Significance
In response to a variety of stress signals, tumor suppressor p53 is activated to influence the cellular outcome. Various stresses result in different levels of p53 activation. We demonstrate here that during mouse development, uniform activation of p53 at a moderate level is well tolerated in the embryos. In contrast, mild activation of p53 in individual cells leads to a growth disadvantage and their out-competition when they are present together with nonactivated cells. These results suggest that p53 may coordinate at a multicellular level for optimal stress response and fitness of the organism.
Keywords: p53, stress response, cell competition, embryogenesis, tissue fitness
Abstract
The function of tumor suppressor p53 has been under intense investigation. Acute stresses such as DNA damage are able to trigger a high level of p53 activity, leading to cell cycle arrest or apoptosis. In contrast, the cellular response of mild p53 activity induced by low-level stress in vivo remains largely unexplored. Murine double minute (MDM)2 and MDM4 are two major negative regulators of p53. Here, we used the strategy of haploinsufficiency of Mdm2 and Mdm4 to induce mild p53 activation in vivo and found that Mdm2+/−Mdm4+/− double-heterozygous mice exhibited normal embryogenesis. However, closer examination demonstrated that the Mdm2+/−Mdm4+/− cells exhibited a growth disadvantage and were outcompeted during development in genetic mosaic embryos that contained wild-type cells. Further study indicated the out-competition phenotype was dependent on the levels of p53. These observations revealed that cells with mild p53 activation were less fit and exhibited altered fates in a heterotypic environment, resembling the cell competition phenomenon first uncovered in Drosophila. By marking unfit cells for elimination, p53 may exert its physiological role to ensure organ and animal fitness.
The protein p53, which is encoded by the Trp53 gene in mouse and by Tp53 gene in human, is regarded as a guardian of the genome (1). As a critical tumor suppressor, p53 mutations were found in about half of human tumor cases (2). Acting predominantly as a transcriptional factor, p53 is able to trigger cell cycle arrest, apoptosis, and senescence under acute stresses (3). In brief, p53 serves as a sensor to integrate the stress signals a cell encounters, and then determines the fate of the cell: live or die (4).
Given the importance of p53 in determining cells’ final fates, its activity must be tightly regulated. Years of research have led to the understanding that posttranslational regulatory mechanisms play a dominant role in regulating p53 activation (5). Among all regulators, murine double minute (MDM)2 and MDM4 are two critical negative regulators that keep p53 activity at a very low level in physiological conditions. MDM2 functions as an E3 ubiquitin ligase responsible for the degradation of p53 (6), whereas MDM4, a homolog of MDM2, appears to inhibit p53 by masking its transcriptional activation domain (7). Deletion of either Mdm2 or Mdm4 resulted in embryonic lethality in mice, which could be rescued by concomitant deletion of p53 (8, 9). Stresses, such as irradiation and aberrant activation of oncogenes, were able to relieve p53 protein from the repression of MDM2 and MDM4, leading to robust p53 activation and subsequent cell cycle arrest or apoptosis (10). In addition to acute stresses, which induce robust p53 activation, organisms are continuously exposed to low-level stresses, including oxidative stress, endoplasmic reticulum stress, inflammation, replication-induced DNA damage, and other constitutive stresses (4), which may result in the induction of varied levels of p53 activity. How an individual cell responds to low levels of p53 activity is not well understood.
Tissue fitness depends on the elimination of unfit cells (11). In contrast to the classical cell autonomous responses of growth arrest or apoptosis, a noncell autonomous mechanism called cell competition, first uncovered in Drosophila, determines whether a cell is to survive or be eliminated based on its relative “fitness” in comparison with its neighboring cells (12, 13). Therefore, otherwise viable but less-fit cells may be eliminated in a competitive environment to ensure the overall fitness of the tissue and, in turn, the organism.
Here, we used an approach of haploinsufficiency of Mdm2 and Mdm4 in mice to mimic mild p53 activation in vivo (14). Interestingly, although the Mdm2+/−Mdm4+/− double-heterozygous mice exhibited normal embryogenesis, Mdm2+/−Mdm4+/− double-heterozygous cells were outcompeted during embryogenesis in the genetically mosaic embryos, in which they were surrounded by wild-type cells. This effect was abrogated when one copy of p53 is deleted, suggesting the “unfit” phenotype of Mdm2+/−Mdm4+/− double-heterozygous cells was mediated by mildly increased p53 activity. Therefore, we found that, rather than inducing cell cycle arrest or apoptosis in a cell-autonomous fashion, mild p53 activity contributes to noncell autonomous cell competition during mammalian embryogenesis to ensure the fitness of the organism.
Results
Mdm2+/−Mdm4+/− Mice Exhibited Normal Embryogenesis.
To mimic daily stress-induced p53 activity, we used the strategy of haploinsufficiency of Mdm2 and Mdm4 to mildly activate p53. The Mdm2+/− and Mdm4+/− mice were backcrossed to C57BL/6J mice for four and five generations, respectively, before they were crossed to generate the Mdm2+/−Mdm4+/− mice. Genetic analysis showed that the double-heterozygous mice were in a nearly complete C57BL/6J strain background (SI Appendix, Tables S1 and S2). We first examined the p53 activity in cultured mouse embryonic fibroblasts (MEFs) generated from the crosses between Mdm2+/− and Mdm4+/− mice. p21, a well-known p53 target gene, was slightly up-regulated at the mRNA level in Mdm2+/− and Mdm4+/− single-heterozygous MEFs while being significantly elevated in Mdm2+/−Mdm4+/− double-heterozygous MEFs compared with that in the wild-type MEFs (SI Appendix, Fig. S1A). Consistently, the p21 protein level was significantly higher in Mdm2+/−Mdm4+/− MEFs compared with that in the single heterozygous MEFs (SI Appendix, Fig. S1B). Moreover, the p21 mRNA level in Mdm2+/−Mdm4+/− MEFs was comparable to that in wild-type MEFs treated with a low dose of either Nutlin-3A (4 μM) or cisplatin (4 μg/mL) (SI Appendix, Fig. S1 C and D). These data indicated that haploinsufficiency of Mdm2 and Mdm4 led to a mild increase of p53 activity in vitro.
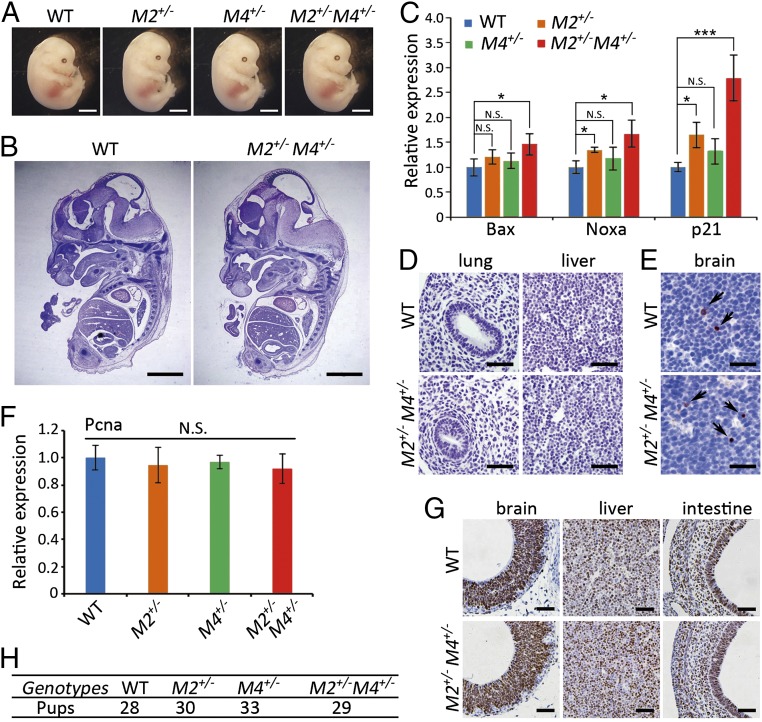
We next studied the physiology of mild p53 activity during embryogenesis. Interestingly, the Mdm2+/−Mdm4+/− embryos at embryonic day (E)12.5 exhibited normal morphology and histology comparable to that of the control embryos at the same stage (Fig. 1 A and B). Given the mild p53 activation in cultured Mdm2+/−Mdm4+/− MEFs, we examined the activity of p53 pathway in these embryos. The expression levels of p21, along with other p53 targets Bax and Noxa, were moderately elevated in the Mdm2+/−Mdm4+/− embryos (Fig. 1C). In addition, the expression of p21 and Noxa were also comparable to those in the age-matched wild-type embryos treated with a low dose of X-ray irradiation (SI Appendix, Fig. S1 E and F). Immunohistochemistry (IHC) for cleaved caspase3 showed no apoptosis in the majority of tissues examined, such as lung and liver from E12.5 Mdm2+/−Mdm4+/− embryos (Fig. 1D). Apoptotic cells were found to be only moderately increased in the brain of these embryos (Fig. 1E). In contrast, Pcna, a marker for cell proliferation, was not altered at the mRNA level (Fig. 1F). Parallel to Pcna expression, IHC for Ki67 in brain, liver, and intestine exhibited comparable proliferation between the Mdm2+/−Mdm4+/− and wild-type embryos (Fig. 1G). Moreover, Mdm2+/−Mdm4+/− mice were born at the Mendelian ratio when Mdm2+/− and Mdm4+/− mice were bred together (Fig. 1H). Collectively, Mdm2+/−Mdm4+/− mice exhibited no detectable defects in embryogenesis despite the mild increase of p53 activity.
Fig. 1.
Mice with haploinsufficiency of Mdm2 and Mdm4 exhibited normal embryogenesis. (A) Images of WT, Mdm2+/−, Mdm4+/−, and Mdm2+/−Mdm4+/− embryos (E12.5). (Scale bars, 2 mm.) (B) H&E staining of the sections from WT and Mdm2+/−Mdm4+/− embryos (E12.5). (Scale bars, 2 mm). (C) Relative mRNA expression of p53 target genes p21, Bax, and Noxa in WT, Mdm2+/−, Mdm4+/−, and Mdm2+/−Mdm4+/− embryos (E12.5; n = 3). Data are presented as mean ± SD. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001 (one-way ANOVA followed by Tukey’s test). N.S., not significant. (D) IHC staining for cleaved-caspase3 in lung and liver of WT and Mdm2+/−Mdm4+/− embryos (E12.5). (Scale bars, 100 μm.) (E) IHC staining for cleaved-caspase3 in brain of WT and Mdm2+/−Mdm4+/− embryos (E12.5). Arrows indicated cleaved caspase3-positive cells. (Scale bars, 100 μm.) (F) Relative mRNA expression of Pcna in WT, Mdm2+/−, Mdm4+/−, and Mdm2+/−Mdm4+/− embryos (E12.5; n = 3). Data are presented as mean ± SD. Statistical significance was determined by one-way ANOVA followed by Tukey’s test. N.S., not significant. (G) IHC staining for Ki67 in E12.5 Mdm2+/−Mdm4+/−, and WT embryos. (Scale bars, 100 μm.) (H) The number of progeny with different genotypes from Mdm2+/− and Mdm4+/− breeding pairs.
Mdm2+/−Mdm4+/− Cells Were Dramatically Reduced in Mosaic Pancreatic Tissue.
Normal embryogenesis of Mdm2+/−Mdm4+/− mice indicated the limited effect of mild p53 activity on cellular fate at the organismal level. To further investigate potential tissue-specific effects of mild p53 activity during embryonic development, we used the strategy of tissue-specific gene knock-out. Ipf1-cre, a Cre line expressed early in pancreatic progenitor cell stage (15), was used to generate pancreas-specific haploinsufficiency of Mdm2 and Mdm4 by crossing with mice harboring Mdm2-conditional alleles in the Mdm4 heterozygous background. Rosa26-LacZ reporter is commonly used to permanently label cells on Cre-mediated recombination. We found that ∼80% of the pancreatic cells expressed β-gal in adult Ipf1-cre; Rosa26-LacZ mice, suggesting a mosaic expression of the Cre recombinase (SI Appendix, Fig. S2 A and B). The cells with the recombined Mdm2 genotype could also be identified by β-gal expression in both Ipf1-cre; Rosa26-LacZ; Mdm2flox/+; Mdm4+/− (herein referred to as Ipf1-cre; M/M) and Ipf1-cre; Rosa26-LacZ; Mdm2flox/+ (herein referred to as Ipf1-cre; M2) mice. To our surprise, X-Gal staining at various postnatal and adult stages showed a dramatic decrease of β-gal–positive cells in Ipf1-cre; M/M mice compared with the β-gal–positive cells in Ipf1-cre; M2 mice, as well as other control mice (SI Appendix, Fig. S2C). This result suggested a selective loss of the cells with the Mdm2+/−Mdm4+/− genotype in the mosaic pancreas during embryogenesis. To quantify the relative contribution of cells with the Mdm2+/−Mdm4+/− genotype to the development of mosaic pancreas, we analyzed LacZ levels in pancreas of 6–8-wk-old Ipf1-cre; M2 and Ipf1-cre; M/M mice and found a dramatic drop in Ipf1-cre; M/M pancreas compared with the controls (SI Appendix, Fig. S2D). Consistently, quantitative PCR analysis of the recombined Mdm2flox/+ allele also revealed a dramatic decrease in the Ipf1-cre; M/M mice (SI Appendix, Fig. S2E). Intriguingly, although cells with the Mdm2+/−Mdm4+/− genotype were rarely found in the mosaic pancreas, the final size and histology of the pancreas were comparable to those of controls in 2-wk-old Ipf1-cre; M/M mice (SI Appendix, Fig. S3 A and B). Moreover, the Ipf1-cre; M/M mice displayed a similar growth rate and blood glucose level as the Ipf1-cre; M2 mice (SI Appendix, Fig. S3 C and D). Collectively, we found that cells with the recombined Mdm2+/−Mdm4+/− genotype were significantly decreased in number during development of mouse pancreas when presented in a mosaic manner, a hallmark of cell competition.
Loss of Mdm2+/−Mdm4+/− Cells Occurred in Multiple Tissues in Mosaic Embryos.
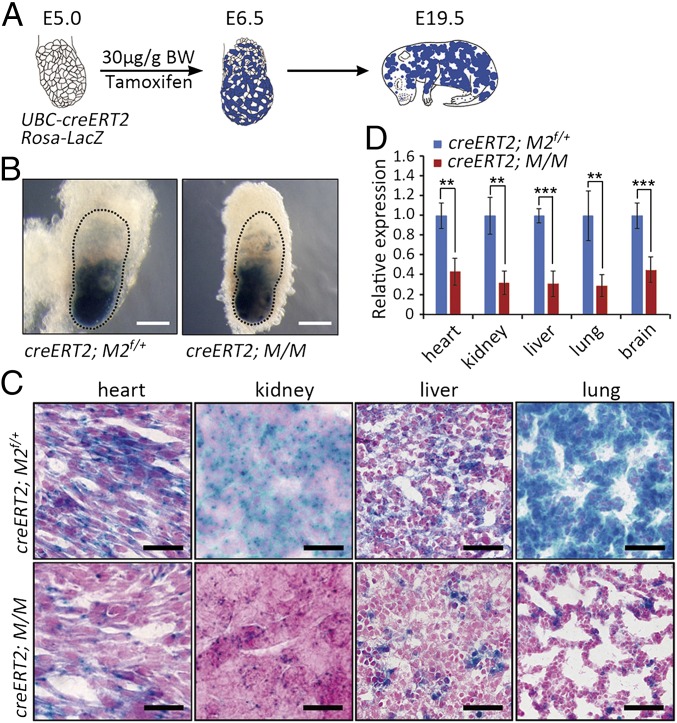
To investigate whether the loss of cells with the recombined Mdm2+/−Mdm4+/− genotype also occurred in other tissues during mouse embryonic development, UBC-creERT2 mice were used to generate mosaic embryos. UBC-creERT2 is a well characterized and broadly expressed Cre line whose activity depends on tamoxifen administration. UBC-creERT2 mice were crossed with the Rosa26-LacZ; Mdm2flox/flox; Mdm4+/− mice to generate embryos with the UBC-creERT2; Rosa26-LacZ, Mdm2flox/+; Mdm4+/− (UBC-creERT2; M/M) and control genotypes; then the pregnant mice were injected with tamoxifen (30 μg/g body weight) at E5.0. The embryos were collected 36 h later (Fig. 2A), followed by X-Gal staining to identify the cells with genetic recombination on Cre activity. Whole-mount X-Gal staining confirmed similar activities of the Cre recombinase in all embryos examined with tamoxifen induction (Fig. 2B). However, at E18.5–19.5, X-Gal staining showed a dramatic loss of β-gal–positive cells in heart, kidney, liver, and lung of tamoxifen-treated UBC-creERT2; M/M embryos compared with β-gal–positive cells in UBC-creERT2; M2 embryos (Fig. 2C), suggesting the loss of cells with the Mdm2+/−Mdm4+/− genotype. Consistently, LacZ mRNA level also decreased dramatically in multiple tissues of tamoxifen-treated UBC-creERT2; M/M embryos (Fig. 2D). Genetic analysis again showed that the UBC-creERT2; M/M mice were highly similar in genotype to the C57BL/6J background (SI Appendix, Tables S1 and S2). Therefore, cell competition that eliminates the Mdm2+/−Mdm4+/− cells occurs in multiple tissues in genetically mosaic embryos.
Fig. 2.
Universal loss of cells with the Mdm2+/−Mdm4+/− genotype in the developing embryos. (A) A schematic diagram of generating the mosaic embryos. BW, body weight. (B) Whole-mount X-Gal staining of the embryos 36 h after tamoxifen injection (E6.5). Embryonic tissues were marked by dotted lines. (Scale bars, 1 mm.) (C) X-Gal staining of heart, kidney, liver, and lung from E19.5 UBC-creERT2; Rosa-lacZ; Mdm2flox/+ (M2f/+) and UBC-creERT2; Rosa-lacZ; M/M embryos. (Scale bars, 100 μm.) (D) QRT-PCR analysis of LacZ levels from tissues of E19.5 UBC-creERT2; Rosa-lacZ; Mdm2flox/+ (M2f/+) and UBC-creERT2; Rosa-lacZ; M/M embryos (n ≥ 3). Data are presented as mean ± SD. **P ≤ 0.01; ***P ≤ 0.001 (two-tailed t test).
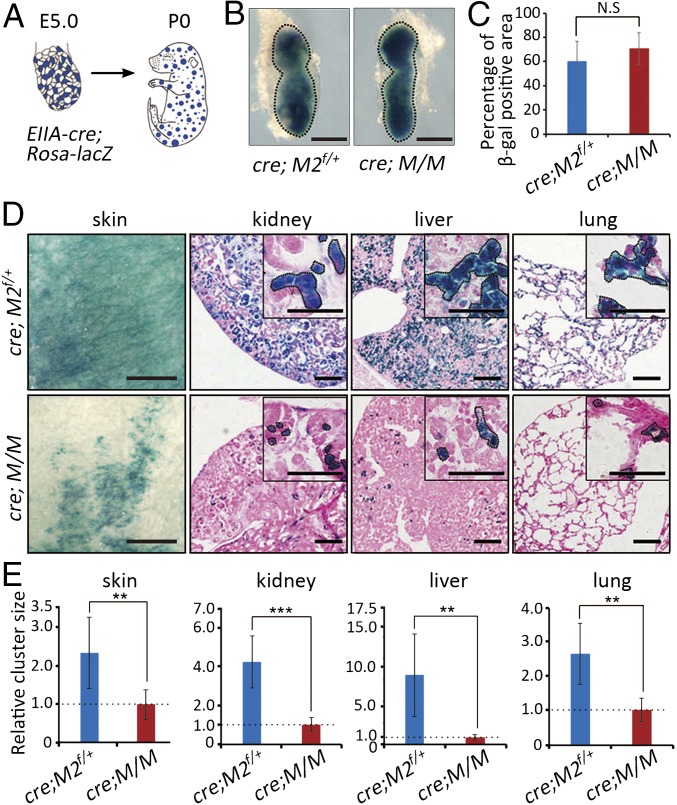
To study the fate of Mdm2+/−Mdm4+/− cells in mosaic embryos without the use of tamoxifen, EIIA-cre that expresses in very early stage embryos (16) was used, and the progeny from male EIIA-cre mice express the Cre recombinase in mosaic patterns (17). After breeding male EIIA-cre mice with the Rosa26-LacZ; Mdm2flox/flox; Mdm4+/− mice, early embryos as well as newborn pups were collected and analyzed with X-Gal staining (Fig. 3 A and B). Although Cre-mediated recombination in the E6.5 EIIA-cre; Rosa26-LacZ, Mdm2flox/+; Mdm4+/− (EIIA-cre; M/M), and control embryos was comparable (Fig. 3 B and C), cells marked by β-gal expression were found to be dramatically reduced in number in the skin, kidney, liver, and lung of the newborn EIIA-cre; M/M mice compared with those in the control mice (Fig. 3D), suggesting a progressive loss of Mdm2+/−Mdm4+/− cells in these tissues during embryonic development. Furthermore, the few residual β-gal–positive cell clusters were much smaller in size in the EIIA-cre; M/M mice (Fig. 3 D and E). Therefore, these observations confirmed the growth disadvantage of cells with Mdm2+/−Mdm4+/− genotype in the mosaic embryos.
Fig. 3.
Cells with the Mdm2+/−Mdm4+/− genotype were outcompeted in EIIA-cre-induced mosaic embryos. (A) A schematic presentation of the mosaic mice. (B) Whole-mount X-Gal staining of E6.5 EIIA-cre; Rosa-lacZ; Mdm2flox/+ (M2f/+) and EIIA-cre; Rosa-lacZ; M/M embryos. Embryonic tissues were marked by dotted lines. (Scale bars, 1 mm.) (C) Percentages of β-gal-expressing cells in E6.5 EIIA-cre; Rosa-lacZ; Mdm2flox/+ (M2f/+) and EIIA-cre; Rosa-lacZ; M/M embryos (n = 6). Data are presented as mean ± SD. Statistical significance was determined by Student’s two-tailed t test. N.S., not significant. (D) X-Gal staining of tissues from neonatal EIIA-cre; Rosa-lacZ; Mdm2flox/+ (M2f/+) and EIIA-cre; Rosa-lacZ; M/M mice. Insets indicated higher magnification. Areas marked with dotted lines indicated cell clusters. (Scale bars, 100 μm.) (E) Quantifications of the size of cell clusters in skin, kidney, liver, and lung of EIIA-cre; Rosa-lacZ; M/M and EIIA-cre; Rosa-lacZ; Mdm2flox/+ (M2f/+) mice (n = 4). Data are presented as mean ± SD. **P ≤ 0.01; ***P ≤ 0.001 (two-tailed t test).
Mdm2+/−Mdm4+/− Cells in Mosaic Embryos Displayed Reduced Proliferation.
To further study the behavior of “unfit” cells with the Mdm2+/−Mdm4+/− genotype in mosaic embryos, clonal analysis, which is commonly used to define cell competition in Drosophila (18) and to investigate the proliferative potential of single cells (19), was performed in the UBC-creERT2; M/M embryos (SI Appendix, Fig. S4A). A very low dose of tamoxifen (2 μg/g body weight) was administered to pregnant mice with E5.0 embryos that resulted from the crosses between UBC-creERT2 and Rosa26-LacZ; Mdm2flox/flox; Mdm4+/− mice. Thirty-six hours later, X-Gal staining indicated that a very small number of cells was initially labeled in the embryos (SI Appendix, Fig. S4B). At E19.5, the β-gal–positive clones formed in the brain, liver, kidney, and skin of the control embryos appeared much larger than those in the UBC-creERT2; M/M embryos (SI Appendix, Fig. S4 C and D), suggesting a growth disadvantage of cells with the Mdm2+/−Mdm4+/− genotype in this setting.
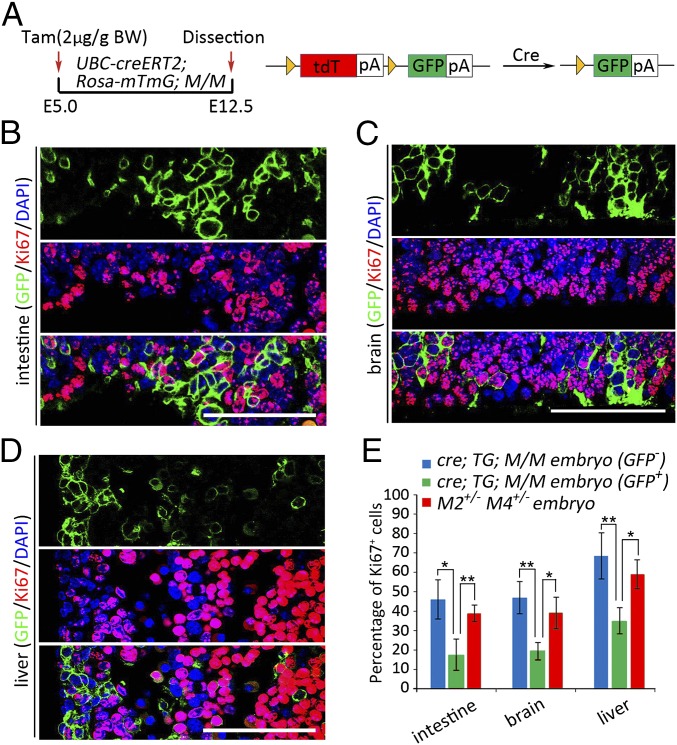
The phenotypes of cells with the Mdm2+/−Mdm4+/− genotype in different neighboring contexts were further investigated in mice with a Rosa26-mTmG background. Rosa26-mTmG mice express a double-fluorescent Cre-reporter in which the original expression of Td-tomato would be switched to that of GFP on Cre activity (20). Therefore, cells with the recombined genotypes in the mosaic embryos would be labeled with membrane-bound GFP. UBC-creERT2 mice and Rosa26-mTmG; Mdm2flox/flox; Mdm4+/− mice were crossed, and the pregnant mice were then treated with 2 μg/g body weight tamoxifen at E5.0 to generate mosaic embryos (Fig. 4A). Immunofluorescent staining for Ki67 showed the GFP+ cells representing the recombined Mdm2+/−Mdm4+/− genotype exhibited significantly less proliferation than GFP-negative cells in tissues of small intestine, brain, and liver from the E12.5 UBC-creERT2; Rosa26-mTmG; M/M mosaic embryos (Fig. 4 B–E). Moreover, immunofluorescent analysis for Ki67 in the Mdm2+/−Mdm4+/− embryos at E12.5 displayed a much higher proliferating index than the GFP+ cells in mosaic embryos (Fig. 4E; SI Appendix, Fig. S5A), indicating the context-dependent nature of the proliferative ability for Mdm2+/−Mdm4+/− cells. In contrast, immunofluorescent staining for cleaved-caspase3 did not show increased apoptotic signals in the GFP-labeled Mdm2+/−Mdm4+/− cells in the mosaic embryos (SI Appendix, Fig. S5B). Collectively, these results suggested that Mdm2+/−Mdm4+/− cells were outcompeted mainly through reduced cell proliferation during the development of mosaic embryos. They also helped explain the distinct fate of Mdm2+/−Mdm4+/− cells in different neighboring contexts.
Fig. 4.
Reduced proliferation of cells with the Mdm2+/−Mdm4+/− genotype in mosaic embryos. (A) A scheme in analyzing the mosaic embryos of the Rosa-mTmG background. BW, body weight. (B–D) Immunofluorescent staining of GFP and Ki67 in fetal intestine (B), brain (C), and liver (D) of E12.5 UBC-creERT2; Rosa-mTmG; M/M embryos. (Scale bars, 100 μm.) (E) Quantifications of cellular proliferation in E12.5 UBC-creERT2; Rosa-mTmG; M/M embryos and E12.5 Mdm2+/−Mdm4+/− embryos (n = 3). Data are presented as mean ± SD. *P ≤ 0.05; **P ≤ 0.01 (one-way ANOVA followed by Tukey’s test).
Long-term or high-dose exposure to Cre recombinase may be toxic because of the possible DNA damage caused by the Cre, which could confound our data interpretation (21). To determine whether there was increased DNA damage in the GFP-positive cells with the recombined Mdm2+/−Mdm4+/− genotype, immunofluorescent analysis for γ-H2AX was performed in the E12.5 UBC-creERT2; Rosa26-mTmG; M/M mosaic embryos. There was no obvious difference in γ-H2AX staining between the GFP+ cells and their neighboring GFP− cells in multiple tissues of the embryos (SI Appendix, Fig. S6 A and C). As a control, X-ray-irradiated E12.5 wild-type embryos exhibited elevated γ-H2AX staining as the dose of irradiation increased (SI Appendix, Fig. S6 B and C). These results suggested that under the regimen of transient and low-dose induction of the Cre activity, DNA damage was unlikely to be responsible for the out-competition of the recombined Mdm2+/−Mdm4+/−cells in the mosaic embryos.
Selective Loss of Mdm2+/−Mdm4+/− Cells in Fast Turnover Tissues of Adult Mice.
Although our data presented here demonstrated cell competition against Mdm2+/−Mdm4+/− cells during embryonic development, we wondered whether the fate of Mdm2+/−Mdm4+/− cells was also determined in a context-dependent manner in mouse adult tissues. Although p21 mRNA or protein levels were significantly up-regulated in multiple neonatal and adult tissues of the Mdm2+/−Mdm4+/− mice compared with their wild-type littermates (SI Appendix, Fig. S7 A–C), all Mdm2+/−Mdm4+/− mice were viable and could grow to adulthood, although exhibiting a moderately reduced growth rate compared with the wild-type littermates (SI Appendix, Fig. S7D). Male Mdm2+/−Mdm4+/− mice carried white belly spots and shorter, occasionally kinked tails (SI Appendix, Fig. S7E), which were reported in the Bst mouse associated with p53 activation (22, 23). Despite the elevated levels of p21, little differences in Pcna mRNA expression and Ki67 staining were observed between Mdm2+/−Mdm4+/− and wild-type mice in the fast-turnover tissues, including bone marrow, small intestine, spleen, testis, and thymus (SI Appendix, Fig. S7 F and G). Furthermore, apoptosis was not detected in these tissues, except for thymus (SI Appendix, Fig. S7H). These results suggested relatively normal homeostasis in most, if not all, the tissues in adult Mdm2+/−Mdm4+/− mice.
Next, we examined the fate of Mdm2+/−Mdm4+/− cells in the UBC-creERT2; M/M mosaic mice induced at adult stage. Eight- to 10-wk-old UBC-creERT2; Rosa26-LacZ; M/M mice were injected with tamoxifen, and the fate of cells with genetic recombination in various tissues was analyzed by LacZ expression 7 d, 3 mo, and 6 mo later (SI Appendix, Fig. S8A). LacZ levels showed little difference in liver, kidney, lung, and brain between these points (SI Appendix, Fig. S8B), suggesting that the cells with the Mdm2+/−Mdm4+/− genotype did not exhibit a disadvantage in relatively quiescent tissues of the adult mosaic mice. However, LacZ levels decreased with time in the spleen, bone marrow, and testis, suggesting a gradual loss of cells with the Mdm2+/−Mdm4+/− genotype (SI Appendix, Fig. S8B). Parallel to the mRNA levels, β-gal protein levels also exhibited gradual decreases in bone marrow and spleen but maintained its levels in liver and kidney of the tamoxifen-treated UBC-creERT2; Rosa26-LacZ; M/M mice (SI Appendix, Fig. S8C). These results suggested that in the adult mosaic mice, cells with the Mdm2+/−Mdm4+/− genotype were gradually outcompeted in fast-turnover tissues, including bone marrow, spleen, and testis. Therefore, the selective loss of cells with the Mdm2+/−Mdm4+/− genotype was found in both embryos and adult tissues in a mosaic setting.
The Out-Competition Phenotype of Mdm2+/−Mdm4+/− Cells in the Mosaic Embryos Was p53 Dependent.
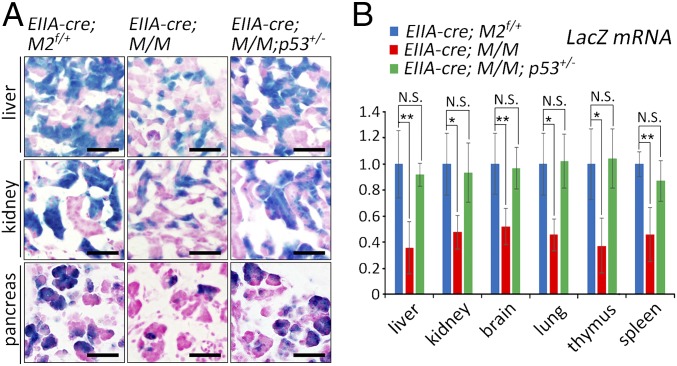
Considering the tight and dose-dependent regulation of p53 activity by MDM2 and MDM4, we examined whether the out-competition of Mdm2+/−Mdm4+/− cells is p53-dependent. The EIIA-cre; p53+/− male mice were crossed with Rosa26-LacZ; Mdm2flox/flox; Mdm4+/− female mice to generate mosaic embryos on either a p53 wild-type or p53+/− background. Newborn pups from the cross were collected and analyzed with X-Gal staining to identify the cells with the recombined Mdm2+/− genotype. Although very few β-gal–positive cells were present in multiple tissues of the neonatal EIIA-cre; M/M mice compared with EIIA-cre; M2 mice, the number of β-gal–positive cells was essentially rescued in EIIA-cre; M/M; p53+/− mice (Fig. 5A). This indicated that even the partial deficiency of p53 was able to rescue the growth disadvantage of the Mdm2+/−Mdm4+/− cells in the mosaic mice. In addition, the LacZ levels in liver, brain, thymus, kidney, lung, and spleen of EIIA-cre; M/M; p53+/− mice were not significantly different from those of EIIA-cre; M2 mice, in sharp contrast to the decreased expression in EIIA-cre; M/M mice (Fig. 5B; SI Appendix, Fig. S9A). Western blot analysis of β-gal expression in liver, kidney, and spleen largely confirmed these results, although some variations existed (SI Appendix, Fig. S9 B and C). Thus, the out-competition phenotype caused by haploinsufficiency of Mdm2 and Mdm4 in the mosaic embryos was mediated by p53 activity.
Fig. 5.
p53 dependency for the out-competition phenotype of the cells with the Mdm2+/−Mdm4+/− genotype in mosaic embryos. (A) X-Gal staining of liver, kidney, and pancreas of neonatal EIIA-cre; Rosa-lacZ; Mdm2flox/+ (M2f/+), EIIA-cre; Rosa-lacZ; M/M, and EIIA-cre; Rosa-lacZ; M/M; p53+/− mice. (Scale bars, 100 μm.) (B) Statistical analysis of LacZ mRNA levels in multiple tissues of neonatal EIIA-cre; Rosa-lacZ; Mdm2flox/+ (M2f/+) (n = 4), EIIA-cre; Rosa-lacZ; M/M (n = 5), and EIIA-cre; Rosa-lacZ; M/M; p53+/− (n = 4) mice. Data are presented as mean ± SD. *P ≤ 0.05; **P ≤ 0.01 (one-way ANOVA followed by Tukey’s test). N.S., not significant.
Discussion
Here we established mouse models with haploinsufficiency of Mdm2 and Mdm4 to study the biological effects of mild p53 activity in vivo. Our results showed that the haploinsufficiency of Mdm2 and Mdm4 in a genetic background close to C57BL/6J was able to induce mild p53 activity in both embryos and postnatal mice that displayed no significant detrimental defects. The Mdm4-deficient allele used in this study, known as Mdm4Δ2 (24, 25), was considered a true null allele (24, 26), possibly accounting for the phenotypic differences of the Mdm2+/−Mdm4+/− mice compared with a previous report (14), in which mice with a different Mdm4 allele (9) and genetic background were analyzed (14). Intriguingly, with the genetic recombination approach, we found the number of Mdm2+/−Mdm4+/− cells was dramatically decreased during embryogenesis in mosaic embryos, suggesting that cells possessing mild p53 activity displayed a different fate when neighbored by genetically distinct cells. Importantly, p53 deficiency rescued the out-competition of Mdm2+/−Mdm4+/− cells in the mosaic embryos. Similar to the observation in Drosophila that cell competition is usually observed in tissues undergoing rapid growth (27), we also found severe under-representation of the cells with the Mdm2+/−Mdm4+/− genotype in the adult tissues of bone marrow, spleen, and testis. Thus, these results suggested that a mild increase in p53 activity is able to direct cell fate through cell competition in mice (SI Appendix, Fig. S10).
Only a few studies reported cell competition phenomenon in mammals (28). Cells with a mutation in the Rpl24 gene (Rpl24Bst/+), which encodes a component of ribosome protein in mouse, were outcompeted in chimeric embryos that were generated by injecting Rpl24Bst/+ ES cells into wild-type blastocysts (22). Because the Bst (belly spot and curly tail) phenotypes in Rpl24Bst/+ mice were largely suppressed by genetic inactivation of a single p53 allele (23), it raised the possibility that p53 may be involved in the out-competition of Rpl24Bst/+ cells. Interestingly, a seminal study by Bondar and Medzhitov described the competition between p53 mutant and wild-type hematopoietic stem and progenitor cells in mice under a low level of irradiation (29). They also used Mdm2+/− hematopoietic stem and progenitor cells to genetically mimic mild p53 activation induced by a low level of irradiation and found that these cells were outcompeted by the wild-type donor in the chimeric hematopoietic system (29). More recently, it was shown that p53 may instruct an out-competition phenotype of the scribKD cells in response to cell crowding (30). However, it remains elusive whether cell competition represents a general mechanism in p53-mediated cellular responses or occurs only under specific circumstances. Our results implicated a more comprehensive and critical role for the p53 pathway in mediating the cell competition process both during embryogenesis and in adult tissues.
In cell competition, the “loser” cells are often eliminated by apoptosis (18, 31) because they are less competitive for the downstream limiting factors such as survival/growth factors (13). However, several studies also suggested that induction of apoptosis was not obligatory in cell competition (32, 33). We found that cells with the Mdm2+/−Mdm4+/− genotype in mosaic embryos displayed reduced proliferation. In contrast, the Mdm2+/−Mdm4+/− cells in a homotypic environment exhibited normal proliferation, clearly demonstrating the distinct behavior and fate of these cells, depending on cell contexts. Therefore, mild increase of p53 activity may affect the competitive status of the cells, rather than directly influence their cell cycle progression per se. p53 is known to play a significant role in influencing the metabolic and redox profiles of cells (34), which might be associated with the cells’ competitive status. In addition, p53 has been shown to be able to antagonize c-Myc (35), whose level is critical in determining cell fitness during cell competition in both Drosophila and mice (18, 36). Thus, it is possible that the relative levels and activities of p53 and c-Myc play instructive roles in the competition of cells in a heterotypic environment.
Various types and degrees of stress and damage in individual cells may result in the accumulation of viable but suboptimal cell populations in the organism. Interestingly, heterogeneous cell populations were found at high frequency even in early human embryos (37). In responses to stress signals that differ in strength and type, p53 could mediate cell-autonomous responses such as apoptosis or cell cycle arrest to directly affect a group of cells, or alternatively, involve cell competition to compare the fitness of cells and to selectively affect individual suboptimal cells. The delicate control in cell fate choices may also help us discern either a general stress response that the whole tissue suffers or the varied responses of individual cells within the tissue. Thus, as a master regulator of stress responses, p53 may play its role by coordinating at a multicellular level for the optimal responses and health of the tissues and organisms. Indeed, p53 is involved in reducing the risk for developmental abnormalities as p53 null mice exhibited increased developmental abnormalities, especially in the presence of teratogens (38). With the emerging roles of cell competition in tissue homeostasis, aging, and cancer, this study suggested a close link between stress response and cell competition and underscored the delicate control of cell fate by p53.
Materials and Methods
Mice.
Mice were bred and maintained under specific pathogen-free conditions. All animal experiments were approved by the Institutional Animal Care and Use Committee of the Model Animal Research Center, Nanjing University, China. The Mdm2flox/flox (39) and Mdm4+/− (25) mice were obtained from G. Lozano from MD Anderson Cancer Center. These mice were backcrossed to C57BL/6J background. Rosa26-LacZ, Rosa26-mTmG, Ipf1-cre, EIIA-cre, UBC-creERT2, and p53+/− mice were from Jackson laboratories and maintained on a C57BL/6J background.
X-Gal Staining.
Tissue and embryo fixation, processing, and staining with X-Gal were performed essentially as previously described (40).
Statistical Analysis.
All experiments were carried out with multiple biological replicates. Assays involving cultured cells were performed with at least three independent replicates, and in vivo data were generated from at least three mice or embryos per genotype unless otherwise indicated. In some instances, representative images or results were shown. Statistical significance for comparison of two groups was determined by Student’s two-tailed t test. The multiple groups’ statistical significance was determined by one-way ANOVA followed by Tukey’s test.
Supplementary Material
Acknowledgments
We thank G. Lozano from the University of Texas MD Anderson Cancer Center for the Mdm2- and Mdm4-deficient mice. This work was supported by Natural Science Foundation of China Grants 31171305 and 31571406 and National High-Tec R&D program of China (863 program) Grant 2014AA021606 (to G.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617414114/-/DCSupplemental.
References
- 1.Lane DP. Cancer. p53, guardian of the genome. Nature. 1992;358(6381):15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 2.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253(5015):49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 3.Vousden KH, Lu X. Live or let die: The cell’s response to p53. Nat Rev Cancer. 2002;2(8):594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 4.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8(4):275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 5.Kruse J-P, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marine J-C, Lozano G. Mdm2-mediated ubiquitylation: p53 and beyond. Cell Death Differ. 2010;17(1):93–102. doi: 10.1038/cdd.2009.68. [DOI] [PubMed] [Google Scholar]
- 7.Marine J-C, et al. Keeping p53 in check: Essential and synergistic functions of Mdm2 and Mdm4. Cell Death Differ. 2006;13(6):927–934. doi: 10.1038/sj.cdd.4401912. [DOI] [PubMed] [Google Scholar]
- 8.Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378(6553):203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 9.Parant J, et al. Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat Genet. 2001;29(1):92–95. doi: 10.1038/ng714. [DOI] [PubMed] [Google Scholar]
- 10.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408(6810):307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 11.Merino MM, et al. Elimination of unfit cells maintains tissue health and prolongs lifespan. Cell. 2015;160(3):461–476. doi: 10.1016/j.cell.2014.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Beco S, Ziosi M, Johnston LA. New frontiers in cell competition. Dev Dyn. 2012;241(5):831–841. doi: 10.1002/dvdy.23783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levayer R, Moreno E. Mechanisms of cell competition: Themes and variations. J Cell Biol. 2013;200(6):689–698. doi: 10.1083/jcb.201301051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terzian T, et al. Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol Cell Biol. 2007;27(15):5479–5485. doi: 10.1128/MCB.00555-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hingorani SR, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4(6):437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 16.Lakso M, et al. Efficient in vivo manipulation of mouse genomic sequences at the zygote stage. Proc Natl Acad Sci USA. 1996;93(12):5860–5865. doi: 10.1073/pnas.93.12.5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heffner CS, et al. Supporting conditional mouse mutagenesis with a comprehensive cre characterization resource. Nat Commun. 2012;3:1218. doi: 10.1038/ncomms2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno E, Basler K. dMyc transforms cells into super-competitors. Cell. 2004;117(1):117–129. doi: 10.1016/s0092-8674(04)00262-4. [DOI] [PubMed] [Google Scholar]
- 19.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488(7412):527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 21.Loonstra A, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98(16):9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliver ER, Saunders TL, Tarlé SA, Glaser T. Ribosomal protein L24 defect in belly spot and tail (Bst), a mouse Minute. Development. 2004;131(16):3907–3920. doi: 10.1242/dev.01268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barkić M, et al. The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol Cell Biol. 2009;29(10):2489–2504. doi: 10.1128/MCB.01588-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tashakori M, Zhang Y, Xiong S, You MJ, Lozano G. p53 Activity Dominates That of p73 upon Mdm4 Loss in Development and Tumorigenesis. Mol Cancer Res. 2016;14(1):56–65. doi: 10.1158/1541-7786.MCR-15-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grier JD, Xiong S, Elizondo-Fraire AC, Parant JM, Lozano G. Tissue-specific differences of p53 inhibition by Mdm2 and Mdm4. Mol Cell Biol. 2006;26(1):192–198. doi: 10.1128/MCB.26.1.192-198.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Migliorini D, et al. Mdm4 (Mdmx) regulates p53-induced growth arrest and neuronal cell death during early embryonic mouse development. Mol Cell Biol. 2002;22(15):5527–5538. doi: 10.1128/MCB.22.15.5527-5538.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de la Cova C, Abril M, Bellosta P, Gallant P, Johnston LA. Drosophila myc regulates organ size by inducing cell competition. Cell. 2004;117(1):107–116. doi: 10.1016/s0092-8674(04)00214-4. [DOI] [PubMed] [Google Scholar]
- 28.Clavería C, Torres M. Cell Competition: Mechanisms and Physiological Roles. Annu Rev Cell Dev Biol. 2016;32(1):411–439. doi: 10.1146/annurev-cellbio-111315-125142. [DOI] [PubMed] [Google Scholar]
- 29.Bondar T, Medzhitov R. p53-mediated hematopoietic stem and progenitor cell competition. Cell Stem Cell. 2010;6(4):309–322. doi: 10.1016/j.stem.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wagstaff L, et al. Mechanical cell competition kills cells via induction of lethal p53 levels. Nat Commun. 2016;7:11373. doi: 10.1038/ncomms11373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moreno E, Basler K, Morata G. Cells compete for decapentaplegic survival factor to prevent apoptosis in Drosophila wing development. Nature. 2002;416(6882):755–759. doi: 10.1038/416755a. [DOI] [PubMed] [Google Scholar]
- 32.Jin Z, et al. Differentiation-defective stem cells outcompete normal stem cells for niche occupancy in the Drosophila ovary. Cell Stem Cell. 2008;2(1):39–49. doi: 10.1016/j.stem.2007.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hogan C, et al. Characterization of the interface between normal and transformed epithelial cells. Nat Cell Biol. 2009;11(4):460–467. doi: 10.1038/ncb1853. [DOI] [PubMed] [Google Scholar]
- 34.Kruiswijk F, Labuschagne CF, Vousden KH. p53 in survival, death and metabolic health: A lifeguard with a licence to kill. Nat Rev Mol Cell Biol. 2015;16(7):393–405. doi: 10.1038/nrm4007. [DOI] [PubMed] [Google Scholar]
- 35.Sachdeva M, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106(9):3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clavería C, Giovinazzo G, Sierra R, Torres M. Myc-driven endogenous cell competition in the early mammalian embryo. Nature. 2013;500(7460):39–44. doi: 10.1038/nature12389. [DOI] [PubMed] [Google Scholar]
- 37.Vanneste E, et al. Chromosome instability is common in human cleavage-stage embryos. Nat Med. 2009;15(5):577–583. doi: 10.1038/nm.1924. [DOI] [PubMed] [Google Scholar]
- 38.Nicol CJ, Harrison ML, Laposa RR, Gimelshtein IL, Wells PG. A teratologic suppressor role for p53 in benzo[a]pyrene-treated transgenic p53-deficient mice. Nat Genet. 1995;10(2):181–187. doi: 10.1038/ng0695-181. [DOI] [PubMed] [Google Scholar]
- 39.Grier JD, Yan W, Lozano G. Conditional allele of mdm2 which encodes a p53 inhibitor. Genesis. 2002;32(2):145–147. doi: 10.1002/gene.10066. [DOI] [PubMed] [Google Scholar]
- 40.Chen L, et al. BAC transgenic mice provide evidence that p53 expression is highly regulated in vivo. Cell Death Dis. 2015;6:e1878. doi: 10.1038/cddis.2015.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.