Significance
The evolution of the brain and of posterior teeth seem to follow parallel trends in hominins. Larger brain size is associated with reduced premolars and molars, but this association is not observed in all hominin species. We have evaluated this association in a quantitative way by measuring lineage-specific rates of dental and cerebral evolution in the different branches of the hominin evolutionary tree. Our results show that different species evolved at different rates and that brain evolution in early Homo was faster than dental evolution. This result points to different ecological and behavioral factors influencing the evolution of hominin teeth and brains.
Keywords: endocast, postcanine teeth, evolutionary rates, selection, paleoanthropology
Abstract
The large brain and small postcanine teeth of modern humans are among our most distinctive features, and trends in their evolution are well studied within the hominin clade. Classic accounts hypothesize that larger brains and smaller teeth coevolved because behavioral changes associated with increased brain size allowed a subsequent dental reduction. However, recent studies have found mismatches between trends in brain enlargement and posterior tooth size reduction in some hominin species. We use a multiple-variance Brownian motion approach in association with evolutionary simulations to measure the tempo and mode of the evolution of endocranial and dental size and shape within the hominin clade. We show that hominin postcanine teeth have evolved at a relatively consistent neutral rate, whereas brain size evolved at comparatively more heterogeneous rates that cannot be explained by a neutral model, with rapid pulses in the branches leading to later Homo species. Brain reorganization shows evidence of elevated rates only much later in hominin evolution, suggesting that fast-evolving traits such as the acquisition of a globular shape may be the result of direct or indirect selection for functional or structural traits typical of modern humans.
In comparison with other hominins, modern humans are characterized by their large brain and small posterior teeth. These traits are among our most distinctive features, and trends in their evolution are well studied because of the phylogenetic and functional implications of variation in dental and cerebral anatomy (1–3). Brain expansion and postcanine reduction appear to follow parallel trends during hominin evolution, and classic views consider that an increase in brain size was linked to more complex behavior that included the manufacture and use of stone tools, which allowed a subsequent dental reduction. A shift toward a higher-quality diet during the evolution of early Homo also has been related to brain size increase and posterior tooth reduction (4, 5). However, it has been suggested recently that brain expansion in early Homo, as inferred from endocranial volume, substantially preceded dental reduction (6). It also has been noted that early in the Neanderthal lineage strong dental reduction preceded the additional brain expansion seen in the later “classic” Neanderthals (7). The suggestion that stone tool use and manufacture substantially predated the increase in brain size observed in early Homo (8) adds further complexity to this scenario.
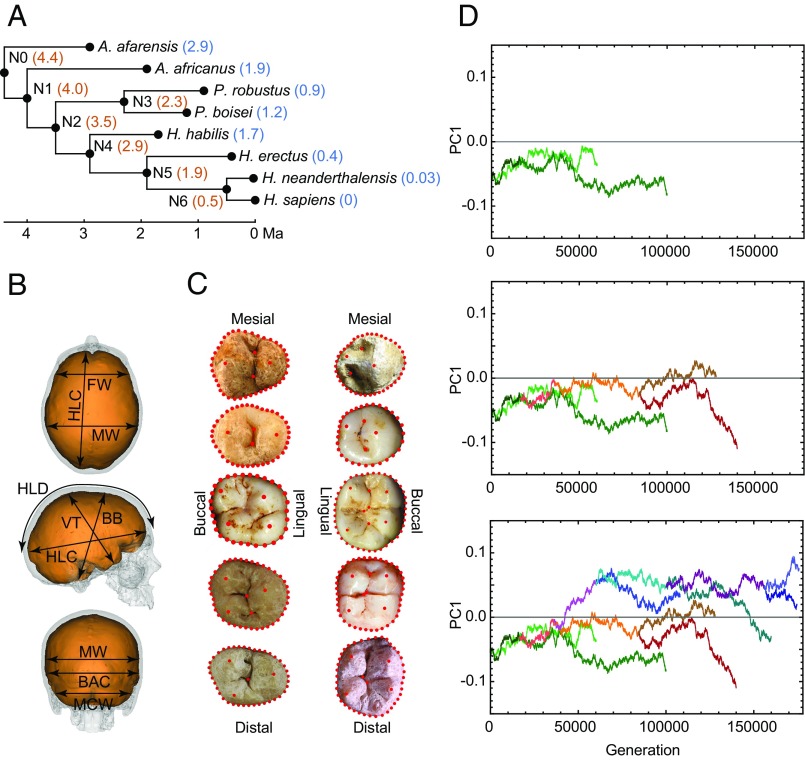
Recent developments in ancestral state reconstruction (9, 10) allow lineage-specific patterns of brain expansion and dental reduction to be quantified and compared. Unlike traditional approaches to ancestral state reconstruction that assume a neutral evolutionary scenario, which is likely unrealistic in most cases, we used a variable rate approach that estimates differences in evolutionary rates across different branches of a given phylogeny. We applied this approach to quantitative data on endocranial and postcanine dental size and shape to develop a comprehensive scenario of trends in endocranial and dental evolution across the hominin clade (Fig. 1). Our assessment used a framework phylogeny based on widely agreed evolutionary relationships and on the currently estimated first and last appearance dates for eight of the most broadly accepted hominin species (Fig. 1 and Table S1) (11). Amounts of change along each branch of the hominin phylogenetic tree estimated through this variable-rate approach were compared with the amount of change observed in evolutionary simulations that used a constant-variance Brownian motion (BM) model (12) in which traits evolve neutrally and at a constant rate without directional trends in any particular branch of the hominin phylogeny (Materials and Methods).
Fig. 1.
Methodological setup of the study. (A) The hominin phylogeny used in our analyses indicating the dates used for terminal species (blue) and nodes (orange). (B) Linear metrics used in the study of endocranial variation. FW, frontal width at Broca's cap; HLC, hemispheric length chord; MW, maximum endocranial width; HLD, hemispheric length dorsal arch; BB, basion–bregma distance; VT, vertex-lowest temporal distance; BAC, biasterionic chord; MCW, maximum cerebellar width. (C) Landmark and semi-landmark datasets used in the study of postcanine dental variation. Upper teeth are on the left, and lower teeth are on the right. Postcanine teeth are represented from top to bottom following the sequence P3, P4, M1, M2, and M3. (D) BM simulation of the evolution of one trait (PC1 score) across the hominin phylogeny. (Top) Green traces show evolution along the A. afarensis and A. africanus branches. (Middle) Simulated evolution along the Paranthropus clade (orange and red traces) is added to the above plot. (Bottom) Simulated evolution along the Homo clade (blue and purple traces) is added to the above graphs.
Table S1.
Specimens used in our study
| Species | Endocasts | Teeth |
| A. afarensis | A.L. 162–28, A.L. 333–45, A.L. 444–2 | A.L. 128–23, A.L. 145–35, A.L. 188–1, A.L. 199–1, A.L. 200–1a, A.L. 207–13, A.L. 266–1, A.L. 288–1, A.L. 333–1, A.L. 333–2, A.L. 333W-57, A.L. 333W-60, A.L. 400–1a, A.L. 486–1, LH 2, LH 3, LH 4, LH 6, LH 17, LH 21 |
| A. africanus | MLD 1*, MLD 37/38*, Sts 5, Sts 19†, Sts 60, Sts 71*, Stw 505*, Taung, Type 2* | MLD 2, MLD 4, MLD 6, MLD 18, MLD 19, MLD 23, MLD 28, Sts 1, Sts 8, Sts 17, Sts 21, Sts 22, Sts 24, Sts 52, Sts 56, Sts 57, Stw 2, Stw 3, Stw 6, Stw 14, Stw 73, Stw 183, Stw 252, Stw 384, Stw 498, TM 1511, Taung |
| P. robustus | SK 54, SK 859, SK 1585 | SK 11, SK 13/14, SK 23, SK 46, SK 48, SK 55, SKW 5, SKW 10, SKX 4446, TM 1517 |
| P. boisei s. l. | KGA-10–525*, KNM-ER 406*, KNM-ER 407*, KNM-ER 732*, KNM-ER 13750, KNM-ER 23000, KNM-WT 17000, KNM-WT 17400*, OH 5, Omo L338y-6* | KNM-CH 1, KNM-ER 729, KNM-ER 3230, KNM-ER 15930, KMN-WT 17400, Omo L51-79, Omo L338X-40, Omo L427-7, OH 5, Peninj 1 |
| H. habilis s. l. | KNM-ER 1805, KNM-ER 1813, KNM-ER 1470, OH 7*, OH 13‡, OH 16‡, OH 24* | KNM-ER 1590, KNM-ER 1802, KNM-ER 1805, KNM-ER 1813, OH 4, OH 6, OH 7, OH 13, OH 16, OH 21, OH 24, OH 39, OH 41, OH 44, SE 255, SK 27, Stw 53, Stw 80§, Stw 151§ |
| H. erectus | Hexian*, Ngawi*, Sangiran 2, 3*, 4, 10, 12, 17, Trinil 2, Yunxian 1*, 2*, Zhoukoudian III E, I L, III L | Sangiran 1a, 1b, 4, 5, 6, 7¶, Trinil 5, Zhoukoudian 19, 20, 25, 28, 29, 35.77, 36, 40, 41, 43, 44, 45, 46, 51, 52, 80, 90, 131, 137, 140, A2, B3.9, F1.5, G1.6, K1.96, PA327 |
| H. neanderthalensis# | Amud 1, Atapuerca-Sima de los Huesos 4*, 5*, 6*, Ganovce 1*, Feldhofer 1, Gibraltar 1, 2*, Krapina B*, 3, 6, La Chapelle-aux-Saints, La Ferrassie 1, La Quina 5, 18*, Le Moustier 1*, Monte Circeo I, Saccopastore 1*, 2*, Shanidar 1*, 5*, Spy 1, 2, Tabun C1* | Amud, Atapuerca-Sima de los Huesos, Arcy-sur-Cure, Ehringsdorf, Hortus, Krapina, Kulna, Le Moustier, Malarnaud, Monsempron, Monte Circeo, Pech-de-l'Aze, Petit-Puymoyen, Pinilla del Valle, St Cesaire, Tabun |
| H. sapiens# | Abri Pataud*, Brno*, Cioclovina‡, Cro-Magnon*, Dolni Vestonice*, Grimaldi*, Herto*, Kostenki*, Minatogawa*, Mladec*, Oberkassel*, Omo*, Predmosti*, Qafzeh*, San Teodoro*, Sungir*, Zhoukoudian Upper Cave*, AMNH recent modern human endocast collection‡ | Abri Pataud, Almonda, Dolni Vestonice, Grimaldi, Isturitz, Les Rois, Mladec, Qafzeh, St Germain-La Riviere, Trou Magritte, AMNH (USA, but collection of European origin), University of Granada (Spain), CENIEH (Spain) |
When no superscript footnote designator is indicated, specimens were used in the study of size and shape.
Specimens used in the study of endocranial size.
Classified by some authors as early Homo.
Specimens used in the study of endocranial shape.
Stw 80 and Stw 151 are also considered part of the A. africanus hypodigm.
Including all isolated teeth in the Sangiran 7 sample.
H. neanderthalensis and H. sapiens dental datasets are listed by site because of the large number of isolated teeth. The H. sapiens endocranial dataset is also listed by site. All other species are listed by specimen.
Results
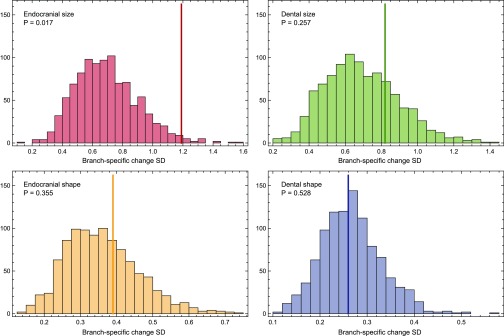
Endocranial volume is the only trait whose evolution has given rise to patterns of variation that are significantly different from those obtained from neutral simulations (Fig. S1). The standard deviation (SD) of the amounts of change per branch observed across the phylogeny is significantly greater than the SDs obtained in constant-rate simulations of the evolution of endocranial size (P = 0.017). This finding indicates that lineage-specific patterns of brain size evolution are more heterogeneous than expected under a neutral model and are unlikely to be explained by genetic drift. In addition, the rates of change for endocranial and dental size and shape through time differ substantially in different parts of the hominin phylogeny (Figs. 2 and 3). These differences are robust to different sample composition (P < 0.001 for all pairwise comparisons of the four traits) and to corrections for small sample size (Fig. S2), and they are substantial for most branches of the hominin phylogeny (Fig. S3 and Table S2). Although we use the term rate to make reference to branch-specific amounts of change, it should be noted that these values are not rates in the strict sense because they do not represent amounts of change per unit of time but rather the ratio of observed to simulated change per branch (Materials and Methods).
Fig. S1.
Comparison of observed and simulated results. Histograms show the distributions of standard deviations (SDs) of the branch-specific amounts of evolutionary change across the hominin phylogeny obtained in 1,000 constant-rate simulations. P values are calculated as the proportion of constant-rate simulations yielding a higher SD than the observed SD for each trait (solid vertical lines).
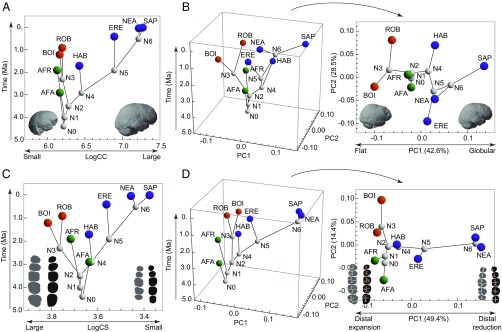
Fig. 2.
Variation in endocranial and dental size and shape through time. (A) Change in endocranial size (logarithm of cranial capacity, LogCC) over time showing extreme examples of variation. (B) PCA of endocranial shape variation over time (Left) and projection of PC1 and PC2 without time (Right). (C) Change in dental size (logarithm of centroid size, LogCS) over time. (D) PCA of dental shape variation over time (Left) and without time (Right). In A and B, the small and flat endocasts are the A. afarensis Sts 5 and P. robustus SK 1585 specimens, respectively. The large, globular endocast is a recent H. sapiens. Endocasts are in the same orientation as in Fig. 1. In C and D dental silhouettes representing large and distally expanded dentitions are based on the P. robustus specimens SK 13/14 (upper teeth) and SK 23 (lower teeth). Small and distally reduced dentitions are based on a recent H. sapiens. The orientation of teeth is the same as in Fig. 1. AFA, A. afarensis; AFR, A. africanus; BOI, P. boisei; ERE, H. erectus; HAB, H. habilis; NEA, H. neanderthalensis; ROB, P. robustus; SAP, H. sapiens.
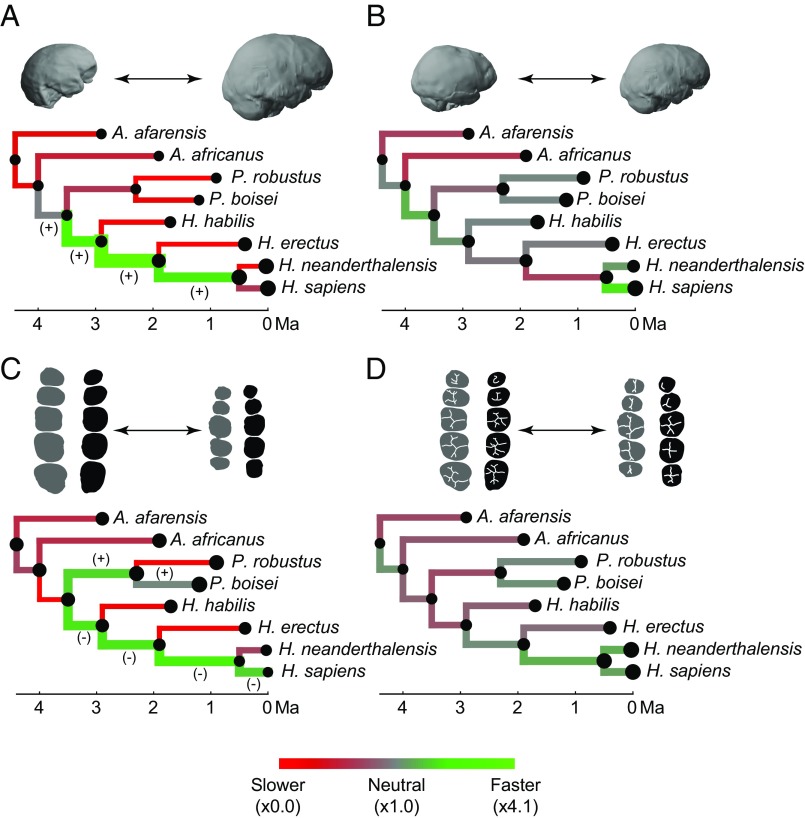
Fig. 3.
Evolution of endocranial and dental size and shape. (A) Comparison of observed and simulated branch-specific amounts of endocranial size variation. (B) Comparison of observed and simulated amounts of endocranial shape variation. (C) Comparison of observed and simulated amounts of dental size variation. (D) Comparison of observed and simulated amounts of dental shape variation. Red represents stasis along a given branch, and green represents fast evolution along a given branch, regardless of the directionality of change. Branch thickness is proportional to the observed amount of change along a given branch. In A and C, the plus sign represents size increase, the minus sign represents size decrease along fast-evolving branches, and tip and node size are proportional to endocranial and dental size. In B and D the amount of change per branch is based on shape distances that include all dimensions of the morphospace, and node and tip size are proportional to the amount of shape change with respect to the ancestral-most node. The specimen examples are the same as in Fig. 2. Orientation of endocasts and teeth is the same as in Figs. 1 and 2.
Fig. S2.
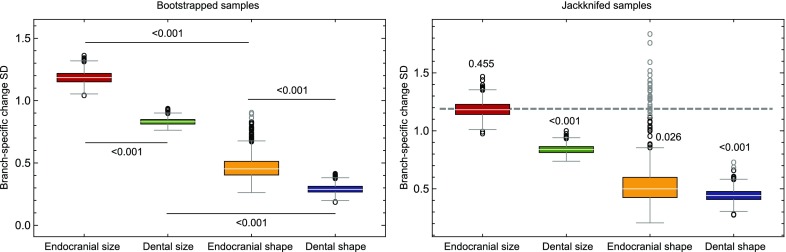
Bootstrapped and jackknifed results. Comparison of the distributions of SDs obtained when bootstrapping and jackknifing (n = 3) original samples showing outliers (black circles) and far outliers (gray circles). Both processes were repeated 1,000 times for each trait. P values on the bootstrap plot correspond to the pairwise comparisons between the traits linked by the extremes of each horizontal line and are based on permutation tests of the SD values between compared traits (1,000 permutations). P values on the jackknife plot correspond to the comparison between the SD of the amounts of change per branch obtained from the original endocranial size sample (SD = 1.19) and jackknifed samples for the four traits. Jackknife-based P values and distributions show that the more heterogeneous pattern of branch-specific rates obtained for endocranial size (dashed line) is not the result of the small sample size of some groups. Bootstrapping and jackknifing also addressed the potential issue that the endocranial size and shape data were obtained from samples that are not identical, whereas dental size and shape values were obtained from the same samples.
Fig. S3.
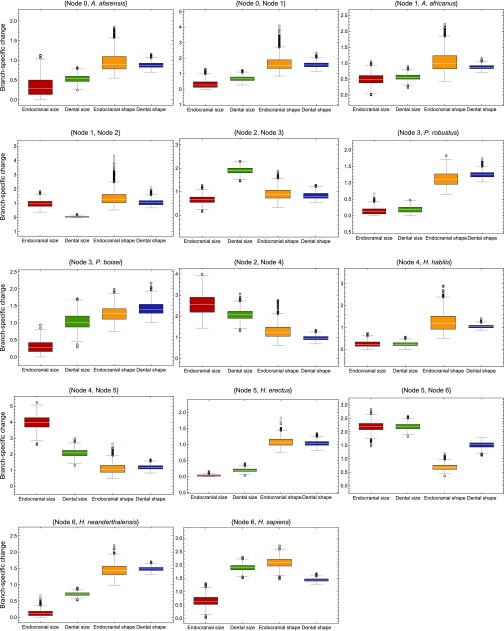
Branch-specific change. Comparison of the amount of change obtained for each branch for the four traits (numerical values are provided in Table S2). Distributions are based on 1,000 random bootstrap iterations of the original samples. Outliers (black circles) and far outliers (gray circles) are represented.
Table S2.
mvBM branch-specific amounts of evolutionary change in endocranial and dental size and shape
| Branch | Size | Shape | ||
| Endocranial | Dental | Endocranial | Dental | |
| N0-A. afarensis | 0.32 | 0.56 | 0.69 | 0.73 |
| N0–N1 | 0.29 | 0.72 | 1.04 | 1.17 |
| N1-A. africanus | 0.48 | 0.58 | 0.60 | 0.80 |
| N1–N2 | 0.97 | 0.04 | 1.55 | 0.85 |
| N2–N3 | 0.64 | 1.93 | 0.87 | 0.74 |
| N3-P. robustus | 0.01 | 0.19 | 1.05 | 1.05 |
| N3-P. boisei | 0.29 | 1.07 | 1.02 | 1.12 |
| N2–N4 | 2.58 | 2.07 | 1.21 | 0.79 |
| N4-H. habilis | 0.22 | 0.24 | 1.02 | 0.84 |
| N4–N5 | 4.01 | 2.09 | 0.94 | 1.10 |
| N5-H. erectus | 0.01 | 0.20 | 0.97 | 0.90 |
| N5–N6 | 2.20 | 2.22 | 0.69 | 1.51 |
| N6-H. neanderthalensis | 0.03 | 0.71 | 1.25 | 1.46 |
| N6-H. sapiens | 0.64 | 1.89 | 2.11 | 1.31 |
Values were obtained with a multiple variance Brownian motion (mvBM) approach and represent the number of times the amount of change per branch is greater or smaller than observed in simulated constant-rate Brownian motion (BM) models. Values smaller than 1 indicate less change than expected under a neutral model. Values greater than 1 indicate more change than expected under a neutral model. Branches showing the greatest amount of evolutionary change for each variable are shown in bold.
Our results show that sustained rapid evolution in brain size started before the separation of Paranthropus and Homo and peaked before the divergence between Homo erectus and the lineage leading to Neanderthals and modern humans (Fig. 3A). That peak rate was more than four times greater than that observed in simulated neutral scenarios (Table S2). Additional rapid brain increase was observed in the lineage immediately predating the Neanderthal–modern human split, but this increase was only twice as fast as that observed in a neutral scenario (Table S2). Other branches within the hominin phylogeny show much slower rates of change than those observed in a pure BM process, as is consistent with stabilizing selection and constrained evolution. These estimates are similar to the ones obtained using a more traditional approach to quantify branch-specific change based on a generalized least squares (GLS) ancestral reconstruction method (Table S3), which detects fast and slow evolutionary rates in the same branches but with less extreme values.
Table S3.
GLS branch-specific amounts of evolutionary change in endocranial and dental size and shape
| Branch | Size | Shape | ||
| Endocranial | Dental | Endocranial | Dental | |
| N0–N1 | 0.61 | 0.52 | 0.79 | 0.86 |
| N0-A. afarensis | 0.61 | 0.52 | 0.79 | 0.86 |
| N1-A. africanus | 0.77 | 0.64 | 0.62 | 0.80 |
| N1–N2 | 1.28 | 0.09 | 0.90 | 0.84 |
| N2–N3 | 0.94 | 1.72 | 0.84 | 0.81 |
| N3-P. robustus | 0.26 | 0.09 | 0.99 | 1.03 |
| N3-P. boisei | 0.60 | 1.40 | 1.13 | 1.13 |
| N2–N4 | 2.15 | 1.69 | 1.01 | 0.86 |
| N4-H. habilis | 0.55 | 0.19 | 1.08 | 0.81 |
| N4–N5 | 2.86 | 2.10 | 0.94 | 1.04 |
| N5-H. erectus | 0.46 | 0.16 | 1.05 | 0.88 |
| N5–N6 | 2.48 | 2.22 | 0.76 | 1.31 |
| N6-H. neanderthalensis | 0.46 | 0.64 | 1.47 | 1.43 |
| N6-H. sapiens | 1.04 | 1.95 | 1.88 | 1.42 |
Values were obtained with a GLS approach and represent the number of times the amount of change per branch is greater or smaller than observed in simulated constant-rate Brownian motion (BM) models. Values smaller than 1 indicate less change than expected under a neutral model. Values greater than 1 indicate more change than expected under a neutral model. The branches showing the greatest amount of evolutionary change for each variable are shown in bold.
Our results support the long-standing hypothesis that within the hominin clade brain organization, as inferred from endocranial shape, evolved independently of brain size (13). The ratios between the change in endocranial shape measured along each branch and those simulated using the BM model were all close to 1, leading to a general scenario that is not statistically different from those observed in constant-rate simulations (P = 0.355) (Fig. S1). This result indicates that endocranial shape evolved according to a quasi-neutral model, as is consistent with a scenario in which genetic drift is predominant (Fig. 3B). Rapid change, about twice that expected under a BM model, was observed only along the branch leading to modern humans from their last common ancestor with Neanderthals (Table S2). This rapid evolutionary change is reflected in the principal component analysis (PCA) of endocranial shape variation, which shows that Homo sapiens strongly diverges from all other species along the first principal component (PC1) (Fig. 2B). The eigenvector of this axis shows that the dorsal arc connecting the frontal and occipital poles is the only variable loading positively on PC1, thus separating flatter from the more globular endocasts that distinguish H. sapiens (Table S4) (14–16). Although frontal changes also can influence this variable, researchers have suggested that globularization is driven by upper parietal reorganization and that this anatomical change can be associated with enhanced visuospatial integration and memory in modern humans (17). The comparatively fast evolution of the dorsal arc trait in the lineage leading to H. sapiens is consistent with such a link between brain anatomy and function, although it could be an indirect result of selection on other craniofacial hard-tissue changes (18). If some individuals that do not show a globular anatomy, such as Jebel Irhoud 1 and 2 and Omo 2, are early members of H. sapiens (19), then the endocranial anatomy typical of modern humans may have evolved within the H. sapiens lineage.
Table S4.
Eigenvectors corresponding to the PCA of endocranial shape data
| Variable | PC1 | PC2 | PC3 | PC4 | PC5 | PC6 | PC7 |
| FW | −0.511 | 0.079 | −0.664 | 0.377 | 0.099 | −0.022 | 0.077 |
| HLC | −0.465 | −0.361 | 0.152 | 0.179 | −0.127 | 0.650 | −0.076 |
| HLD | 0.490 | −0.700 | −0.419 | −0.080 | 0.189 | 0.190 | −0.023 |
| MW | −0.385 | −0.413 | −0.131 | −0.340 | −0.088 | −0.608 | 0.163 |
| VT | −0.070 | 0.224 | −0.287 | −0.714 | −0.369 | 0.379 | 0.230 |
| BAC | −0.108 | −0.263 | 0.427 | 0.065 | 0.053 | 0.025 | 0.726 |
| MCW | −0.322 | −0.211 | 0.279 | −0.335 | 0.156 | −0.040 | −0.597 |
| BB | −0.120 | 0.196 | −0.022 | −0.279 | 0.876 | 0.160 | 0.161 |
BAC, biasterionic chord; BB, basion-bregma distance; FW, frontal width at Broca's cap; HLC, hemispheric length chord; HLD, hemispheric length dorsal arch; MCW, maximum cerebellar width; MW, maximum endocranial width; VT, vertex-lowest temporal distance.
Although there are differences in branch-specific evolutionary rates for dental size, they are still within the expectations of a constant-rate model (P = 0.257) (Fig. S1). Sustained reduction in the posterior dentition began in the branches antedating the origin of the genus Homo and continued along the sequence of branches leading to H. sapiens (Figs. 2C and 3C). Dental reduction along all these branches occurred at a rate that was approximately twice as fast as expected under a neutral evolutionary model (Table S2). Although the posterior teeth of Homo habilis and Australopithecus afarensis are similar in size, a fast evolutionary rate is inferred before the evolution of early Homo because this change is calculated with respect to the last common ancestor of Paranthropus and Homo, and this last common ancestor is inferred to have had larger posterior teeth than A. afarensis (Fig. 2C). A rapid rate of dental reduction is observed in the lineage leading to modern humans but not in Neanderthals, resulting in the comparatively small postcanine dentition of our species (Fig. 2C) (20). Contrary to our results, a previous quantitative study of molar size found that molar reduction observed in H. erectus, Neanderthals, and modern humans occurred at a faster rate than in early Homo (21). That study, however, used the area of the second molar (M2) as a proxy for molar size without considering variation in molar proportions across the molar row. Those proportions are known to change in the genus Homo in concert with absolute molar size, thus making M2s and M3s disproportionately small in species with overall small dental size (22, 23). Reduction in the dentition was not the only rapidly evolving trend, because dental expansion occurred at similarly high rates in the lineage leading to Paranthropus species (Fig. 3C and Table S2). Our data suggest that posterior tooth size in Paranthropus robustus stabilized after its divergence from the Paranthropus boisei lineage, whereas P. boisei continued its dental expansion but in a way consistent with quasi-neutral evolution. Assuming that the Paranthropus clade is monophyletic, which is the most common assumption even if other explanations are possible (24), these observations suggest that the postcanine megadontia of this genus is the result of long-term selective pressures that predate the divergence of the Paranthropus species.
As with endocranial shape, the shape of tooth crowns also evolved under a quasi-neutral model in which the evolutionary change along each branch is close to and statistically indistinguishable from that expected from a pure BM model (P = 0.528) (Fig. 3D and Fig. S1). The difference that drives PC1 of dental crown shape is a preferential reduction of the distal areas of premolars and molars in Neanderthals and modern humans (Fig. 2D and Fig. S4). The most rapid evolutionary change on the tree (1.5× greater than expected in a neutral scenario) is associated with this change along the branch antedating the separation of Neanderthals and modern humans (Table S2). Although the distal regions of posterior teeth are strongly reduced in both species, they have their own species-specific configurations. The characteristically derived dentition of Neanderthals (25, 26) is reflected in the relatively fast rate of evolution of dental shape in this lineage (Table S2).
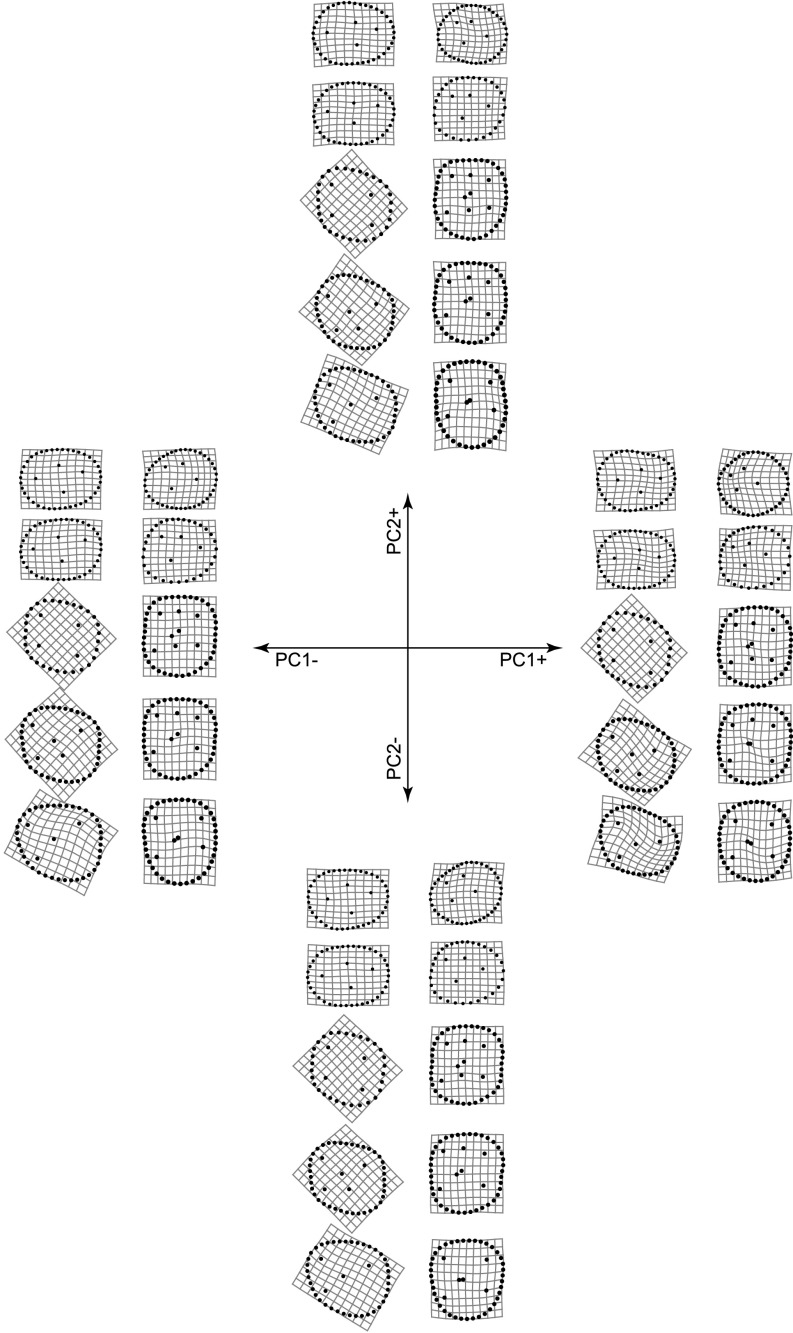
Fig. S4.
Thin plate spline (TPS) grids showing the most extreme patterns of tooth shape variation along PC1 and PC2. Represented grids are those corresponding to the most extreme PC scores in our dataset. The position and orientation of the TPS grids are the same as those of the teeth in Fig. 1.
Discussion
Our results show clear differences in evolutionary patterns corresponding to endocranial and dental size and shape during hominin evolution. Endocranial volume evolved at relatively heterogeneous rates that differ significantly from those observed under a constant-rate neutral model (Fig. S1). Endocranial shape and dental size and shape evolved at comparatively more uniform rates, with shape traits evolving under a quasi-neutral model. Although the evolution of these traits does not differ significantly from the expectations of a constant-rate scenario, endocranial shape, dental size, and dental shape still show significantly different evolutionary patterns. Given similar genetic variance, drift is expected to affect all traits in the same population equally (27). However, studies of brain anatomy in chimpanzees and modern humans have shown that brain size and brain organization have substantially different heritabilities (28) representing the proportion of total phenotypic variance in a population that has a genetic basis. Likewise, genetic variances of the traits included in our study can plausibly be different and might explain their different evolutionary behavior even if neither significantly differs from neutrality.
The observed patterns of branch-specific variation are consistent regardless of sample size and composition (Fig. S2), but they could be affected by changes in the phylogenetic scenario. We have chosen to deal with phylogenetic uncertainty by removing from our analyses those species whose phylogenetic position is particularly controversial, such as Homo ergaster, Homo antecessor, and Homo heidelbergensis. The resulting phylogenetic topology generally agrees with most quantitative and qualitative assessment of hominin phylogenetic relationships (21, 29, 30), but new fossil findings resulting in different relationships or branch lengths could potentially modify some of our findings.
Our results, which indicate that the evolution of hominin brain organization and brain size are decoupled, are consistent with larger brain size being positively selected across the entire genus Homo (31). Strong selection for larger brains has been linked to the selective advantages associated with the enhanced computational abilities of a larger neocortex with more neurons (32), but it also can be linked to other neural modifications such as an increased level of developmental plasticity arising from changes in the developmental patterns associated with larger brains (28, 33, 34). Selection for certain aspects of brain organization, particularly in the upper parietal reorganization that is arguably associated with modern human-specific functional modifications (17), is confined primarily to the branch leading directly to H. sapiens. No other aspects of brain reorganization as described by our set of variables show evidence of fast evolution across the hominin clade. However, many aspects of brain reorganization are not captured by those endocranial metrics, particularly those related to finer-grained organization such as sulcal variation, brain asymmetries, and volumetric changes of certain areas, among others. The predominant role of neutral mechanisms in the evolution of endocranial shape is consistent with previously published work reporting a major role of genetic drift in craniofacial evolution during the Australopithecus–Homo transition (35, 36) and during the divergence of Neanderthals and modern humans (37). Although our study focuses on endocranial variation, our findings are consistent with a general neutral scenario for the evolution of craniofacial shape in hominins.
The evolution of tooth crown size and shape is more closely linked than the evolution of brain size and shape. The branch antedating the separation of Neanderthals and modern humans is characterized by strong reduction in overall dental size associated with strong localized reduction of the distal areas of the crown of all postcanine teeth (20, 26). However, this anatomical change took place over a long period and does not show evidence of particularly fast evolution indicating strong selection. Although H. sapiens shows substantially faster reduction in dental size than Neanderthals, the two species share similar evolutionary rates of crown shape evolution, thus demonstrating that their species-specific dental traits have been subject to similar selection intensities. Our results show that crown shape evolution does not depart radically from a BM model, and that postcanine dental shape evolved at very similar rates within most branches of the hominin phylogeny. This observation lends quantitative support to dental shape as a useful proxy for reconstructing phylogenetic relationships in hominin fossil species. Indeed, the utility of dental shape for inferring evolutionary relationships is also supported by recent DNA analyses that confirmed a relationship of Middle Pleistocene European fossils to Neanderthals (38, 39) as initially proposed using fossil evidence (7, 26).
If branch-specific trends are not quantified, the sustained brain expansion found in some branches of the genus Homo may appear to be associated with sustained dental reduction. However, our results, which show that teeth and brains evolved at different rates in different hominin species, suggest that the two trends were decoupled. Our analysis shows that the apparent coupling of the traits is confined to the three branches that connect the last common ancestor of Paranthropus and Homo with the last common ancestor of Neanderthals and modern humans and that, even in those cases, brain evolution occurred at faster rates than dental evolution. We suggest that the context-specific ecological and behavioral factors that influenced the evolution of teeth and brains were not the same for the two morphological regions, nor were the combinations of those factors the same at different stages during hominin evolution.
Materials and Methods
Materials.
We used four datasets to evaluate postcanine and endocranial size and shape (Table S1 and Datasets S1–S4). The dataset for dental size and shape was assembled by A.G.-R. as part of quantitative descriptions of occlusal postcanine morphology (26, 40). Those samples were pruned to include only species with relatively uncontroversial phylogenetic positions (see below) and for which data on endocranial size and shape were also available. Endocranial size was studied using species-specific endocranial volumes based on values listed in ref. 41. This dataset does not reflect the reduction in endocranial volume seen in recent H. sapiens. Mean cranial capacity in H. erectus was estimated from a subsample of Asian H. erectus with a geographical and chronological origin similar to that of the dental sample (41). Endocranial shape was evaluated in a smaller sample of complete or partial hominin endocasts.
Quantitative Description of Dental and Endocranial Size and Shape.
Postcanine dental shape was characterized with configurations of landmarks and sliding semi-landmarks on the occlusal surface of tooth crowns (26, 40), and dental size was quantified as the centroid size of those configurations (defined as the square root of the sum of the squared distances between each landmark and the center of gravity of the configuration). Procrustes superimposition (42) was used to remove variation in position, size, and orientation, and species-specific mean shapes were obtained by averaging Procrustes-superimposed coordinates for each species (26). PCAs of Procrustes coordinates were used to obtain the principal component (PC) scores used in subsequent analyses (12). When all dimensions of shape variation are considered, as we did throughout all our analyses, PC scores contain the same information as original variables but are mathematically more convenient (12).
Tooth-specific size and shape data were pooled to analyze the complete postcanine dentition. For shape analyses, landmark coordinates corresponding to the 10 postcanine teeth (upper and lower premolars and molars) were subjected to different Procrustes superimpositions and then were combined in the same PCAs. Overall dental size was estimated by summing up centroid sizes across all the postcanine teeth. Analyses of dental size therefore reflect increases or decreases of total postcanine occlusal areas but not changes in dental proportions among teeth.
Endocranial size was evaluated using species-specific mean endocranial volumes. Endocranial shape was quantified using a set of classic linear metrics measured by R.L.H. These metrics included eight variables used in other studies of hominin endocranial variation (Fig. 1) (43). Size variation was removed from these analyses by dividing each of these metrics by the cube root of cranial capacity in each individual. Species-specific mean values for each of these variables were subjected to PCA, and PC scores were used in ancestral reconstructions of endocranial shape.
The robustness of our results to sample composition was evaluated by bootstrapping the original samples 1,000 times and then recalculating species-specific mean values and running all the analyses in bootstrapped samples. Likewise, we assessed if the more heterogeneous evolutionary rates obtained for endocranial evolution with respect to dental evolution result from differences in sample size. Because some of the species in our samples are represented by only three endocasts, we jackknifed all the samples to three individuals per species. This down-sampling process was also repeated 1,000 times. Resampling rounds for both approaches were performed independently for each tooth position because most individuals in the dental samples do not preserve all postcanine teeth.
Hominin Phylogeny.
Because our methodological approach requires the use of an a priori phylogeny, we used only species whose phylogenetic positions are relatively uncontroversial. Following the most widely accepted view, we considered Homo and Paranthropus as two monophyletic clades (29, but also see ref. 44). Australopithecus africanus was considered to be a sister group to both Paranthropus and Homo clades following ref. 45, although some analyses have suggested other phylogenetic positions for this species (29), including a recent classification as a sister group only to Homo (30). We chose not to use a pruned version of the recently published Bayesian phylogeny proposed in ref. 30 for two reasons. First, the supermatrix on which this analysis is based pools traits and character states based on different studies, criteria, and scoring systems; this approach may bias results by recovering nodes that have little or no support or by failing to recover nodes that do have high support (46). Second, posterior probabilities yielded by this analysis for most of the nodes included in our phylogeny are very low. Although they are unquestionably valuable for considering alternative scenarios for hominin evolution, we believe that evolutionary relationships reflected in the summary of best trees presented in ref. 30 have weaker support in general than the relationships used in our study.
Times of node divergence and ages of terminal species followed ref. 11. Tips were dated to the last appearance date (LAD) for each species listed in table 1 of ref. 11, whereas nodes were dated to the corresponding first appearance date (FAD). Assuming that FADs and LADs observed in the fossil record are unlikely to represent the actual FADs and LADs for each species, we used the nonconservative version of these dates, which incorporate “the age, and the published error of the age, of the nearest underlying dated horizon in the case of the FAD, and the age, and the published error of the age, of the nearest overlying dated horizon in the case of the LAD” (11, p. 55).
To account for some phylogenetic patterns that are not reflected in these values, we dated the oldest ancestor in our tree to 4.4 Ma assuming an evolutionary continuity between Australopithecus anamensis and A. afarensis (47), which was dated to 2.9 Ma. The divergence between P. robustus and P. boisei was established at 2.3 Ma. To account for the recent early Homo findings that have pushed the FAD of the genus Homo back to at least 2.8 Ma (48), we set the origin of this genus at 2.9 Ma. The divergence of the Paranthropus and Homo clades was estimated at 3.5 Ma. Because our samples do not include late H. erectus fossils, we dated H. erectus to 400 ka. An early Neanderthal status for the Middle Pleistocene hominins from Sima de los Huesos is strongly supported by both the paleontological and molecular evidence (7, 38, 49), so we established the divergence date of Neanderthals and modern humans at 0.5 Ma, although morphological studies suggest that an earlier divergence time for these species is likely (26, 30). The averaging of data points at the LADs used for each species is likely to provide conservative estimates of branch-specific amounts of change. However, the use of data at time points that are closer to individual values would artificially inflate the measured amounts of change per branch because of the uncertainty regarding finer-grained population-specific dates and their particular relationships.
Ancestral Estimation.
A multiple-variance Brownian motion (mvBM) framework was used to estimate ancestral values in the hominin phylogeny (10). Most ancestral estimation approaches assume a standard BM model of character evolution (50). In standard BM the rate of evolution is assumed to have a single mean and variance across all branches, and trait divergence is proportional to the square root of time. Biologically, these assumptions imply there is no sustained difference in the direction and rate of change among the different lineages of the phylogeny. In many cases we expect this assumption to be unrealistic because selection may be associated with environments that differ systematically between subclades or with particular evolutionary or environmental events that occurred on only one branch of the tree, thus producing different evolutionary rates and directions in different lineages. Our approach relaxes the pure BM model to capture different patterns of trait variation along each branch of the phylogeny (10).
Specifically, ancestral values were estimated using a two-step process. The first step infers branch-specific patterns of change based on a model that assumes that trait values for ancestral nodes are a compromise between global and local effects. The baseline assumption that phylogenetic relatedness accurately reflects how traits evolve is hereby leveraged against local deviations from this expectation. Specifically, a global estimate (a weighted estimate based on the phylogenetic tree and the tip values) is combined with a local estimate (accounting for information from a node’s closest relatives without taking tree structure into account). Measures of the rate of evolution then are estimated by dividing the squared trait difference by the branch length for each ancestor–descendant pair. Rates hereby represent the extent to which lineage-specific changes are found to align with the baseline expectation that phylogenetic relatedness is an accurate proxy for trait evolution. Each branch rate can be considered to be a point estimate of the rate of change along each individual branch under an mvBM model.
In the second step, the branch lengths of the original phylogenetic tree are rescaled according to the estimated rates of evolution. The model with the rescaled branches is then parameterized using a standard BM model to produce ancestral estimates. This procedure makes use of the analytical power of BM estimation techniques while allowing local variation in evolutionary rates. This method, which is explained in greater detail in ref. 10 and implemented in the R package evomap (51), was applied to the hominin phylogeny and endocranial and dental datasets.
Evolutionary Simulations.
Results obtained through the previously described process were compared with results obtained through a simulated pure BM scenario. For size traits, evolutionary variation was simulated on log-transformed size values, whereas for shape variation, PC scores were used (12). Simulations were initiated at the ancestral-most values estimated through the mvBM approach. A per-generation variance rate (per-generation σ2) was estimated after rescaling the hominin phylogeny to generations using a constant generation time of 25 y (52). A GLS approach (53) implemented in the package Phylogenetics for Mathematica (54) was used to estimate a constant per-generation variance rate for each variable (log-size and PC scores) based on available data.
Using trait-specific constant per generation rates, evolutionary change was simulated as a uni- or multidimensional random walk (12) on the hominin phylogeny. Simulations were run 1,000 times, and the mean change between all ancestors and descendants was used as the expectation of the amount of change if each branch had evolved neutrally under a pure BM model. For endocranial and dental shape, this simulation was performed in PC morphospace. Shape distances between ancestors and descendants were calculated as the square root of the sum of the squared differences in all PC scores between two given species, which is equivalent to the definition of Procrustes distance for landmark data. For dental and endocranial size, branch-specific amounts of change were calculated simply as the difference between descendants and ancestors. Transformations between landmark coordinates and PC morphospace were done with the package Geometric Morphometrics for Mathematica (55).
The mvBM branch-specific changes were compared with the pure BM changes as the mvBM/BM ratio. A value larger than 1 indicates that a given branch has experienced more change than expected under a BM model (i.e., that branch has evolved faster than expected under a neutral model regardless of the directionality of the change). A value smaller than 1 is indicative of slower evolution than expected under a neutral model, which in turn is indicative of stabilizing selection along a certain branch. As we emphasized earlier, although we refer to these values as rates, we recognize that they are not rates in the strict sense but are the ratios of observed to simulated changes per branch. These values were color coded and overlaid on the original phylogeny.
Supplementary Material
Acknowledgments
Images of endocranial models were provided by José Manuel de la Cuétara (H. sapiens endocast), Antoine Balzeau (P. robustus endocast), and Simon Neubauer (A. africanus endocast, which is based on a CT scan from the University of Vienna database). We thank the following people for discussion, facilitating access to material, constructive peer review, or technical support: C. Sherwood, J. M. Bermúdez de Castro, J. L. Arsuaga, E. Carbonell, O. Kullmer, B. Denkel, F. Schrenk, M. A. de Lumley, A. Vialet, I. Tattersall, G. Sawyer, G. García, Y. Haile-Selassie, L. Jellema, M. Botella, P. Gunz, and D. Sánchez-Martín.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.D.W. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1608798114/-/DCSupplemental.
References
- 1.Pilbeam D, Gould SJ. Size and scaling in human evolution. Science. 1974;186(4167):892–901. doi: 10.1126/science.186.4167.892. [DOI] [PubMed] [Google Scholar]
- 2.McHenry HM. The pattern of human evolution: Studies on bipedalism, mastication, and encephalization. Annu Rev Anthropol. 1982;11:151–173. [Google Scholar]
- 3.Jiménez-Arenas JM, Pérez-Claros JA, Aledo JC, Palmqvist P. On the relationships of postcanine tooth size with dietary quality and brain volume in primates: Implications for hominin evolution. BioMed Res Int. 2014;2014:406507. doi: 10.1155/2014/406507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aiello LC, Wheeler P. The expensive-tissue hypothesis: The brain and the digestive system in human and primate evolution. Curr Anthropol. 1995;36(2):199–221. [Google Scholar]
- 5.Ungar PS. Dental evidence for the reconstruction of diet in African early Homo. Curr Anthropol. 2012;53(S6):S318–S329. [Google Scholar]
- 6.Spoor F, et al. Reconstructed Homo habilis type OH 7 suggests deep-rooted species diversity in early Homo. Nature. 2015;519(7541):83–86. doi: 10.1038/nature14224. [DOI] [PubMed] [Google Scholar]
- 7.Arsuaga JL, et al. Neandertal roots: Cranial and chronological evidence from Sima de los Huesos. Science. 2014;344(6190):1358–1363. doi: 10.1126/science.1253958. [DOI] [PubMed] [Google Scholar]
- 8.Harmand S, et al. 3.3-million-year-old stone tools from Lomekwi 3, West Turkana, Kenya. Nature. 2015;521(7552):310–315. doi: 10.1038/nature14464. [DOI] [PubMed] [Google Scholar]
- 9.Venditti C, Meade A, Pagel M. Multiple routes to mammalian diversity. Nature. 2011;479(7373):393–396. doi: 10.1038/nature10516. [DOI] [PubMed] [Google Scholar]
- 10.Smaers JB, Mongle CS, Kandler A. A multiple variance Brownian motion framework for estimating variable rates and inferring ancestral states. Biol J Linn Soc Lond. 2016;118(1):78–94. [Google Scholar]
- 11.Wood B, K Boyle E. Hominin taxic diversity: Fact or fantasy? Am J Phys Anthropol. 2016;159(Suppl 61):S37–S78. doi: 10.1002/ajpa.22902. [DOI] [PubMed] [Google Scholar]
- 12.Polly PD. On the simulation of the evolution of morphological shape: Multivariate shape under selection and drift. Palaeontol Electronica. 2004;7(2):1–28. [Google Scholar]
- 13.Holloway RL. Cranial capacity, neural reorganization, and hominid evolution: A search for more suitable parameters. Am Anthropol. 1966;68(1):103–121. [Google Scholar]
- 14.Lieberman DE, McBratney BM, Krovitz G. The evolution and development of cranial form in Homosapiens. Proc Natl Acad Sci USA. 2002;99(3):1134–1139. doi: 10.1073/pnas.022440799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruner E, Manzi G, Arsuaga JL. Encephalization and allometric trajectories in the genus Homo: Evidence from the Neandertal and modern lineages. Proc Natl Acad Sci USA. 2003;100(26):15335–15340. doi: 10.1073/pnas.2536671100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gunz P, Neubauer S, Maureille B, Hublin J-J. Brain development after birth differs between Neanderthals and modern humans. Curr Biol. 2010;20(21):R921–R922. doi: 10.1016/j.cub.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Bruner E, Iriki A. Extending mind, visuospatial integration, and the evolution of the parietal lobes in the human genus. Quat Int. 2016;405(Part A):98–110. [Google Scholar]
- 18.Martínez-Abadías N, et al. Pervasive genetic integration directs the evolution of human skull shape. Evolution. 2012;66(4):1010–1023. doi: 10.1111/j.1558-5646.2011.01496.x. [DOI] [PubMed] [Google Scholar]
- 19.Stringer C. The origin and evolution of Homo sapiens. Phil Trans R Soc Lond B Biol Sci. 2016;371(1698):20150237. doi: 10.1098/rstb.2015.0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wolpoff MH. 1971. Metric Trends in Hominid Dental Evolution Case Western Reserve Univ Press, Cleveland)
- 21.Organ C, Nunn CL, Machanda Z, Wrangham RW. Phylogenetic rate shifts in feeding time during the evolution of Homo. Proc Natl Acad Sci USA. 2011;108(35):14555–14559. doi: 10.1073/pnas.1107806108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans AR, et al. A simple rule governs the evolution and development of hominin tooth size. Nature. 2016;530(7591):477–480. doi: 10.1038/nature16972. [DOI] [PubMed] [Google Scholar]
- 23.Gómez-Robles A. Palaeoanthropology: What teeth tell us. Nature. 2016;530(7591):425–426. doi: 10.1038/530425a. [DOI] [PubMed] [Google Scholar]
- 24.Wood B, Constantino P. Paranthropus boisei: Fifty years of evidence and analysis. Am J Phys Anthropol. 2007;134(Suppl 45):106–132. doi: 10.1002/ajpa.20732. [DOI] [PubMed] [Google Scholar]
- 25.Bailey SE, Weaver TD, Hublin J-J. Who made the Aurignacian and other early Upper Paleolithic industries? J Hum Evol. 2009;57(1):11–26. doi: 10.1016/j.jhevol.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Gómez-Robles A, Bermúdez de Castro JM, Arsuaga J-L, Carbonell E, Polly PD. No known hominin species matches the expected dental morphology of the last common ancestor of Neanderthals and modern humans. Proc Natl Acad Sci USA. 2013;110(45):18196–18201. doi: 10.1073/pnas.1302653110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lande R. Natural selection and random genetic drift in phenotypic evolution. Evolution. 1976;30(2):314–334. doi: 10.1111/j.1558-5646.1976.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 28.Gómez-Robles A, Hopkins WD, Schapiro SJ, Sherwood CC. Relaxed genetic control of cortical organization in human brains compared with chimpanzees. Proc Natl Acad Sci USA. 2015;112(48):14799–14804. doi: 10.1073/pnas.1512646112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strait D, Grine FE, Fleagle JG. Analyzing hominin phylogeny: Cladistic approach. In: Henke W, Tattersall I, editors. Handbook of Paleoanthropology. Springer; Berlin: 2015. pp. 1989–2014. [Google Scholar]
- 30.Dembo M, Matzke NJ, Mooers AØ, Collard M. Bayesian analysis of a morphological supermatrix sheds light on controversial fossil hominin relationships. Proc R Soc B Biol Sci. 2015;282(1812):20150943. doi: 10.1098/rspb.2015.0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rightmire GP. Brain size and encephalization in early to Mid-Pleistocene Homo. Am J Phys Anthropol. 2004;124(2):109–123. doi: 10.1002/ajpa.10346. [DOI] [PubMed] [Google Scholar]
- 32.Herculano-Houzel S. The human brain in numbers: A linearly scaled-up primate brain. Front Hum Neurosci. 2009;3:31. doi: 10.3389/neuro.09.031.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosenberg KR. The evolution of modern human childbirth. Am J Phys Anthropol. 1992;35(S15):89–124. [Google Scholar]
- 34.Dunsworth HM, Warrener AG, Deacon T, Ellison PT, Pontzer H. Metabolic hypothesis for human altriciality. Proc Natl Acad Sci USA. 2012;109(38):15212–15216. doi: 10.1073/pnas.1205282109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackermann RR, Cheverud JM. Detecting genetic drift versus selection in human evolution. Proc Natl Acad Sci USA. 2004;101(52):17946–17951. doi: 10.1073/pnas.0405919102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schroeder L, Roseman CC, Cheverud JM, Ackermann RR. Characterizing the evolutionary path(s) to early Homo. PLoS One. 2014;9(12):e114307. doi: 10.1371/journal.pone.0114307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weaver TD, Roseman CC, Stringer CB. Were neandertal and modern human cranial differences produced by natural selection or genetic drift? J Hum Evol. 2007;53(2):135–145. doi: 10.1016/j.jhevol.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 38.Meyer M, et al. Nuclear DNA sequences from the Middle Pleistocene Sima de los Huesos hominins. Nature. 2016;531(7595):504–507. doi: 10.1038/nature17405. [DOI] [PubMed] [Google Scholar]
- 39.Meyer M, et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature. 2014;505(7483):403–406. doi: 10.1038/nature12788. [DOI] [PubMed] [Google Scholar]
- 40.Gómez-Robles A, Polly PD. Morphological integration in the hominin dentition: Evolutionary, developmental, and functional factors. Evolution. 2012;66(4):1024–1043. doi: 10.1111/j.1558-5646.2011.01508.x. [DOI] [PubMed] [Google Scholar]
- 41.Holloway RL, Broadfield DC, Yuan MS. The Human Fossil Record, Brain Endocasts: The Paleoneurological Evidence. Wiley-Liss; New York: 2004. [Google Scholar]
- 42.Rohlf FJ, Slice D. Extension of the Procrustes method for the optimal superimposition of landmarks. Syst Zool. 1990;39(1):40–59. [Google Scholar]
- 43.Bruner E, Grimaud-Hervé D, Wu X, de la Cuétara JM, Holloway R. A paleoneurological survey of Homo erectus endocranial metrics. Quat Int. 2015;368:80–87. [Google Scholar]
- 44.Wood B, Collard M. The human genus. Science. 1999;284(5411):65–71. doi: 10.1126/science.284.5411.65. [DOI] [PubMed] [Google Scholar]
- 45.Strait DS, Grine FE. Inferring hominoid and early hominid phylogeny using craniodental characters: The role of fossil taxa. J Hum Evol. 2004;47(6):399–452. doi: 10.1016/j.jhevol.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 46.Kluge AG. A concern for evidence and a phylogenetic hypothesis of relationships among Epicrates (Boidae, Serpentes) Syst Biol. 1989;38(1):7–25. [Google Scholar]
- 47.Kimbel WH, et al. Was Australopithecus anamensis ancestral to A. afarensis? A case of anagenesis in the hominin fossil record. J Hum Evol. 2006;51(2):134–152. doi: 10.1016/j.jhevol.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 48.Villmoare B, et al. Paleoanthropology. Early Homo at 2.8 Ma from Ledi-Geraru, Afar, Ethiopia. Science. 2015;347(6228):1352–1355. doi: 10.1126/science.aaa1343. [DOI] [PubMed] [Google Scholar]
- 49.Gómez-Robles A, Bermúdez de Castro JM, Martinón-Torres M, Prado-Simón L, Arsuaga JL. A geometric morphometric analysis of hominin lower molars: Evolutionary implications and overview of postcanine dental variation. J Hum Evol. 2015;82:34–50. doi: 10.1016/j.jhevol.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 50.Pagel M. Modelling the evolution of continuously varying characters on phylogenetic trees: The case of hominid cranial capacity. In: MacLeod N, Forey PL, editors. Morphology, Shape and Phylogeny. Taylor & Francis; London: 2002. pp. 269–286. [Google Scholar]
- 51.Smaers JB. 2014 evomap: R package for the evolutionary mapping of continuous traits (Github: https://github.com/JeroenSmaers/evomap). Accessed April 7, 2016.
- 52.Weaver TD, Roseman CC, Stringer CB. Close correspondence between quantitative- and molecular-genetic divergence times for Neandertals and modern humans. Proc Natl Acad Sci USA. 2008;105(12):4645–4649. doi: 10.1073/pnas.0709079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martins EP, Hansen TF. Phylogenies and the comparative method: A general approach to incorporating phylogenetic information into the analysis of interspecific data. Am Nat. 1997;149(4):646–667. [Google Scholar]
- 54.Polly PD. Phylogenetics for Mathematica. Version 3.0. Department of Geological Sciences, Indiana University; Bloomington, Indiana: 2014. [Google Scholar]
- 55.Polly PD. Geometric Morphometrics for Mathematica. Version 12.0. Department of Geological Sciences, Indiana University; Bloomington, Indiana: 2016. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.