Abstract
Proteolytic cascades are widely implicated in signaling between cellular compartments. In Escherichia coli, accumulation of unassembled outer membrane porins (OMPs) in the envelope leads to expression of σE-dependent genes in the cytoplasmic cellular compartment. A proteolytic cascade conveys the OMP signal by regulated proteolysis of RseA, a membrane-spanning anti-sigma factor whose cytoplasmic domain inhibits σE-dependent transcription. Upon activation by OMP C termini, the membrane localized DegS protease cleaves RseA in its periplasmic domain, the membrane-embedded protease RseP (YaeL) cleaves RseA near the inner membrane, and the released cytoplasmic RseA fragment is further degraded. Initiation of RseA degradation by activated DegS makes the system sensitive to a wide range of OMP concentrations and unresponsive to variations in the levels of DegS and RseP proteases. These features rely on the inability of RseP to cleave intact RseA. In the present report, we demonstrate that RseB, which binds to the periplasmic face of RseA, and DegS each independently inhibits RseP cleavage of intact RseA. Thus, the function of RseB, widely conserved among bacteria using the σE pathway, and the second role of DegS (in addition to RseA proteolysis initiation) is to improve the performance characteristics of this signal transduction system.
Keywords: RseB, DegS, RseP (YaeL), regulated intramembrane proteolysis, σE, stress response
Intracellular communication is an essential feature of living cells, permitting them to mount a coordinated cellular response to changing conditions. In Escherichia coli, physiological stress in the envelope compartment induced either by overproduction of outer membrane porins (OMPs) or by temperature upshift, is communicated through the inner membrane to the cytoplasmic compartment of the cell (Mecsas et al. 1993; Hiratsu et al. 1995; Raina et al. 1995; Rouviere et al. 1995). The stress signal originates from accumulation of immature OMP species and possibly from other unfolded proteins in the extracytoplasmic space (Mecsas et al. 1993; Missiakas et al. 1996; Rouviere and Gross 1996; Walsh et al. 2003) and is transduced to the cytoplasm to activate σE, the bacterial transcription initiation factor that governs the response to envelope stress. The gene encoding σE, rpoE, is essential for viability under all conditions tested, indicating that σE transcriptional activity is required during normal growth as well as under stress (De Las Penas et al. 1997a). In this work, we dissect the elements of the transmembrane signaling pathway that contribute to the sensitivity of the pathway to inducing signals and make the pathway unresponsive to noise.
The signal transduction cascade conveys envelope stress signals to the cytoplasmic compartment by altering the stability of RseA, a negative regulator of σE activity (Ades et al. 1999, 2003). RseA is a membrane-spanning anti-sigma factor that binds to σE with its cytoplasmic domain, preventing σE from interacting with RNA polymerase (De Las Penas et al. 1997b; Missiakas et al. 1997; Campbell et al. 2003). In response to stress signals generated in the envelope, a protease cascade, consisting of DegS, RseP (YaeL), and cytoplasmic proteases including ClpX, is activated to degrade RseA, thereby releasing σE from its inhibitory interaction with RseA and thus transmitting the signal through the inner membrane (Ades et al. 1999; Alba et al. 2002; Kanehara et al. 2002; Flynn et al. 2004; R. Chaba, unpubl.). Both the DegS and RseP proteases are essential for E. coli viability; their essential function is to provide E. coli with active σE via proper proteolysis of the anti-sigma factor (Alba et al. 2001, 2002; Kanehara et al. 2002).
The inner-membrane-anchored DegS protease initiates degradation by cleaving RseA in its periplasmic domain ∼30 amino acids C-terminal to the transmembrane domain of RseA (Fig. 7A, below) (Ades et al. 1999; Alba et al. 2002; Kanehara et al. 2002). Until DegS receives an activating signal, it exists in a proteolytically inactive conformation (that we call here unactivated) (Walsh et al. 2003; Wilken et al. 2004). Both in vivo and in vitro evidence is consistent with the idea that DegS is activated when OMP C termini bind to its PDZ domain (Fig. 7A,C, below) (Karshikoff et al. 1994; Cowan et al. 1995; Walsh et al. 2003; Wilken et al. 2004). The build-up of exposed OMP C termini signals that the normal OMP folding pathway is impaired, as OMP C termini are likely to be buried in the trimer interface in the native protein (Karshikoff et al. 1994; Cowan et al. 1995). RseP then cleaves the DegS-generated membrane-localized fragment of RseA to release the cytoplasmic domain of RseA (Fig. 7A, below) (Ades et al. 1999; Alba et al. 2002; Kanehara et al. 2002). This cleavage is supported by genetic, physiological, and biochemical evidence (Alba et al. 2002; Kanehara et al. 2002, 2003) and has been shown to occur within the transmembrane sequence of RseA in vivo and in vitro (Y. Akiyama, K. Kanehara, and K. Ito, pers. comm.). Two Gln-rich regions (Q1 and Q2) in the periplasmic domain of RseA inhibit RseP from cleaving the intact protein (Kanehara et al. 2003). When DegS cleaves RseA, it removes these Gln-rich regions, thereby creating an attractive substrate for RseP (Alba et al. 2002; Kanehara et al. 2002; Walsh et al. 2003). The periplasmically located PDZ domain of RseP is required for the inhibitory reaction that prevents RseP cleavage of intact RseA because RsePΔPDZ can perform that reaction (Kanehara et al. 2003; Bohn et al. 2004).
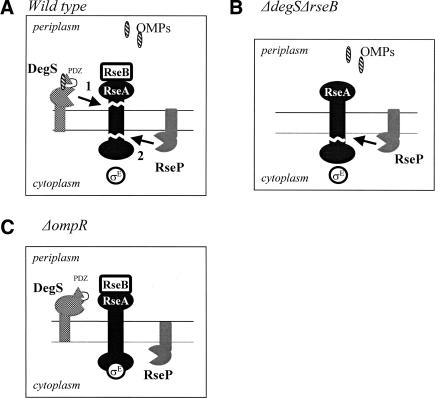
Figure 7.
Various modes of RseA degradation. (A) Wild type: DegS-dependent proteolysis of RseA. OMP monomers activate DegS by binding to the PDZ domain of DegS. DegS initiates proteolysis of RseA. RseP can cleave the RseA fragment generated by DegS cleavage. (B) ΔdegS ΔrseB: RseP can initiate cleavage of the full-length RseA in the absence of RseB and DegS, which block RseP from cleaving RseA in wild-type cells. (C) ΔompR: In the absence of the OMP signal, DegS is catalytically inactive and therefore does not initiate degradation of RseA. RseA is not degraded.
The transmembrane σE activation pathway has two principal design features. First, σE activity is very sensitive to the periplasmic OMP signal, varying greatly from cells expressing low OMPs to those expressing high OMPs (Mecsas et al. 1993). Second, σE activity is relatively unresponsive to variations in the levels of the DegS and RseP proteases themselves (Alba et al. 2002). We have investigated construction features of this pathway that contribute to these characteristics. We find that the OMP signal is sensed only by DegS. Thus, coordination of the magnitude of the σE response to the extent of the OMP inducing signal requires that RseA cleavage be initiated only by activated DegS and not by RseP. We show here that two additional players reinforce the inability of RseP to cleave intact RseA: RseB, a periplasmic protein that binds to the periplasmic domain of RseA, and DegS itself. Whereas in the absence of DegS, the transmembrane signal transduction pathway completely loses its sensitivity to the OMP signal, in the absence of RseB, sensitivity is suboptimal in that it does not respond to the full range of OMP signals and is affected by the levels of the proteases. Sequential proteolytic cascades are used for transmembrane signal transduction by several organisms (Brown et al. 2000; Alba and Gross 2004). We suggest that the regulatory and construction principles that we describe here are likely to be general design features for these signal transduction circuits.
Results
RseB inhibits DegS-independent proteolysis of RseA
Previous work indicated that RseA degradation increased 1.5- to 2.0-fold in the absence of RseB (Ades et al. 1999) (Fig. 1B). This result could be explained if RseB partially shields RseA from cleavage either by DegS and/or other proteases. If other proteases are able to degrade RseA, cells lacking both DegS and RseB should have increased σE activity relative to a strain lacking only DegS. We tested this prediction by comparing the activity of a chromosomal lacZ reporter under σE control in the two strains (see Materials and Methods). As DegS is essential, these experiments were performed in a strain that suppressed the requirement for DegS (ΔdegSsup+) (Alba et al. 2001). To our surprise, σE activity was 6.5-fold higher in a ΔdegSsup+ ΔrseB derivative than in the original ΔdegSsup+ strain (Fig. 1A), suggesting the possibility that other proteases do degrade RseA when RseB was missing.
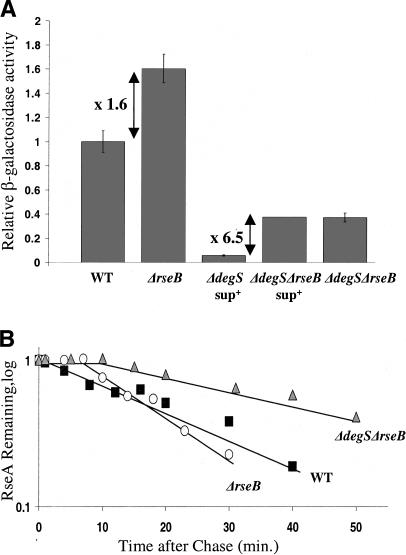
Figure 1.
σE activity and RseA degradation in ΔrseB strains. (A) Relative σE activity in the wild-type (CAG16037), ΔrseB (CAG22951), ΔdegSsup+ (CAG33315), ΔdegSsup+ ΔrseB (CAG51021), and ΔdegS ΔrseB (CAG51025) strains grown in LB at 30°C. Samples were assayed for σE activity by monitoring β-galactosidase activity produced from a single-copy [ΦλrpoH P3::lacZ] fusion. The differential rate of lacZ synthesis was quantified as described in Materials and Methods. (B) RseA stability in the wild-type (filled squares), ΔrseB (open circles), and ΔdegS ΔrseB (gray triangles) strains. Cells were grown in supplemented M9 media at 30°C to O.D.450 ∼ 0.3, pulse-labeled with [35S]methionine followed by chase of cold methionine. The stability of RseA was determined as described in Materials and Methods.
As the essential function of DegS is to provide active σE (Alba et al. 2001), we considered the possibility that DegS would no longer be essential in ΔrseB strains. We therefore transduced ΔdegS into a ΔrseB strain by selecting for the closely linked argR:Tn5 (KanR) marker. When tested by PCR, ∼50% of the KanR transductants in the ΔrseB strain had acquired the ΔdegS marker, indicating that DegS is no longer essential in a ΔrseB strain (Table 1). As previously reported, ΔdegS could not be cotransduced with this KanR marker in the wild-type strain (Table 1) (Alba et al. 2001). The σE activity of the ΔdegS ΔrseB strain was 6.5-fold higher than that of the ΔdegSsup+ strain and was equivalent to that exhibited by the ΔdegSsup+ ΔrseB strain (Fig. 1A), indicating that the elevation in σE activity was not caused by the suppressor mutation. We directly tested whether elevated σE activity resulted from DegS-independent proteolysis of RseA by measuring RseA stability using a pulse-chase immunoprecipitation protocol. Whereas RseA was completely stable in a ΔdegSsup+ strain (Ades et al. 1999), it was unstable in the ΔdegS ΔrseB strain, exhibiting a half-life several-fold slower than that of RseA in the wild-type (WT) strain (Fig. 1B). A rate of RseA degradation slower than wild type was expected because the σE activity of the ΔdegS ΔrseB strain was lower than wild type (Fig. 1A; data not shown). In conclusion, in the absence of RseB, DegS is no longer essential to cellular viability, and both σE activity and RseA degradation are significantly increased in the ΔdegS ΔrseB strain relative to the ΔdegSsup+ strain. These observations are consistent with the idea that other proteases can degrade RseA in the absence of RseB.
Table 1.
P1 transduction experiments demonstrate that DegS is dispensable in ΔrseB cells, whereas RseP is essential in all backgrounds tested
| Donor P1 strain
|
|||||
|---|---|---|---|---|---|
| ΔdegS argR::Tn5 (CAG43081)
|
rseP::kan (CAG43445)
|
||||
| Recipient | Stain | Number of KanR colonies | Number tested by colony PCR | % linkage | Number of KanR colonies |
| ΔrseB | (CAG22951) | ∼70a | 9 | 67 | 0 |
| ∼35b | 11 | 36 | n/dc | ||
| wt | (CAG16037) | ∼10 | 9 | 0 | 0 |
| ΔdegSsup+ ΔrseB | (CAG51021) | n/d | n/d | n/d | 0a |
| n/d | n/d | n/d | 0b | ||
Stand for two separate experiments. Control transductions showed that each recipient strain is transducible (data not shown).
Stand for two separate experiments. Control transductions showed that each recipient strain is transducible (data not shown).
(n/d) Not determined.
RseP is essential in the ΔrseB and ΔdegSsup+ ΔrseB strains
Our finding that RseA degradation is initiated in a DegS-independent manner in the ΔdegS ΔrseB strain raised the possibility that RseA was degraded by an alternative pathway that bypassed RseP as well as DegS. To test this, we asked whether rseP was dispensable in strains lacking RseB. The essential function of RseP is to provide active σE (Alba et al. 2002; Kanehara et al. 2002); therefore, RseP should not be essential if it is not required for RseA degradation in ΔrseB strains. Contrary to this expectation, we could not transduce rseP::kan either into a ΔrseB strain, or a ΔdegSsup+ ΔrseB strain, although control experiments indicated that these strains were fully transducible (Table 1). (We had previously reported that rseP::kan could be transduced into ΔdegSsup+ [Alba et al. 2002]; however, further investigation of that strain indicated that rseP was still present and the DegS suppressor could not itself substitute for RseP function [I. Grigorova, unpubl.].) Thus, rseP is still essential in strains lacking both DegS and RseB. We verified that rseP was required to generate active σE in this background by depleting plasmid-borne RseP under Para control carried in a ΔdegSsup+ ΔrseB ΔrseP strain. Upon transfer from inducing medium (arabinose) to noninducing (glucose) medium, the RseP protein was diluted out by cell growth and division, σE activity decreased, and growth ceased after three dilutions (data not shown). This phenotype is essentially the same as that observed after depletion of RseP from wild-type strains (Alba et al. 2002), indicating that RseP is required for RseA degradation in the ΔdegSsup+ ΔrseB strain background, just as it is in wild-type cells.
RseP can cleave full-length RseA in a ΔdegS ΔrseB strain
There are two potential alternative routes for the degradation of RseA observed in cells lacking both DegS and RseB. First, other periplasmic proteases could substitute for DegS, thereby creating an attractive substrate for RseP cleavage. Second, RseP itself might recognize and cleave full-length RseA. As RseP does not cleave intact RseA in wild-type cells, this finding would imply that RseB and/or DegS actively inhibit that cleavage. The idea that RseP initiates cleavage of full-length RseA in the ΔdegS ΔrseB cells makes several explicit predictions. First, an increased level of RseP should result in increased σE activity. Second, altering the proteolytic activity of RseP should alter the capacity of overexpressed RseP to increase σE activity. Finally, the proteolytic target of RseP should be full-length RseA rather than an RseA fragment generated by other proteases. We tested these predictions.
A moderate twofold increase in RseP level (as estimated from quantitative Westerns) caused a twofold increase in σE activity in the ΔdegS ΔrseB strain but did not change σE activity in the wild-type background (data not shown). Overproduction of RseP from a pTrc promoter gave a 60-fold increase in σE activity in the ΔdegS ΔrseB background but less than a twofold increase in wild-type cells (Fig. 2A). The increase in RseP level was roughly comparable in both cases (∼20- to 40-fold), indicating that the dramatic difference in σE activity cannot be explained by differential induction (Fig. 2B). Finally, comparable overexpression of RseP-E23D (Fig. 2B), an RseP active-site mutant, which cleaves the DegS-generated RseA fragment significantly slower than wild-type RseP (Alba et al. 2002), gave an eightfold increase in σE activity in ΔdegS ΔrseB cells, only 13% as much as overproducing wild-type RseP (Fig. 2A). These experiments show that the level and catalytic activity of RseP are directly reflected in altered σE activity, thereby providing evidence that RseP is rate-limiting for RseA degradation in the ΔdegS ΔrseB background.
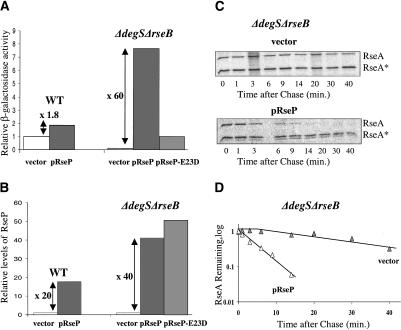
Figure 2.
Effects of RseP overexpression on σE activity and RseA stability in the wild-type and ΔdegS ΔrseB strain backgrounds. Wild-type cells containing the plasmid pRseP (CAG51091) or vector alone (CAG43604) and ΔdegS ΔrseB cells with vector (CAG51093), pRseP (CAG51092), and pRseP-E23D (CAG51122) were grown in supplemented M9 media in the presence of IPTG at 30°C. (A) Relative σE activity in the wild-type and ΔdegS ΔrseB cells with vector alone (white bar), with overexpressed RseP (gray bar), and with overexpressed RseP-E23D (light-gray bar) was assayed by monitoring β-galactosidase activity produced from a single-copy [ΦλrpoH P3::lacZ] fusion. The differential rate of lacZ synthesis was quantified as described in Materials and Methods. (B) Relative RseP levels in the wild-type and ΔdegS ΔrseB cells with vector alone (white bar), with overexpressed RseP (gray bar), and with overexpressed RseP-E23D (light-gray bar). At O.D.450 ∼ 0.3, cells were collected and TCA-precipitated. RseP levels were determined by Western blot analysis as described in Materials and Methods with polyclonal antibodies to RseP. (C,D) RseA stability in ΔrseB ΔdegS cells with overexpressed RseP from pRseP plasmid, or with vector alone. Cells were grown to O.D.450 ∼ 0.3, pulse-labeled with [35S]methionine, and chased with cold methionine. At various time points after the chase, cells were collected and TCA-precipitated. Equal amounts of [35S]methionine-labeled periplasmic domain of RseA, used as the standard (RseA*), were added to the samples, and then intracellular RseA and the standard were immunoprecipitated with antibodies against peri-RseA. The stability of RseA was determined as described in Materials and Methods. Representative data are shown in C and plotted in D.
We next tested whether the increased σE activity of ΔdegS ΔrseB cells overexpressing RseP (Fig. 2A,B) was accompanied by very rapid disappearance of full-length RseA using a pulse-chase immunoprecipitation protocol. Indeed, full-length RseA disappears much faster (∼20-fold) in cells overexpressing RseP than in the vector control (Fig. 2C,D). Thus, the substrate of RseP is full-length RseA, rather than a smaller RseA fragment, generated by some periplasmic protease. Taken together, these experiments strongly support the idea that RseP is able to cleave full-length RseA in the absence of RseB and DegS (Fig. 7B, below). The small (less than or equal to twofold) increase in σE activity in wild-type cells upon dramatic overproduction of RseP (Fig. 2A,B) could indicate the normal, very low rate of RseP cleavage of intact RseA in wild-type cells or could indicate escape from RseB and/or DegS inhibition as a consequence of massive overproduction of RseP.
RseP missing its PDZ domain is able to cleave full-length RseA (Kanehara et al. 2003; Bohn et al. 2004). We asked whether RseB inhibits cleavage of intact RseA by RsePΔPDZ, just as it inhibits cleavage by wild-type RseP. Removing RseB from a ΔdegSsup+ strain with wild-type RseP increased σE activity at least sixfold (Fig. 1A), as a consequence of impaired inhibition of RseP cleavage of intact RseA. In sharp contrast, removing RseB from the ΔdegSsup+ strain with RsePΔPDZ gave little or no increase in σE activity over the isogenic strain containing RseB (Fig. 3A, cf. lanes 2 and 3). The inability of RseB to inhibit RsePΔPDZ cleavage of intact RseA does not result from the fact that the rate of RseA degradation is already maximal in this strain: increasing the amount of RsePΔPDZ present in the cell leads to a significant increase in σE activity (Fig. 3B). We conclude that the ability of RseB to inhibit RseP cleavage of intact RseA is significantly impaired by the absence of the PDZ domain of RseP.
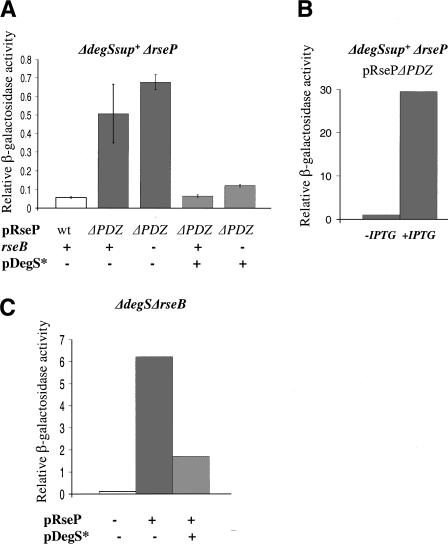
Figure 3.
The role of the RseP PDZ domain and DegS in inhibiting σE activity. (A) Effects of the presence of RseB and overexpression of a catalytically dead DegS mutant DegS-S201A (pDegS*) on σE activity in cells with RseP ΔPDZ. σE activity in various strains grown in LB at 30°C was determined as described in Figure 2. (White bar) ΔdegSsup+ ΔrseP cells carrying pRseP plasmid (CAG51143); (gray bars) ΔdegSsup+ ΔrseP (CAG51149) and ΔdegSsup+ ΔrsePΔrseB (CAG51147) cells carrying pRsePΔPDZ plasmid and pSU21 vector; (light-gray bars) ΔdegSsup+ ΔrseP (CAG51150) and ΔdegSsup+ ΔrsePΔrseB (CAG51148) cells carrying pRsePΔPDZ and pLC261 plasmids. (B) σE activity increases when RsePΔPDZ is induced. σE activity in the ΔdegSsup+ ΔrseP strain carrying pRsePΔPDZ plasmid and pSU21 vector (CAG51149) grown in LB at 30°C determined as described in Figure 2A, with (+IPTG) or without (-IPTG) overexpression of RsePΔPDZ. (C) Effect of overexpression of DegSS201A on σE activity induced by overexpression of RseP. σE activity in cells grown in supplemented M9 medium in the presence of IPTG at 30°C was determined as described in Figure 2A. (White bar) ΔdegS ΔrseB cells carrying pTrc99a and pSU21 vectors (CAG51120); (gray bar) ΔdegS ΔrseB cells carrying the pRseP plasmid and pSU21 vector (CAG51115); (light-gray bar) ΔdegS ΔrseB cells carrying pRseP and pLC261 encoding DegSS201A (CAG 51116)
DegS itself inhibits DegS-independent proteolysis of RseA by RseP
The above experiments were performed in the absence of both DegS and RseB, raising the possibility that DegS also contributes to the inability of RseP to cleave full-length RseA in wild-type cells. We tested this idea by measuring the inhibitory effect of DegS in the absence of its contribution to RseA proteolysis using a catalytically dead DegS mutant, DegS-S201A (Ades et al. 1999; Walsh et al. 2003). Simultaneous overexpression of DegS-S201A and RseP decreases σE activity three- to fourfold in a ΔdegS ΔrseB strain compared with overexpression of RseP alone, indicating that DegS inhibits RseP cleavage of full-length RseA independently from RseB (Fig. 3C). Importantly, the RseP PDZ domain is unnecessary for this inhibitory mechanism as overexpression of DegSS201A inhibited cleavage of RseA by RsePΔPDZ as well as wild-type RseP (Fig. 3A, cf. lanes 2 and 4). DegS-S201A inhibits RsePΔPDZ whether or not RseB is present (Fig. 3A, cf. lanes 3 and 5). DegS-mediated inhibition is not an artifact of using the catalytically dead mutant as we can demonstrate inhibition by wild-type DegS in a circumstance in which constitutive cleavage by RseP is likely to contribute to σE activity. In ΔrseB cells, RseA degradation will be initiated by RseP as well as by activated DegS. This may be the reason that ΔrseB cells exhibit a 1.6-fold increase in σE activity. Interestingly, overexpression of wild-type DegS decreased σE activity of ΔrseB cells to that of wild-type cells (∼1.6-fold) but did not affect σE activity in wild-type cells (data not shown), consistent with the idea that DegS can inhibit constitutive cleavage of RseA by RseP.
The alternative degradation pathway is not induced by OmpC
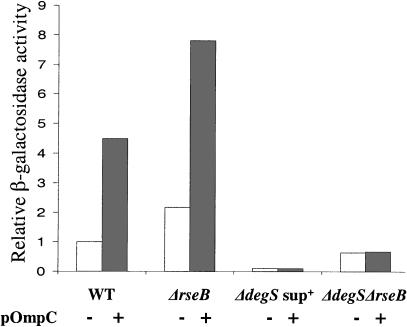
In the usual RseA degradation pathway, OMPs directly activate DegS, thereby initiating RseA proteolysis and activating σE (Ades et al. 1999; Walsh et al. 2003). We tested whether OMP overproduction also induces σE activity via the DegS-independent pathway, either by titrating RseB from RseA or by directly activating RseP cleavage of intact RseA. In ΔdegSsup+ cells, removal of RseB increased σE activity about sixfold (Fig. 1A). Therefore, if OmpC removed RseB from RseA, overexpression of OmpC would significantly increase σE activity. It was previously observed (Ades et al. 1999) and we show here that OmpC overexpression did not increase σE activity in ΔdegSsup+ cells (Fig. 4, cf. lanes 5 and 6), indicating that OMPs cannot remove RseB from RseA. We then tested whether OmpC overexpression activated RseP. OmpC overexpression did not increase σE activity in the ΔdegS ΔrseB background, where σE activity is dependent on the rate of RseP cleavage (Fig. 4, cf. lanes 7 and 8). Therefore OmpC does not activate RseP. Control experiments demonstrated appropriate induction when OmpC overexpression was performed in wild-type or ΔrseB cells (Fig. 4, lanes 1-4). In addition, overexpression of OmpC in the ΔdegSsup+ and ΔdegS ΔrseB backgrounds was confirmed by monitoring OmpC levels in the outer membranes of the cells, as described in Mecsas et al. (1993; data not shown). We conclude that DegS remains the only identified sensor of the OMP signal.
Figure 4.
Induction of σE activity by overexpression of OmpC in various strains. Relative σE activity in cells grown in LB with arabinose at 30°C, determined as described in Figure 2A. (White bar) Cells carrying vector; (gray bar) cells carrying pOmpC in the following backgrounds: wild-type (CAG43256 and CAG43216), ΔrseB (CAG51033 and CAG51032), ΔdegSsup+ (CAG43278 and CAG43629), and ΔdegS ΔrseB (CAG51083 and CAG51108).
Appropriate down-regulation of σE activity requires RseB
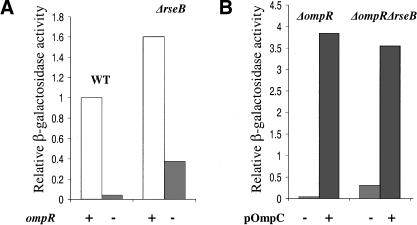
Removal of OmpR, an activator of ompC and ompF transcription, decreases OMP expression (Forst et al. 1988; Mizuno and Mizushima 1990). This results in fewer OMP intermediates to bind to the PDZ domain of DegS and activate the initiating protease, thereby down-regulating σE activity (Mecsas et al. 1993). As cells lacking RseB have lost one mechanism for inhibiting RseP cleavage of intact RseA, we suspected that RseP would make a significant contribution to initiating RseA cleavage in such cells. As RseP cleavage does not depend on the OMP signal, it should not be down-regulated in response to decreased concentration of OMP intermediates. We therefore tested whether a ΔrseB strain was partially defective in down-regulating σE activity in response to decreased OMP expression. Whereas deletion of ompR in wild-type cells resulted in an ∼20-fold drop in σE activity, only a fourfold decrease in σE activity was observed in ΔrseB cells (Fig. 5A). The ΔompR derivatives have not lost their sensitivity to the OMP signal as overexpressing OmpC from a plasmid strongly induced σE activity in both strains (Fig. 5B). These results indicate that RseB is required for appropriate down-regulation of σE activity in the absence of the OMP inducing signal, and thus is necessary for the full range of response of the system.
Figure 5.
Down-regulation of σE activity by deletion of ompR. Relative σE activity was assayed as described in Figure 1A. (A) Down-regulation of σE activity upon deletion of ompR in the wild-type and ΔrseB strains. Wild-type and ΔrseB cells with wild-type ompR (CAG16037 and CAG51050; white bar) or with deleted ompR (CAG43217 and CAG 51055; light-gray bar) were grown in LB at 30°C. (B) Induction of σE activity in the ΔompR and ΔompR ΔrseB strains by overexpression of OmpC. ΔompR and ΔompR ΔrseB cells carrying vector alone (CAG51057 and CAG51059; light-gray bar) and pOmpC plasmid (CAG51058 and CAG51060; gray bar) were grown in LB with arabinose at 30°C.
A possible second role for RseB?
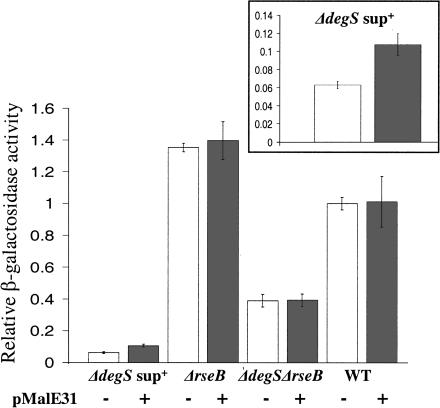
Because RseB inhibits DegS-independent proteolysis of RseA by RseP, RseB titration by unfolded proteins could be exploited as an alternative way to induce the σE pathway. RseB has been shown to colocalize with the periplasmic inclusion bodies, formed by the MalE31 unstable mutant (Betton and Hofnung 1996; Collinet et al. 2000). We therefore tested whether overexpression of MalE31 would activate σE in the ΔdegSsup+ background, where complete removal of RseB should result in a sixfold induction. Upon overproduction, σE activity increased ∼1.7-fold, indicating that MalE31 is likely to titrate a small fraction of RseB (25%-30%) from RseA (Fig. 6, cf. lanes 1 and 2, and see the inset, which presents these data in an expanded scale). We confirmed that induction resulted from removal of RseB rather than activation of DegS or RseP by showing that no detectable σE induction was observed in ΔrseB or ΔdegS ΔrseB backgrounds (Fig. 6, lanes 3-6). MalE31 overexpression did not perceptibly induce wild-type cells (Fig. 6, cf. lanes 7 and 8). As complete removal of RseB results in only a 1.6-fold increase in σE activity in wild-type cells, the limited RseB titration by MalE31 is not expected to result in significant induction in this strain background. This latter finding is in accord with a recent report that the previously observed increase in DegP synthesis upon accumulation of MalE31 in wild-type cells is caused by activation of the Cpx pathway, rather than the σE pathway (Hunke and Betton 2003).
Figure 6.
Induction of σE activity by overexpression of MalE31 in various strains. Relative σE activity of cells grown in LB with IPTG at 30°C, determined as described in Figure 2A. Shown are the average values from three experiments. (White bar) Cells carrying vector alone; (gray bar) cells carrying pMalE31 in the following strain backgrounds: wild-type (CAG43604 and CAG51070), ΔrseB (CAG43341 and CAG51072), ΔdegSsup+ (CAG43605 and CAG51074), and ΔdegS ΔrseB (CAG51080 and CAG51079). (Inset) Data for the ΔdegSsup+ background on an expanded scale.
Discussion
The signal transduction pathway linking periplasmic stress with σE activity converts the accumulation of OMP intermediates into activation of the DegS protease. DegS, RseP, and ClpX, together with as-yet-unidentified cytoplasmic proteases, then degrade RseA, the membrane-spanning anti-sigma factor that inhibits σE activity (Fig. 7) (Mecsas et al. 1993; Alba et al. 2002; Kanehara et al. 2002; Walsh et al. 2003; Flynn et al. 2004; Wilken et al. 2004). The principal goal of this work was to elucidate design features that make the σE pathway sensitive to the OMP signal and unresponsive to variations in the levels of DegS and RseP proteases themselves. We find that only DegS senses the OMP signal. Thus, effective coupling of signal to degradation necessitates that only DegS initiates the proteolytic cascade. We show here that RseB and, to a lesser extent, DegS itself inhibit RseP-mediated degradation of RseA, thereby contributing to the sensitivity and robustness of the σE pathway (Fig. 7).
Examples of protease cascades that carry out intercompartmental signaling are common from bacteria to humans (Brown et al. 2000). The σE signal transduction pathway itself is broadly present in Gram-negative bacteria. In addition, similar protease cascades have been identified in Gram-positive bacteria (Raivio and Silhavy 2001; Alba and Gross 2004). For example, it was recently shown that upon alkaline shock in Bacillus subtilis, RsiW (an RseA ortholog) is degraded from the extracytoplasmic side to a 14-kDa fragment that is further degraded by YluC, an ortholog of RseP (Schobel et al. 2004). Finally, the DegS family of proteases is widely distributed among prokaryotic and eukaryotic organisms, where they have been implicated in stress signaling pathways (Clausen et al. 2002). For example, murine HtrA2 has been suggested to sense mitochondrial stress (Li et al. 2002). Although the signals inducing these other responses may be entirely different (Brown et al. 2000), these cascades will need to use mechanisms comparable to those described here to ensure signal-dependent initiation of proteolysis. Strategies similar to those elucidated here may be used by many such cascades to block the degradation of intact regulator by proteases meant to function after the initiating event.
RseP is able to cleave intact RseA in cells lacking DegS and RseB
In wild-type cells, cleavage of RseA is overwhelmingly initiated by DegS, as demonstrated by the fact that in the absence of this protease, σE activity is very low and RseA is a stable protein (Fig. 7A) (Ades et al. 1999; Alba et al. 2002; Kanehara et al. 2002). In the present work, we demonstrate that removal of RseB in addition to DegS significantly increases σE activity and RseA cleavage. Elimination of RseB could either expose RseA to proteases that substitute for DegS in performing initial cleavage of RseA or allow RseP to cleave intact RseA more efficiently. Our data support the latter idea.
DegS cleavage of RseA is the rate-limiting proteolytic step in wild-type cells. Thus, increasing the amount or altering the activity of RseP has little effect on σE activity (Alba et al. 2002). It is probably the case because RseP cleavage of intact RseA is effectively inhibited, whereas DegS-cleaved RseA is a very attractive substrate for RseP (Kanehara et al. 2003). In cells lacking RseB and DegS, the situation is very different. We have conclusively demonstrated that σE activity correlates with the level and activity of RseP, indicating that RseP cleavage is now the rate-limiting step in generating active σE in such cells. This finding rules out the idea that some other protease cleaves in the vicinity of the DegS cleavage site to generate an attractive RseP substrate. In that case, the other protease, not RseP, would be rate-limiting for the reaction. In further validation of the idea that RseP itself performs the initial cleavage event, we show that in ΔdegS ΔrseB cells, RseP overexpression increases the rate of degradation of full-length RseA rather than an intermediate formed by preliminary cleavage by other proteases. Together these data support the idea that removing both RseB and DegS partially relieves the inhibitory mechanisms that prevent RseP cleavage of intact RseA (Fig. 7B).
Our data suggest that at least two independent mechanisms inhibit RseP from cleaving intact RseA. The first mechanism requires the presence of the PDZ domain of RseP. Both the two Gln-rich regions of RseA (Kanehara et al. 2003) and RseB inhibit cleavage of intact RseA by RseP but not by RsePΔPDZ. The most parsimonious interpretation of these data is that the RseP PDZ domain, RseB, and the Gln-rich RseA region all participate in the same inhibitory reaction. We tested whether RseB binding to the Gln-rich regions of RseA might make RseA refractory to RseP cleavage. As periplasmic RseA variants with amino acid substitutions in one or both Glnrich regions (Gln to Ala) bind RseB indistinguishably from wild-type RseA, this idea is incorrect (data not shown) (Kanehara et al. 2003). As an alternative, RseB binding to RseA might facilitate a conformational change in RseA that makes its Gln-rich regions more accessible to binding by the PDZ domain of RseP, thereby facilitating the inhibitory reaction. Of course, we cannot eliminate the possibility that RseB binds independently to the RseP PDZ domain to inhibit cleavage. We note that bacterial RseP orthologs all contain PDZ domains. Therefore, it is likely that these domains play a similar role(s) in other systems.
The second mechanism for inhibiting RseP cleavage of intact RseA involves a reaction mediated by unactivated DegS and is independent of the PDZ domain of RseP. DegS could negatively regulate RseP by forming a complex with RseA, thereby either blocking or altering the RseA recognition sites for RseP. Because overexpression of catalytically dead DegS-S201A does not inhibit σE activity in wild-type cells (data not shown), we believe it is unlikely that DegS-S201A occludes RseA, as this binding should also reduce the ability of wild-type DegS to initiate cleavage. Alternatively, unactivated DegS could form a complex with RseP, thereby reducing its ability to cleave intact RseA. Interestingly, whereas unactivated DegS-S201A inhibits RseP cleavage, DegS-S201A activated by overexpression of Omp C termini increases the rate of RseP cleavage (data not shown). Together these experiments suggest that complex interactions between DegS and RseP may promote cleavage of RseA in response to the activation signal.
RseB and DegS increase both the sensitivity and robustness of the signal transduction pathway activating σE
To provide adequate response to signals, signal transduction pathways must sense inducing signals over a wide concentration range, and to be unresponsive to variations in the concentrations of the signal transduction molecules themselves. The work reported here documents the roles of RseB and DegS in enhancing these two properties of the σE signal transduction pathway.
In wild-type cells, the signal transduction pathway activating σE is sensitive to a wide range of OMP concentrations. σE activity changes >40-fold from its low point in a ΔompR strain, which should have a very low concentration of unassembled OMPs, to its high point in a wild-type strain with overproduced OMPs (Fig. 5A,B). Cells can modulate σE activity over such a broad range because the system is designed so that the rate-limiting step in activation is sensitive to the OMP signal. This is achieved as follows. OMP binding to the PDZ domain of DegS activates a corresponding fraction of the DegS molecules (Fig. 7A). Because DegS-dependent initiation of RseA degradation is the rate-limiting step in proteolysis, the rate of RseA degradation is set to be proportional to the amount of active DegS and thus to the OMP signal. Therefore, a graded σE response over a wide range of OMP signals requires initiation of RseA proteolysis via active DegS. In the present work, we showed that RseB is required to make RseA proteolysis completely dependent on DegS. In the absence of RseB, RseP, which ordinarily degrades only DegS-cleaved RseA, is able to cleave intact RseA (Fig. 7B). As RseP is not responsive to the OMP signal, this decreases the extent to which σE activity reflects the concentration of OMP intermediates. This deficit is clearly seen in a ΔompR strain, where ΔrseB cells show a fourfold higher activity than wild-type cells. Additionally, DegS plays a second role by reinforcing sequential cleavage. It not only senses the OMP signal but also, in its unactivated form, inhibits RseP cleavage of intact RseA, thereby reinforcing the dependence on activated DegS for initiating proteolysis (Fig. 7C). Together, these two mechanisms increase the sensitivity of the system to OMP signal.
In wild-type cells, σE activity is unaffected by variations in the levels of RseP or DegS (Alba et al. 2002). This situation results from the fact that the rate of RseA degradation is determined solely by the amount of active DegS, which is defined by the extent of the OMP signal. In contrast, when cells lack RseB, changes in either RseP or DegS levels are translated into changes in σE activity. Because RseP can initiate RseA degradation constitutively in cells lacking RseB, σE activity increases with increased amounts of RseP. For this same reason, σE activity decreases with increased amounts of DegS, as accumulation of DegS inhibits RseP cleavage of intact RseA. The σE pathway might be unresponsive to the protease levels to suppress the contribution of “noise” in the system, which could arise from stochastic variation, lack of tight control, or alteration in the levels of these proteases as part of another physiological pathway. In conclusion, by turning off RseP-initiated proteolysis of RseA, RseB and DegS make the system both sensitive to the OMP signal, and insensitive (robust) to variations in the absolute levels of DegS and RseP.
RseB is a possible sensor of other periplasmic stress signals
It is rather curious that cells use a separate protein, RseB, in addition to interactions between RseA and RseP to dampen RseP activity and adjust the sensitivity and robustness of the system, and that RseB is not removed by OmpC overexpression. One possibility is that RseB plays an additional role in the signal transduction pathway. The full extent of signals inducing the σE pathway is currently unknown, as is the essential activity of this system. Based on our indication that overexpression of MalE31 could partially titrate RseB from RseA, we speculate that in addition to the OMP signal, other signals may exist that activate σE by titrating RseB from RseA and thus activate RseP-dependent proteolysis of the anti-sigma factor. In this scenario, RseB would act as a “switch” between the two modes of RseA degradation: DegS-dependent and OMP-sensitive versus RseP-dependent and OMP-insensitive. RseB would then be responsible for adjusting the sensitivity of the pathway to these different types of signals. We are currently determining whether physiologically relevant signals of this type exist.
Materials and methods
Media and antibiotics
Luria-Bertani (LB) and M9 minimal medium were prepared as described (Sambrook et al. 1989). M9 was supplemented with 0.2% glucose, 1 mM MgSO4, 2 μg/mL thiamine, and all amino acids (40 μg/mL), except methionine. When required, the media was supplemented with 30 μg/mL kanamycin (Kan), 20 μg/mL chloramphenicol (Cm), and/or 100 μg/mL ampicillin (Ap). A final concentration of 0.2% L-(+)-arabinose was used to induce the expression of rseP and ompC from the arabinose-inducible promoter Para. 0.2% glucose was used to repress expression of rseP from Para. Isopropyl-β-D-galactoside (IPTG) at a final concentration of 0.1 mM was added to induce the expression of rseP, rseP-E23D, malE31, and degS from the PTrc promoter.
Strains
Bacterial strains used in this study are described in Table 2.
Table 2.
Strains and plasmids used in this study
| Strains/plasmids | Relevant genotype | Source/reference/P1 transduction donor strains |
|---|---|---|
| Strains | ||
| MC1061 | araD Δ(ara-leu)7697 Δ(codB-lacI) galK16 galE15 mcrAO relA1 rpsL150 spoT1 mcrB9999 hsdR2 | Casadaban and Cohen 1980; E. coli Genetic Stock Center |
| CAG16037 | MC1061 ϕλ[rpoH P3::lacZ] | Mecsas et al. 1993 |
| CAG22951 | 16037 ΔrseB nadB-3140::Tn10, TetR | De Las Penas et al. 1997b |
| CAG22955 | WT ΔrseB nadB::Tn10, KanR | A. De Las Penas, unpubl. |
| CAG33315 | MC1061 ΔdegS ϕλ[rpoH P3::lacZ] | Ades et al. 1999 |
| CAG41001 | MC1061 rpoE+ with suppressor of rpoE::ΩCm | Alba et al. 2001 |
| CAG43081 | MC1061 ΔdegS arg::Tn5, KanR | Alba et al. 2001 |
| CAG43216 | 16037 pBA114, CmR | Walsh et al. 2003 |
| CAG43217 | 16037 ompR::Tn10, TetR | This work |
| CAG43256 | 16037 pBAD33, CmR | This work |
| CAG43263 | 33315 pTrc99a, ApR | This work |
| CAG43278 | 33315 pBAD33, CmR | This work |
| CAG43341 | 22951 pTrc99a, ApR, TetR | This work |
| CAG43445 | rseP::kanR pJAH184, KanR, CmR | Jennifer Leeds |
| CAG43509 | 16037 rseP::kanR pJAH184, KanR, CmR | Alba et al. 2002 |
| CAG43586 | 16037 pBA191, ApR | This work |
| CAG43604 | 16037 pBA169, ApR | Walsh et al. 2003 |
| CAG43605 | 33315 pBA169, ApR | Walsh et al. 2003 |
| CAG43629 | 33315 pBA114, CmR | This work |
| CAG51021 | 33315 ΔrseB nadB::Tn10, TetR | This work; P1 donor CAG22951 |
| CAG51025 | 22951 ΔdegS arg::Tn5, KmR, TetR | This work; P1 donor CAG43081 |
| CAG51032 | 22951 pBA114, CmR, TetR | This work |
| CAG51033 | 22951 pBAD33, CmR, TetR | This work |
| CAG51034 | 51021 pJAH184, CmR, TetR | This work |
| CAG51035 | 51021 pBAD45, CmR, TetR | This work |
| CAG51036 | 51034 resP::kanR pJAH184, KanR, CmR, TetR | This work; P1 donor CAG43445 |
| CAG51050 | 16037 ΔrseB nadB::Tn10, KmR | This work; P1 donor CAG22955 |
| CAG51055 | 51050 ompR::Tn10, TetR, KmR | This work; P1 donor CAG43217 |
| CAG51057 | 43217 pBAD33, CmR, TetR | This work |
| CAG51058 | 43217 pBA114, CmR, TetR | This work |
| CAG51059 | 51055 pBAD33, CmR, TetR, KmR | This work |
| CAG51060 | 51055 pBA114, CmR, TetR, KmR | This work |
| CAG51070 | 16037 pIG02, ApR | This work |
| CAG51072 | 51050 pIG02, ApR, KmR | This work |
| CAG51074 | 33315 pIG02, ApR | This work |
| CAG51076 | 16037 pJAH184, CmR | This work |
| CAG51077 | 51025 pJAH184, CmR, KmR, TetR | This work |
| CAG51079 | 51025 pIG20, ApR, KmR, TetR | This work |
| CAG51083 | 51025 pBAD33, CmR, KmR, TetR | This work |
| CAG51084 | 51025 pBAD45, CmR, KmR, TetR | This work |
| CAG51085 | 16037 pBAD45, CmR | This work |
| CAG51091 | 16037 pRseP, ApR | This work |
| CAG51092 | 51025 pRseP, ApR, KmR, TetR | This work |
| CAG51093 | 51025 pBA169, ApR, KmR, TetR | This work |
| CAG51098 | 22951 pBA191, ApR, TetR | This work |
| CAG51108 | 51025 pBA114, CmR, KmR, TetR | This work |
| CAG51115 | 51092 pRseP, pSU21, ApR, CmR, KmR, TetR | This work |
| CAG51116 | 51092 pRseP, pLC261, ApR, CmR, KmR, TetR | This work |
| CAG51120 | 51093 pBA169, pSU21, ApR, CmR, KmR, TetR | This work |
| CAG51122 | 51025 pRseP-E23D, ApR, KmR, TetR | This work |
| CAG51138 | 51091 rseP::kanR KanR, ApR | This work; P1 donor CAG43445 |
| CAG51139 | 16037 rseP::kanR pRsePΔPDZ, ApR, KanR | This work; P1 donor CAG43445 |
| CAG51140 | 22951 rseP::kanR pRsePΔPDZ, ApR, TetR, KanR | This work; P1 donor CAG43445 |
| CAG51143 | 33315 rseP::kanR pRseP, ApR, KanR | This work; P1 donor CAG43445 |
| CAG51144 | 51021 rseP::kanR pRsePΔPDZ, ApR, TetR, KanR | This work; P1 donor CAG43445 |
| CAG51146 | 33315 rseP::kanR pRsePΔPDZ, ApR, KanR | This work; P1 donor CAG43445 |
| CAG51147 | 51144 pSU21, CmR, ApR, TetR, KanR | This work |
| CAG51148 | 51144 pLC261, CmR, ApR, TetR, KanR | This work |
| CAG51149 | 51146 pSU21, CmR, ApR, KanR | This work |
| CAG51150 | 51146 pLC261, CmR, ApR, KanR | This work |
| Plasmids | ||
| pLC261 | degS-S201A and degS promoter in pSU21, CmR | Ades et al. 1999 |
| pJAH184 | rseP in pBAD45, CmR | Alba et al. 2002 |
| pBA169 | pTrc99A ΔNcoI, ApR | Walsh et al. 2003 |
| pBA191 | DegS-6His in pBA169, ApR | Walsh et al. 2003 |
| pBA114 | ompC in pBAD33, CmR | Alba et al. 2002 |
| pBAD33 | Vector, pACYC ori, Para, CmR | Guzman et al. 1995 |
| pSU21 | Vector, p15a ori, lac promoter, CmR | Bartolome et al. 1991 |
| pTrc99a | Vector, pBR322 ori, ApR | Amersham Pharmacia Biotech |
| pBAD45 | Vector, p15A ori, Para, CmR | Beckwith lab |
| pRseP | rseP in pTrc99a, ApR | This work |
| PRseP-E23D | rseP-E23D in pTrc99a, ApR | This work |
| pRsePΔPDZ | rsePΔPDZ in pTrc99a, ApR | This work |
| pIG02 | malE31 in pBA169, ApR | This work |
Plasmids
pRseP was constructed in two steps. The rseP gene was amplified from E. coli MG1655 DNA with primers 5′-CCGGAATTCATGCTGAGTTTTCTCTGGGATTTGGC-3′ and 5′-GCGGGATCCTCATAACCGAGAGAAATCATTGAAAAGTGCAAG-3′. The product was then digested with restriction enzymes BamHI/EcoRI and cloned at the corresponding sites of vector pTrc99a.
pRseP-E23D (glutamic acid 23 changed to aspartic acid) was constructed by quick-change mutagenesis with primers 5′yaeLig02 (5′-CTTATCACCGTGCATGATTTTGGTCATTTCTGG-3′) and 3′yaeLig02 (5′-CCAGAAATGACCAAAATCATGCACGGTGATAAG-3′) using pRseP as the template.
pRsePΔPDZ was obtained from pRseP by deleting the PDZ domain of RseP (glutamic acid E203 through glutamine Q279) by quick-change mutagenesis with primers 5′yaeLig04 (5′-GTAAAGCTCGATTTACGTCACTGGGCGTTTGGGAGTCCCTTGTCTTTGACATTAATCCCG-3′) and 3′yaeLig04 (5′-CGGGATTAATGTCAAAGACAAGGGACTCCCAAACGCCCAGTGACGTAAATCGAGCTTTAC-3′).
For the construction of pIG02, malE was PCR-amplified from E. coli MG1655 DNA with primers malE1 (5′-GGGGTACCAGGACCATAGATTATGAAAATAAAAACAGGTGCA-3′) and malE2 (5′-GGAAGCTTTTACTTGGTGATACGAGTC-3′) followed by digestion at KpnI/HindIII and ligation at the corresponding sites of pBA169. The malE double mutant (Gly 32 changed to aspartic acid and Ile 33 changed to proline) was generated by quick-change mutagenesis using primers malE3 (5′-TTCGAGAAAGATACCGATCCGAAAGTCACCGTTGAG-3′) and malE4 (5′-CTCAACGGTGACTTTCGGATCGGTATCTTTCTCGAA-3′).
β-Galactosidase assays
Overnight cultures were diluted to an O.D.600 ∼ 0.03 (in LB) or O.D.450 ∼ 0.02 (in supplemented M9 minimal medium) and grown at 30°C. In experiments with rseP, rseP-E23D, rsePΔPDZ, degS, degS-S201A, ompC, and malE31 overproduction, arabinose or IPTG was added immediately after dilution to turn on transcription from Para or PTrc promoters, respectively. σE activity was measured by monitoring β-galactosidase expression from a single-copy σE-dependent lacZ reporter gene. β-Galactosidase activity/0.5 mL cells was plotted versus O.D.600 of the culture. The observed plots showed two linear regions: the first linear region, at O.D.600 <0.25-0.3, had a smaller slope, and the second, at O.D.600 between 0.3 and 0.6, had a bigger slope (data not shown). Existence of the two phases implied that there was a growth-phase-dependent increase in σE activity at O.D.600 around 0.3. Interestingly, in the cells lacking wild-type degS, the second slope was not observed. β-Galactosidase activity/0.5 mL cells was plotted against O.D.600 (O.D.450) ranging from 0.3 to 0.6. The slope of the data, representing the differential rate of β-galactosidase synthesis and a measure of σE activity, was calculated. All assays were performed at least twice reproducibly, and data from a single experiment are shown. In some cases, where differences were small, assays were performed at least three times, and data from all samples with error are shown. Assays were performed as described (Miller 1972; Mecsas et al. 1993; Ades et al. 1999).
Determination of RseA stability by pulse-chase immunoprecipitation
Cells were grown in supplemented M9 minimal medium lacking methionine (with added antibiotics and arabinose/IPTG when necessary) at 30°C. At O.D.450 ∼ 0.3, the cells were pulse-labeled for 1 min by L-[35S]methionine, followed by a chase of 0.1% cold methionine, and samples were processed as described (Ades et al. 2003).
RseP depletion in vivo
CAG43509 and CAG51036 were grown at 30°C in LB/Cm/arabinose to an O.D.600 ∼ 0.3. The culture was poured onto a 0.45 μm Millipore filter (Millipore) in a Nalgene filtering system and washed with 10 mL of 30°C LB. The cells were resuspended in 30°C LB/Cm containing glucose to an O.D.600 ∼ 0.03. The culture was maintained in exponential growth phase by periodically diluting the culture (to O.D.600 ∼ 0.03) into a flask with fresh, prewarmed media. Aliquots were sampled for Western blots.
Western blotting (RseP, cyto-RseA)
Western blotting of RseP and RseA was performed as described (Alba et al. 2002). The Western blots were developed with the SuperSignal West Dura Extended Duration Substrate from Pierce. Epi Chemi II Darkroom (UVP Laboratory Products) was used to capture the light emitted from the blots. The band's intensity was quantified using associated software (Labworks).
Acknowledgments
We thank Tania Baker and Akiyama Yoshinori for communicating unpublished results. We also thank Eric Guisbert and Svetlana Makovets for critically reading the manuscript. This work was supported by U.S. Public Health Service Grant GM36278-18 from the NIH to C.A.G. and a Burroughs Well-come Predoctoral Fellowship awarded to I.L.G.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1238604.
References
- Ades S.E., Connolly, L.E., Alba, B.M., and Gross, C.A. 1999. The Escherichia coli σE-dependent extracytoplasmic stress response is controlled by the regulated proteolysis of an anti-sigma factor. Genes & Dev. 13: 2449-2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ades S.E., Grigorova, I.L., and Gross, C.A. 2003. Regulation of the alternative sigma factor σE during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J. Bacteriol. 185: 2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alba B.M. and Gross, C.A. 2004. Regulation of the Escherichia coli σ-dependent envelope stress response. Mol. Microbiol. 52: 613-619. [DOI] [PubMed] [Google Scholar]
- Alba B.M., Zhong, H.J., Pelayo, J.C., and Gross, C.A. 2001. degS (hhoB) is an essential Escherichia coli gene whose indispensable function is to provide σE activity. Mol. Microbiol. 40: 1323-1333. [DOI] [PubMed] [Google Scholar]
- Alba B.M., Leeds, J.A., Onufryk, C., Lu, C.Z., and Gross, C.A. 2002. DegS and YaeL participate sequentially in the cleavage of RseA to activate the σE-dependent extracytoplasmic stress response. Genes & Dev. 16: 2156-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolome B., Jubete, Y., Martinez, E., and de la Cruz, F. 1991. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene 102: 75-78. [DOI] [PubMed] [Google Scholar]
- Betton J. and Hofnung, M. 1996. Folding of a mutant maltose-binding protein of Escherichia coli which forms inclusion bodies. J. Biol. Chem. 271: 8046-8052. [DOI] [PubMed] [Google Scholar]
- Bohn C., Collier, J., and Bouloc, P. 2004. Dispensable PDZ domain of Escherichia coli YaeL essential protease. Mol. Microbiol. 52: 427-435. [DOI] [PubMed] [Google Scholar]
- Brown M.S., Ye, J., Rawson, R.B., and Goldstein, J.L. 2000. Regulated intramembrane proteolysis: A control mechanism conserved from bacteria to humans. Cell 100: 391-398. [DOI] [PubMed] [Google Scholar]
- Campbell E.A., Tupy, J.L., Gruber, T.M., Wang, S., Sharp, M.M., Gross, C.A., and Darst, S.A. 2003. Crystal structure of Escherichia coli σE with the cytoplasmic domain of its anti-sigma RseA. Mol. Cell 11: 1067-1078. [DOI] [PubMed] [Google Scholar]
- Casadaban M.J. and Cohen, S.N. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138: 179-207. [DOI] [PubMed] [Google Scholar]
- Clausen T., Southan, C., and Ehrmann, M. 2002. The HtrA family of proteases: Implications for protein composition and cell fate. Mol. Cell 10: 443-455. [DOI] [PubMed] [Google Scholar]
- Collinet B., Yuzawa, H., Chen, T., Herrera, C., and Missiakas, D. 2000. RseB binding to the periplasmic domain of RseA modulates the RseA:σE interaction in the cytoplasm and the availability of σE:RNA polymerase. J. Biol. Chem. 275: 33898-33904. [DOI] [PubMed] [Google Scholar]
- Cowan S.W., Garavito, R.M., Jansonius, J.N., Jenkins, J.A., Karlsson, R., Konig, N., Pai, E.F., Pauptit, R.A., Rizkallah, P.J., Rosenbusch, J.P., et al. 1995. The structure of OmpF porin in a tetragonal crystal form. Structure 3: 1041-1050. [DOI] [PubMed] [Google Scholar]
- De Las Penas A., Connolly, L., and Gross, C.A. 1997a. σE is an essential σ factor in Escherichia coli. J. Bacteriol. 179: 6862-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1997b. The σE-mediated response to extracytoplasmic stress in Escherichia coli is transduced by RseA and RseB, two negative regulators of σE. Mol. Microbiol. 24: 373-385. [DOI] [PubMed] [Google Scholar]
- Flynn J.M., Levchenko, I., Sauer, R.T., and Baker, T.A. 2004. Modulating substrate choice: The SspB adaptor delivers a regulator of the extracytoplasmic-stress response to the AAA+ protease ClpXP for degradation. Genes & Dev. 18: 2292-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forst S., Delgado, J., Ramakrishnan, G., and Inouye, M. 1988. Regulation of ompC and ompF expression in Escherichia coli in the absence of envZ. J. Bacteriol. 170: 5080-5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin, D., Carson, M.J., and Beckwith, J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177: 4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratsu K., Amemura, M., Nashimoto, H., Shinagawa, H., and Makino, K. 1995. The rpoE gene of Escherichia coli, which encodes σE, is essential for bacterial growth at high temperature. J. Bacteriol. 177: 2918-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunke S. and Betton, J.M. 2003. Temperature effect on inclusion body formation and stress response in the periplasm of Escherichia coli. Mol. Microbiol. 50: 1579-1589. [DOI] [PubMed] [Google Scholar]
- Kanehara K., Ito, K., and Akiyama, Y. 2002. YaeL (EcfE) activates the σE pathway of stress response through a site-2 cleavage of anti-σE, RseA. Genes & Dev. 16: 2147-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. YaeL proteolysis of RseA is controlled by the PDZ domain of YaeL and a Gln-rich region of RseA. EMBO J. 22: 6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karshikoff A., Spassov, V., Cowan, S.W., Ladenstein, R., and Schirmer, T. 1994. Electrostatic properties of two porin channels from Escherichia coli. J. Mol. Biol. 240: 372-384. [DOI] [PubMed] [Google Scholar]
- Li W., Srinivasula, S.M., Chai, J., Li, P., Wu, J.W., Zhang, Z., Alnemri, E.S., and Shi, Y. 2002. Structural insights into the pro-apoptotic function of mitochondrial serine protease HtrA2/Omi. Nat. Struct. Biol. 9: 436-441. [DOI] [PubMed] [Google Scholar]
- Mecsas J., Rouviere, P.E., Erickson, J.W., Donohue, T.J., and Gross, C.A. 1993. The activity of σE, an Escherichia coli heat-inducible σ-factor, is modulated by expression of outer membrane proteins. Genes & Dev. 7: 2618-2628. [DOI] [PubMed] [Google Scholar]
- Miller J.H. 1972. Experiments in molecular genetics, pp. 274-281. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- Missiakas D., Betton, J.M., and Raina, S. 1996. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 21: 871-884. [DOI] [PubMed] [Google Scholar]
- Missiakas D., Mayer, M.P., Lemaire, M., Georgopoulos, C., and Raina, S. 1997. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24: 355-371. [DOI] [PubMed] [Google Scholar]
- Mizuno T. and Mizushima, S. 1990. Signal transduction and gene regulation through the phosphorylation of two regulatory components: The molecular basis for the osmotic regulation of the porin genes. Mol. Microbiol. 4: 1077-1082. [DOI] [PubMed] [Google Scholar]
- Raina S., Missiakas, D., and Georgopoulos, C. 1995. The rpoE gene encoding the σ E (σ 24) heat shock σ factor of Escherichia coli. EMBO J. 14: 1043-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raivio T.L. and Silhavy, T.J. 2001. Periplasmic stress and ECF σ factors. Annu. Rev. Microbiol. 55: 591-624. [DOI] [PubMed] [Google Scholar]
- Rouviere P.E. and Gross, C.A. 1996. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes & Dev. 10: 3170-3182. [DOI] [PubMed] [Google Scholar]
- Rouviere P.E., De Las Penas, A., Mecsas, J., Lu, C.Z., Rudd, K.E., and Gross, C.A. 1995. rpoE, the gene encoding the second heat-shock σ factor, σE, in Escherichia coli. EMBO J. 14: 1032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Schobel S., Zellmeier, S., Schumann, W., and Wiegert, T. 2004. The Bacillus subtilis σW anti-sigma factor RsiW is degraded by intramembrane proteolysis through YluC. Mol. Microbiol. 52: 1091-1105. [DOI] [PubMed] [Google Scholar]
- Walsh N.P., Alba, B.M., Bose, B., Gross, C.A., and Sauer, R.T. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113: 61-71. [DOI] [PubMed] [Google Scholar]
- Wilken C., Kitzing, K., Kurzbauer, R., Ehrmann, M., and Clausen, T. 2004. Crystal structure of the DegS stress sensor: How a PDZ domain recognizes misfolded protein and activates a protease. Cell 117: 483-494. [DOI] [PubMed] [Google Scholar]