two of 12 strains (PAL-36, PAL-38) showed high-molecular-weight bands, which might indicate palindromes formed close to the chromosome end (Fig. 5F). Consistent with this, microarray analysis detected large duplications close to this chromosome end (Supplementary Fig. 2; data not shown).
Two of 12 strains (PAL-2, PAL-11) showed novel high-molecular-weight bands (Fig. 5F). These bands are difficult to classify, because it is not possible to design specific probes closer to this end of chromosome II, due to 9 kb of repetitive DNA (subtelomeric regions and LTRs).
The remaining four of 12 strains (PAL-52, PAL-55, PAL-3, PAL-6) showed diffuse bands (Fig. 5F,G; data not shown), characteristic of freely degradable chromosome ends, and they may form palindromes with time. To test whether this was the case, we analyzed the left arm of chromosome II in PAL-55 at different time points, between 60 and 400 d (Fig. 5H). Consistent with the diffuse band being a freely degradable chromosome end, the band shortened over time. Between 60 and 300 d, the chromosome had lost ∼8.5 kb, on average 28 bp/d, which can be explained solely by the end-replication problem. At 370 d, three subpopulations of palindrome-containing cells were visible (main palindrome bands are labeled with A, B, and C and corresponding half-sized bands with a, b, and c). Palindrome A appeared by 290 d, then increased in intensity. Interestingly, the subpopulation containing Palindrome B appeared to be overgrown with time, while the subpopulation containing Palindrome C remained constant. These data show that even in a single culture different subpopulations evolve and compete with each other and that palindromes can form at different locations.
In summary, there is evidence that palindrome formation is an important mechanism to protect the left end of chromosome II, occurring in 7/12 survivors (PAL-15, PAL-22, PAL-5, PAL-1, PAL-36, PAL-38, PAL-55). Another 3/12 showed diffuse bands (PAL-52, PAL-3, PAL-6) and, like PAL-55, will probably form palindromes with time.
Palindromes originate at inverted repeats
To better understand how palindromes form in PAL-survivors, we amplified and sequenced the junctions between palindrome arms. Palindromes are difficult to amplify by PCR, because they fold back rapidly through intrastrand base pairing between the arms. Tanaka et al. (2002) have shown that bisulfite treatment of palindromic DNA converts cytosine to uracil, disrupting intrastrand base pairing and permitting PCR amplification. We used this method to sequence a total of nine palindrome junctions on four different chromosomes, in several independent PAL-survivors.
We found that all PCR-amplified palindromes contained single copies of short wild-type inverted repeats (IRs) at the junction between palindrome arms (Fig. 6A-D). These data indicate that palindromes are formed when degradation reaches a wild-type IR. Importantly, the wild-type IR was not itself duplicated. The duplication started exactly after the IR, consistent with the mechanism we describe in the Discussion. Interestingly, we found that the spacer and a few additional base pairs have been deleted from the IR found on chromosome V in four independent strains (Fig. 6D). Similar loss of DNA at the center of palindromes has been recently observed in mouse cells (Cunningham et al. 2003). Several other palindrome junctions were intact (Fig. 6A-C). In conclusion, short inverted repeats, naturally present on chromosome arms, catalyze palindrome formation in PAL-survivors.
Metamorphosis of large palindromes
Microarray analysis of PAL-22 at 240 d and later at 317 d allowed us to monitor the stability of large palindromes over a 77-d period. We found that seven duplications (palindromes) were relatively stable (Supplementary Fig. 1). We also noted at least one new duplication, on the right arm of chromosome IX in PAL-22 at 317 d (Fig. 6E). Interestingly, part of the duplication detected on chromosome XIII at 240 d has been lost by 317 d, so that the terminal duplication evolved into what appears to be an internal duplication (Fig. 6F). Although at this stage we do not understand the mechanism underlining this change, it is probably responsible for other “internal duplications” observed in all PAL-survivors (e.g., three internal duplications in PAL-5 and PAL-38; Supplementary Fig. 2 and in other strains; data not shown).
Finally, we sometimes saw more than twofold amplification of DNA in PAL-survivors (triplications, quadruplications), as on the right arm of chromosome IX in PAL-2 at 170 d (Fig. 6G). This is most likely caused by reduplication of a palindrome, because the end of one palindrome arm is exposed to degradation/end-replication defect and also contains many short inverted repeats that could trigger a reamplification process.
Discussion
The PAL-mechanism
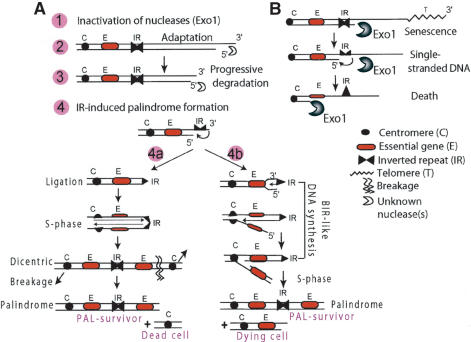
We have discovered a way to immortalize budding yeast cells in the absence of telomerase and recombination. The PAL mechanism (for palindrome-dependent mechanism) allows cells to overcome senescence and immortalize. This mechanism involves four phases (Fig. 7A): (1) Inactivation of Exo1 (or functionally equivalent nucleases). (2) Adaptation to telomere defects. Adaptation is resumption of cell cycle progression without repair of telomere defects. Inactivation of Mre11, another nuclease/checkpoint protein, significantly increases adaptation of exo1Δ strains, but is not itself sufficient to permit phase 2. (3) Early postsenescent phase, characterized by progressive degradation of chromosome ends. (4) Late postsenescent phase, characterized by palindrome formation, initiated at inverted repeats. Palindromes may further evolve to reduplicate and/or delete regions within arms.
Figure 7.
A model for telomerase- and recombination-independent immortalization of yeast cells. (A) The PAL mechanism. (B) Exo1 opposes adaptation and PAL mechanism. See Discussion for further details.
How inverted repeats catalyze palindrome formation has been previously modeled by others (Cavalier-Smith 1974; Bateman 1975; Butler et al. 1995, 1996, 2002; Qin and Cohen 2000, 2002; Lobachev et al. 2002; Tanaka et al. 2002). These models, summarized in 4a, Figure 7A, are based on the idea that once an IR becomes single-stranded, it flips back at the junction between its complementary halves and base pairs. Ligase seals the nick between the 3′-end and the 5′-end. After DNA replication, a dicentric chromosome forms, because two identical chromosomes remain joined together through the inverted repeat. Because a dicentric chromosome has two active centromeres, it can be pulled in opposite directions during mitosis and break. If dicentric chromosomes break asymmetrically between centromeres, one daughter cell will contain a palindrome and be alive, whereas the other may lose essential genes and die.
This model (4a in Fig. 7A) does not explain why the 300-kb duplication detected on chromosome XIII contained two centromeres (Fig. 3D), because every palindrome should form after breakage of a dicentric chromosome between centromeres. Therefore, we propose another model of palindrome formation that does not necessarily require breakage of dicentric chromosomes. In this model, the 3′-end of the IR serves as a primer for DNA polymerases to initiate DNA synthesis as a form of DNA repair, similar to break-induced replication (4b in Fig. 7A). The break-induced replication-like (BIR-like) process starts at the chromosome end and may stop before duplicating the entire chromosome. The result is a chromosome with a duplicated region and, perhaps, a stalled replication fork. After S phase, the result of replicating such a chromosome would be a chromosome with a duplicated region, as in PAL-survivors and another chromosome that loses the inverted repeat (4b in Fig. 7A). Our data show that Rad52 is not necessary for these BIR-like events.
We found that EXO1 strongly interferes with the PAL mechanism, because EXO1+ senescent cells do not generate survivors (Table 1). We presume this is because Exo1 generates single-stranded DNA (Fig. 7B), a potent activator of cell cycle arrest, as it does in yku70Δ mutants (Maringele and Lydall 2002) and ultimately kills cells by degrading essential genes. Presumably, palindromes cannot form in arrested cells because the passage through S phase is necessary (4a, 4b in Fig. 7A). Interestingly, deletion of MRE11 increases the rate at which exo1Δ tlc1Δ rad52Δ strains escape senescence. Mre11 plays many roles: It is a nuclease, DNA-repair protein, checkpoint protein, and telomere protection protein (D'Amours and Jackson 2002). Mre11 also makes palindromes unstable (Lobachev et al. 2002). In this case, we believe it is most likely that Mre11 is behaving primarily as a checkpoint protein, rather than a palindrome-degrading protein, since its deletion allows tlc1Δ exo1Δ cells to escape senescence rapidly, within 2 or 3 d (Maringele and Lydall 2004), whereas palindromes take longer to form. Additionally, microarray and Southern blots showed similar numbers of palindromes in cells with and without Mre11. However, we do not exclude the possibility that MRE11 or its interacting partners have been mutated in exo1Δ tlc1Δ rad52Δ survivors. Although we establish here some of the genetic requirements essential for adaptation to senescence, the mechanism of adaptation will be addressed separately.
The role of inverted repeats in natural palindrome formation
Our experiments suggest that short inverted repeats catalyze palindrome formation. Previous experiments demonstrated that IRs initiate palindromes, when situated close to experimentally induced double-strand breaks (Yasuda and Yao 1991; Butler et al. 1995; Qin and Cohen 2000; Lobachev et al. 2002; Tanaka et al. 2002). However, there was doubt whether IRs play a role in naturally occurring palindromes, since natural palindromes contained large deletions that made it difficult to interpret their origin (Okuno et al. 2004).
Our data are the first evidence that IRs are at the junction of several naturally formed palindromes (Fig. 6) and strongly suggest that the palindromes we identified were caused by IR-assisted replication, as presented in Figure 7A, and not by sister-chromatid fusion. If IRs catalyzed sister-chromatid fusions, so that a chromatid with an IR at one end would fuse to its sister (also terminating in an IR), this would most probably result in duplicated wild-type IRs at the junction. Importantly, we found that wild-type IRs were not duplicated at palindrome junctions, and this supports the argument against sister-chromatid fusion as a mechanism. However, we cannot eliminate the possibility of sister-chromatid fusions by ligation of two identical 3′-single-stranded and base-paired IRs, as shown in Supplementary Figure 3C. It is clear that the process of palindrome generation did not require Rad52-dependent recombination, because all PAL-survivors were deleted for RAD52.
Are palindromes essential for immortalization?
The fact that many chromosome ends in PAL-survivors showed no detectable duplications raised the question of whether palindrome formation was essential for immortalization of these cells. One hypothesis to explain why not all chromosomes contained palindromes is that mature PAL-survivors down-regulate degradation, by mutating genes encoding nucleases or by increasing protection against degradation. In this case, the selective pressure to maintain essential genes would certainly decrease and palindromes might take longer to form (e.g., on the left arm of chromosome II in PAL-55; Fig. 5H).
An alternative hypothesis is that there are other ways to protect chromosome ends and solve the end-replication problem, such as chromosome end-fusions, patching the ends with mitochondrial, ribosomal DNA (Yu and Gabriel 1999) or transposons. However, we have not detected such events. We believe that proteins that bind chromosome ends or specific structural modifications of chromosome ends (e.g., t-loops) are relevant for adaptation and for continued cell cycle progression without telomeres; however, they do not stop degradation.
In conclusion, in the absence of telomerase and recombination, palindrome formation is a naturally selected way to stop chromosomal degradation and rescue essential genes. We believe that while at first, palindromes were an effect of early postsenescent proliferation (phases 1-3 of the PAL mechanism; Fig. 7A), they then became an important cause of immortalization in PAL-survivors (phase 4).
The PAL mechanism and its potential relevance to cancer
The PAL mechanism described here may be relevant to cancer cells. First, palindrome formation might be responsible for maintaining chromosome ends in precancerous cells, before telomerase activation. It is known that in many cancers, telomerase activation is a late event, occurring after widespread karyotypic changes (Blasco and Hahn 2003). Second, the PAL mechanism leads to amplification of genes, and oncogene amplification is involved in tumorigenesis, whereas amplification of drug-resistance genes is a problem in cancer therapy. Third, the PAL mechanism may lead to amplification of whole chromosomes, that is, aneuploidy, the most common genetic change in cancer. Fourth, it may be responsible for chromosome maintenance in a fraction of those tumor cells that do not express telomerase (ALT cells).
According to one hypothesis, senescent cells do not transform directly into cancer cells, but they induce malign transformation of their neighbors (Krtolica et al. 2001). Here we show that senescent yeast cells deficient in nucleases immortalize and have many genetic similarities to cancer cells (deletions, gene amplifications, palindromes). Like yeast cells, senescent mammalian cells might be able to escape senescence (by mutation in EXO1, MRE11, or other nucleases with functions at telomeres), and a PAL-type mechanism might be responsible, at least in part, for their malignant transformation.
Interestingly, in PAL-survivors we have not so far observed chromosome fusions that would lead to translocations and other aspects of genetic instability found in cancer cells. We propose two hypotheses to explain why not: (1) Genes involved in chromosome fusions in cancer cells were mutated in PAL-survivors. (2) Chromosome ends in PAL-survivors, although degradable, might have special protection (proteins?) against fusions, and this protection is absent in cancer cells. Further investigation of the PAL mechanism may lead to further insights into carcinogenesis.
The PAL mechanism and genomic evolution
The recent sequencing of the human Y chromosome identified eight massive palindromes on this chromosome (Skaletsky et al. 2003). The P1 palindrome, situated adjacent to one of the telomeres of the Y chromosome, contains many genes specifically expressed in testes. Therefore, it may be that the Y chromosome, which is unable to undergo some of the recombination mechanisms available to autosomes and X chromosomes, has used a PAL-type mechanism to protect genes that are important for sex determination.
A palindrome theory of end-replication was proposed in 1974 by Cavalier-Smith (1974) and later modified by Bateman (1975). They imagined that telomeres of eukaryotic cells consisted of palindromes. However, the sequencing of telomeres and the discovery of telomerase made this theory redundant.
We suggest that palindrome formation might have been a primordial pathway for chromosome end-replication, long before telomerase ancestors became responsible for end-replication. This is because the PAL mechanism is based on the existence of short inverted repeats, which are universal. Today, PAL-like-mechanisms may replicate prokaryotic chromosomes (Qin and Cohen 2000), linear plasmids (Qin and Cohen 2000), mitochondrial DNA (Nosek et al. 1998), chloroplast DNA (Ellis and Day 1986), and parvoviruses (Cotmore and Tattersall 2003) and amplify specific genes (Yasuda and Yao 1991; Butler et al. 1995). Here we show that the PAL mechanism can also be activated on eukaryotic chromosomes, as a consequence of postsenescence growth in the absence of telomerase, and it allows PAL-survivors to immortalize. Thus, the PAL mechanism is an end-replication solution for linear chromosomes that unifies eukaryotes, prokaryotes, organelles, and viruses and has the potential to become active in every genome that has inverted repeats, theoretically in any genome.
Materials and methods
Yeast strains
All strains used in this study were in the W303 background and RAD5+. To construct strains, standard genetic procedures of transformation and tetrad analysis were followed (Adams et al. 1997). Because W303 strains contain an ade2-1 mutation, YPD (yeast extract, peptone, and dextrose) medium was routinely supplemented with adenine at 50 mg/L. yku70Δ mre11Δ exo1Δ rad52Δ strains and their controls were made by crossing DLY2041 (mre11Δ::hisG::URA3) with DLY1708 (yku70Δ::HIS3 exo1Δ::LEU2 rad52::TRP1). tlclΔ exo1Δ rad52Δ strains came from dissection of diploid DLY2151 (tlc1Δ::HIS3/+ pTLC1::URA3) (pSD120 from D. Gottschling, Fred Hutchinson Cancer Research Center, Seattle, WA), exo1Δ::LEU2/EXO1 rad52Δ::TRP1/RAD52, after loss of the pTLC1 plasmid or from crossing DLY1950 (mre11Δ::hisG::URA3 rad52Δ::TRP1) with an early passage tlc1Δ::HIS3 exo1Δ::LEU2 strain (1a/767). tlclΔ exo1Δ mre11Δ rad52Δ strains came from crossing DLY1950 (mre11Δ::hisG::URA3 rad52Δ::TRP1) with an early passage tlc1Δ::HIS3 exo1Δ::LEU2 strain (1a/767).
Yeast propagation and growth assay
Cells from a fresh germination plate were passaged every 4 d on YPD plates at 25°C by pooling colonies on a toothpick (∼107 cells) and spreading them onto fresh plates. Serial dilutions and growth rates were performed as previously described (Maringele and Lydall 2002), except that we grew cells at 25°C and inoculated populations of cells, rather than colony-purified strains. Sonication was avoided, to reduce stress on PAL-survivors.
Quantitative PCR
Taqman assays, using 5′-Fam- and 3′-Tamra-labeled probes and an ABI7700 real-time PCR machine were used to quantify the amount of DNA at different genomic loci (Supplementary List 1). To calculate the relative amount of DNA in survivors, 10-fold dilution series of wild-type genomic DNA were used to prepare standard curves. DNA was prepared by a zymolyase-based method, and DNA levels in survivors and wild type were equalized at centromeric loci as previously described (Booth et al. 2001).
Southern blots
Southern blot analyses were performed using nonradioactive, fluorescein-labeled probes and detection kits from Amersham according to the manufacturer's instructions. Alkaline Southern blots were performed as previously described (Lydall et al. 1996), at 4°C, with buffer recirculation, except that transfer of DNA was in 1 M NH4OAc to a Magna nylon membrane.
Pulsed Field Gel Electrophoresis (PFGE)
DNA plugs were prepared according to a protocol kindly provided by Liti and Louis (2003). Manufacturer-recommended conditions were used to separate chromosomes on a Bio-Rad Chef DRIII apparatus, and gels were stained with ethidium bromide to visualize chromosomes.
Microarray analysis
DNA was prepared and labeled for microarray analysis on Affymetrix S98 chips exactly as described (Winzeler et al. 2003).
Amplification and sequencing of palindrome junctions
To facilitate PCR amplification of palindrome junctions, QIAGEN yeast DNA preps were treated with sodium metabisulfite to convert cytosine to uracil (Paulin et al. 1998; Tanaka et al. 2002). This destroyed the symmetry of the palindromes and also changed the sequence of wild-type DNA. PCR primers were designed to amplify one strand of the converted DNA from PAL-survivors and wild-type DNA. Thus, forward primers comprised only A, T, and G bases and reverse primers A, T, and C bases. In each case, no-template controls and wild-type DNA amplified with palindrome-specific primers gave no significant product. PCR products were sequenced directly and also cloned into Topo-vectors, then sequenced using primers directed to the vector.
Acknowledgments
We thank S. Cohen, A. Day, J. Haber, S. Jackson, E. Louis, J. Murnane, P. Miezckowski, T. Petes, G. Saretzki, S. Taylor, V. Zakian, and T. von Zglinicki for input. We are grateful to L. Lockhart and G. Liti for advice on PFGE; to D. Gardner, A. Hayes, S. Oliver, COGEME, and the BBSRC for microarray analysis; and to H. Tanaka, M. Yao, and A. Piper for advice on meta-bisulfite treatment. The work was funded by the Wellcome Trust.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.316504.
References
- Adams A., Gottshcling, D.E., Kaiser, C.A., and Stearns, T. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Bateman A.J. 1975. Letter: Simplification of palindromic telomere theory. Nature 253: 379-380. [DOI] [PubMed] [Google Scholar]
- Blasco M.A. and Hahn, W.C. 2003. Evolving views of telomerase and cancer. Trends Cell Biol. 13: 289-294. [DOI] [PubMed] [Google Scholar]
- Booth C., Griffith, E., Brady, G., and Lydall, D. 2001. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucleic Acids Res. 29: 4414-4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler D.K., Yasuda, L.E., and Yao, M.C. 1995. An intramolecular recombination mechanism for the formation of the rRNA gene palindrome of Tetrahymena thermophila. Mol. Cell. Biol. 15: 7117-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1996. Induction of large DNA palindrome formation in yeast: Implications for gene amplification and genome stability in eukaryotes. Cell 87: 1115-1122. [DOI] [PubMed] [Google Scholar]
- Butler D.K., Gillespie, D., and Steele, B. 2002. Formation of large palindromic DNA by homologous recombination of short inverted repeat sequences in Saccharomyces cerevisiae. Genetics 161: 1065-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. 1974. Palindromic base sequences and replication of eukaryote chromosome ends. Nature 250: 467-470. [DOI] [PubMed] [Google Scholar]
- Cervantes R.B. and Lundblad, V. 2002. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 14: 351-356. [DOI] [PubMed] [Google Scholar]
- Cotmore S.F. and Tattersall, P. 2003. Resolution of parvovirus dimer junctions proceeds through a novel heterocruciform intermediate. J. Virol. 77: 6245-6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham L.A., Cote, A.G., Cam-Ozdemir, C., and Lewis, S.M. 2003. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol. 23: 8740-8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D. and Jackson, S.P. 2002. The MRE11 complex: At the crossroads of DNA repair and checkpoint signalling. Nat. Rev. Mol. Cell. Biol. 3: 317-327. [DOI] [PubMed] [Google Scholar]
- DuBois M.L., Haimberger, Z.W., McIntosh, M.W., and Gottschling, D.E. 2002. A Quantitative assay for telomere protection in Saccharomyces cerevisiae. Genetics 161: 995-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunham M.A., Neumann, A.A., Fasching, C.L., and Reddel, R.R. 2000. Telomere maintenance by recombination in human cells. Nat. Genet. 26: 447-450. [DOI] [PubMed] [Google Scholar]
- Ellis T.H.N. and Day, A. 1986. A hairpin plastid genome in barley. EMBO J. 5: 2769-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M.G., Miller, K.M., and Cooper, J.P. 2004. Indecent exposure: When telomeres become uncapped. Mol. Cell 13: 7-18. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Comeau, L., Rosenfield, S., Stansel, R.M., Bianchi, A., Moss, H., and de Lange, T. 1999. Mammalian telomeres end in a large duplex loop. Cell 97: 503-514. [DOI] [PubMed] [Google Scholar]
- Hara E., Tsurui, H., Shinozaki, A., Nakada, S., and Oda, K. 1991. Cooperative effect of antisense-Rb and antisense-p53 oligomers on the extension of life span in human diploid fibroblasts, TIG-1. Biochem. Biophys. Res. Commun. 179: 528-534. [DOI] [PubMed] [Google Scholar]
- Harrington L. 2004. Those dam-aged telomeres! Curr. Opin. Genet. Dev. 14: 22-28. [DOI] [PubMed] [Google Scholar]
- Henson J.D., Neumann, A.A., Yeager, T.R., and Reddel, R.R. 2002. Alternative lengthening of telomeres in mammalian cells. Oncogene 21: 598-610. [DOI] [PubMed] [Google Scholar]
- Krtolica A., Parrinello, S., Lockett, S., Desprez, P.Y., and Campisi, J. 2001. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: A link between cancer and aging. Proc. Natl. Acad. Sci. 98: 12072-12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.E., Bressan, D.A., Petrini, J.H.J., and Haber, J.E. 2002. Complementation between N-terminal Saccharomyces cerevisiae mre11 alleles in DNA repair and telomere length maintenance. DNA Repair 1: 27-40. [DOI] [PubMed] [Google Scholar]
- Lewis L.K., Karthikeyan, G., Westmoreland, J.W., and Resnick, M.A. 2002. Differential suppression of DNA repair deficiencies of yeast rad50, mre11 and xrs2 mutants by EXO1 and TLC1 (the RNA component of telomerase). Genetics 160: 49-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G. and Louis, E.J. 2003. NEJ1 prevents NHEJ-dependent telomere fusions in yeast without telomerase. Mol. Cell 11: 1373-1378. [DOI] [PubMed] [Google Scholar]
- Lobachev K.S., Gordenin, D.A., and Resnick, M.A. 2002. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell 108: 183-193. [DOI] [PubMed] [Google Scholar]
- Lundblad V. and Blackburn, E.H. 1993. An alternative pathway for yeast telomere maintenance rescues est1-senescence. Cell 73: 347-360. [DOI] [PubMed] [Google Scholar]
- Lundblad V. and Szostak, J.W. 1989. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell 57: 633-643. [DOI] [PubMed] [Google Scholar]
- Lydall D. 2003. Hiding at the ends of yeast chromosomes: Telomeres, nucleases and checkpoint pathways. J. Cell Sci. 116: 4057-4065. [DOI] [PubMed] [Google Scholar]
- Lydall D., Nikolsky, Y., Bishop, D.K., and Weinert, T. 1996. A meiotic recombination checkpoint controlled by mitotic checkpoint genes. Nature 383: 840-843. [DOI] [PubMed] [Google Scholar]
- Maringele L. and Lydall, D. 2002. EXO1-dependent single-stranded DNA at telomeres activates subsets of DNA damage and spindle checkpoint pathways in budding yeast yku70D mutants. Genes & Dev. 16: 1919-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2004. EXO1 plays a role in generating type I and type II survivors in budding yeast. Genetics 166: 1641-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutomi K., Yu, E.Y., Khurts, S., Ben-Porath, I., Currier, J.L., Metz, G.B., Brooks, M.W., Kaneko, S., Murakami, S., DeCaprio, J.A., et al. 2003. Telomerase maintains telomere structure in normal human cells. Cell 114: 241-253. [DOI] [PubMed] [Google Scholar]
- Moreau S., Morgan, E.A., and Symington, L.S. 2001. Overlapping functions of the Saccharomyces cerevisiae Mre11, Exo1 and Rad27 nucleases in DNA metabolism. Genetics 159: 1423-1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T.M., Cooper, J.P., and Cech, T.R. 1998. Two modes of survival of fission yeast without telomerase. Science 282: 493-496. [DOI] [PubMed] [Google Scholar]
- Nosek J., Tomaska, L., Fukuhara, H., Suyama, Y., and Kovac, L. 1998. Linear mitochondrial genomes: 30 years down the line. Trends Genet. 14: 184-188. [DOI] [PubMed] [Google Scholar]
- Okuno Y., Hahn, P.J., and Gilbert, D.M. 2004. Structure of a palindromic amplicon junction implicates microhomology-mediated end joining as a mechanism of sister chromatid fusion during gene amplification. Nucleic Acids Res. 32: 749-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov A.M. 1973. A theory of marginotomy. The incomplete copying of template margin in enzymic synthesis of polynucleotides and biological significance of the phenomenon. J. Theor. Biol. 41: 181-190. [DOI] [PubMed] [Google Scholar]
- Paques F. and Haber, J.E. 1999. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63: 349-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin R., Grigg, G.W., Davey, M.W., and Piper, A.A. 1998. Urea improves efficiency of bisulphite-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res. 26: 5009-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Z. and Cohen, S.N. 2000. Long palindromes formed in Streptomyces by nonrecombinational intra-strand annealing. Genes & Dev. 14: 1789-1796. [PMC free article] [PubMed] [Google Scholar]
- ____. 2002. Survival mechanisms for Streptomyces linear replicons after telomere damage. Mol. Microbiol. 45: 785-794. [DOI] [PubMed] [Google Scholar]
- Shay J.W. and Wright, W.E. 1989. Quantitation of the frequency of immortalization of normal human diploid fibroblasts by SV40 large T-antigen. Exp. Cell Res. 184: 109-118. [DOI] [PubMed] [Google Scholar]
- Singer M.S. and Gottschling, D.E. 1994. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science 266: 404-409. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi, T., Minx, P.J., Cordum, H.S., Hillier, L., Brown, L.G., Repping, S., Pyntikova, T., Ali, J., Bieri, T., et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825-837. [DOI] [PubMed] [Google Scholar]
- Tanaka H., Tapscott, S.J., Trask, B.J., and Yao, M.C. 2002. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. 99: 8772-8777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.T., Erdenez, N., Dudley, S., and Liskay, R.M. 2002. Characterization of nuclease-dependent functions of Exo1p in Saccharomyces cerevisiae. DNA Repair 1: 895-912. [DOI] [PubMed] [Google Scholar]
- Tsubouchi H. and Ogawa, H. 2000. Exo1 roles for repair of DNA double-strand breaks and meiotic crossing over in Saccharomyces cerevisiae. Mol. Biol. Cell 11: 2221-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley H., Pickett, H.A., Foxon, J.L., Reddel, R.R., and Royle, N.J. 2002. Molecular characterization of inter-telomere and intra-telomere mutations in human ALT cells. Nat. Genet. 30: 301-305. [DOI] [PubMed] [Google Scholar]
- Watson J.D. 1972. Origin of concatemeric T7 DNA. Nat. New Biol. 239: 197-201. [DOI] [PubMed] [Google Scholar]
- Winzeler E.A., Castillo-Davis, C.I., Oshiro, G., Liang, D., Richards, D.R., Zhou, Y., and Hartl, D.L. 2003. Genetic diversity in yeast assessed with whole-genome oligonucleotide arrays. Genetics 163: 79-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda L.F. and Yao, M.C. 1991. Short inverted repeats at a free end signal large palindromic DNA formation in Tetrahymena. Cell 67: 505-516. [DOI] [PubMed] [Google Scholar]
- Yu X. and Gabriel, A. 1999. Patching broken chromosomes with extranuclear cellular DNA. Mol. Cell 4: 873-881. [DOI] [PubMed] [Google Scholar]