Significance
Signal transduction through Tropomyosin-related kinase A (TrkA), a receptor tyrosine kinase, is a target for inhibition of chronic pain and could lead to a new class of drugs against pain. Selectivity against kinases can be difficult to achieve, especially against members of the same kinase family. Structures of the compounds bound to TrkA show a binding site comprised of the kinase, which is conserved among the Trk family, and the juxtamembrane (JM), which is not well conserved. Depending on their chemical substructure, the region of the juxtamembrane that interacts with the compounds can be different, leading to differences in specificity. This study emphasizes the importance of including residues beyond the catalytic domain for small-molecule screening, importance of screening by affinity, and structural characterization to understand binding interactions.
Keywords: TrkA, kinase, pain, inhibition, selectivity
Abstract
Current therapies for chronic pain can have insufficient efficacy and lead to side effects, necessitating research of novel targets against pain. Although originally identified as an oncogene, Tropomyosin-related kinase A (TrkA) is linked to pain and elevated levels of NGF (the ligand for TrkA) are associated with chronic pain. Antibodies that block TrkA interaction with its ligand, NGF, are in clinical trials for pain relief. Here, we describe the identification of TrkA-specific inhibitors and the structural basis for their selectivity over other Trk family kinases. The X-ray structures reveal a binding site outside the kinase active site that uses residues from the kinase domain and the juxtamembrane region. Three modes of binding with the juxtamembrane region are characterized through a series of ligand-bound complexes. The structures indicate a critical pharmacophore on the compounds that leads to the distinct binding modes. The mode of interaction can allow TrkA selectivity over TrkB and TrkC or promiscuous, pan-Trk inhibition. This finding highlights the difficulty in characterizing the structure-activity relationship of a chemical series in the absence of structural information because of substantial differences in the interacting residues. These structures illustrate the flexibility of binding to sequences outside of—but adjacent to—the kinase domain of TrkA. This knowledge allows development of compounds with specificity for TrkA or the family of Trk proteins.
The current standards of care for chronic pain include the nonsteroidal anti-inflammatory drugs, Gabapentinoids, serotonin-norepinephrine reuptake inhibitors, and opioids. However, patients often report inadequate pain relief, inability to tolerate side effects, and in the case of opiates have other risks associated with diversion and drug abuse (1, 2). As a result, it is critical to find alternative targets to treat pain, such as the signaling cascade mediated by the receptor tyrosine kinase, Tropomyosin-related kinase A (TrkA) (3–5).
In addition to its role in pain signaling, chromosomal rearrangement of Trk receptors have been found in tumors of multiple cancers (6, 7). Development of inhibitors against TrkA could play a role in pain relief and oncology.
TrkA is a member of a family of type I transmembrane receptors (8, 9). This family also includes TrkB and TrkC, all of which bind to neurotrophins (10). Although originally identified as a proto-oncogene, TrkA was determined to be the primary receptor for NGF (11, 12). TrkA is critical for the development of neurons, including those involved in pain signaling, and mutations of TrkA have been found to be the bases for the genetic disorder, congenital insensitivity to pain with anhydrosis (13). It has been found that administration of NGF to patients causes pain and that neutralization of NGF blocks chronic pain (14, 15).
There are 518 kinases in the human genome and kinases have proven to be a fruitful target for small-molecule drugs (16). Numerous structures have been determined for kinases bound to inhibitors. From these, inhibitors have been classified as different types, depending on their interactions with the kinase domain (17). Inhibitors that bind to the active site (type I) make interactions with the conserved backbone atoms of the kinase hinge, thus making it difficult to gain selectivity over other kinases unless interactions are achieved with less-conserved regions of the active site. To improve selectivity, inhibitors have been designed to bind in the active site but extend to less-conserved residues outside the active site (known as type II inhibitors). Alternatively, inhibitors have been found to bind the kinase outside the active site (type III) or with other domains of the protein (type IV).
The intracellular portion of Trk proteins are largely comprised of the kinase domain (8, 9). In TrkA, between the transmembrane region and the kinase domain, there are ∼60 residues that are not homologous to other proteins and lack predicted secondary structure. At the C-terminal end of this juxtamembrane (JM) region is the NPXY motif, which upon phosphorylation of Tyr-496, forms the binding site for the phosphotyrosine binding domain of the adaptor protein, Shc (18). Following the kinase domain is a short stretch of ∼10 residues that contain the YXXL motif, which binds to the SH2 domain of PLCγ1, upon phosphorylation of Tyr-791 (19, 20). Signaling downstream of TrkA has been well established to lead to differentiation and proliferation of neuronal cells. Possibly less studied are the signaling events that lead to potentiation of a pain response through increased expression of nociceptors, like TrpV1, on the cell surface and release of pain-signaling peptides, like substance P and CGRP (21). However, clinical proof-of-concept studies with the antibody Tanezumab, which binds to NGF and prohibits activation of TrkA, have demonstrated positive results for relief of osteoarthritis pain, chronic lower back pain, diabetic peripheral neuropathic pain, and cancer pain (22, 23).
Structures of the isolated TrkA kinase domain have been published by us and others (5, 24–27). Compounds found to inhibit TrkA have all bound to the kinase active site. Although selectivity can be achieved for the Trk family of kinases by developing type II kinase inhibitors that reach into the back pocket behind the active site, selectivity of these type I and type II active site inhibitors have not been achieved for TrkA over the other Trk family members.
Here, we describe the identification of nonactive site compounds that bind to TrkA, which demonstrate TrkA selectivity over TrkB and TrkC in biochemical and cellular assays. Structures of the compounds bound to TrkA show a binding site comprised of the kinase, which is conserved among the Trk family, and the JM, which is not well conserved. Depending on their chemical substructure, the region of the JM that interacts with the compounds can be different, leading to differences in specificity.
Results
Hits and Generating the Constructs.
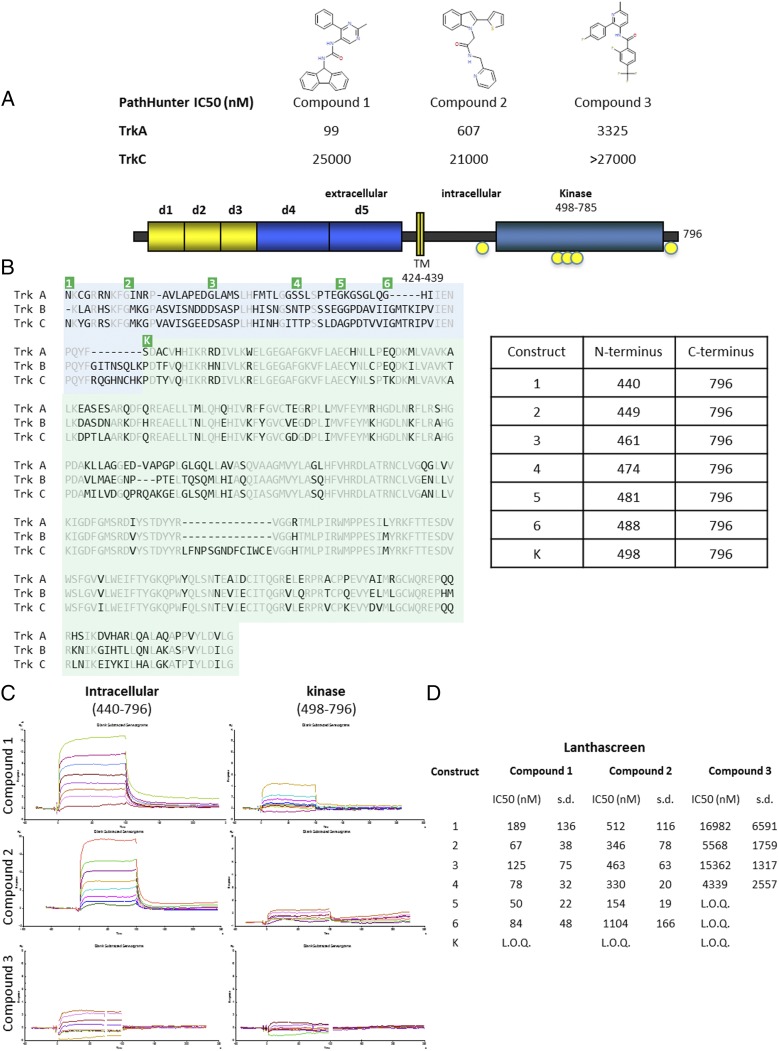
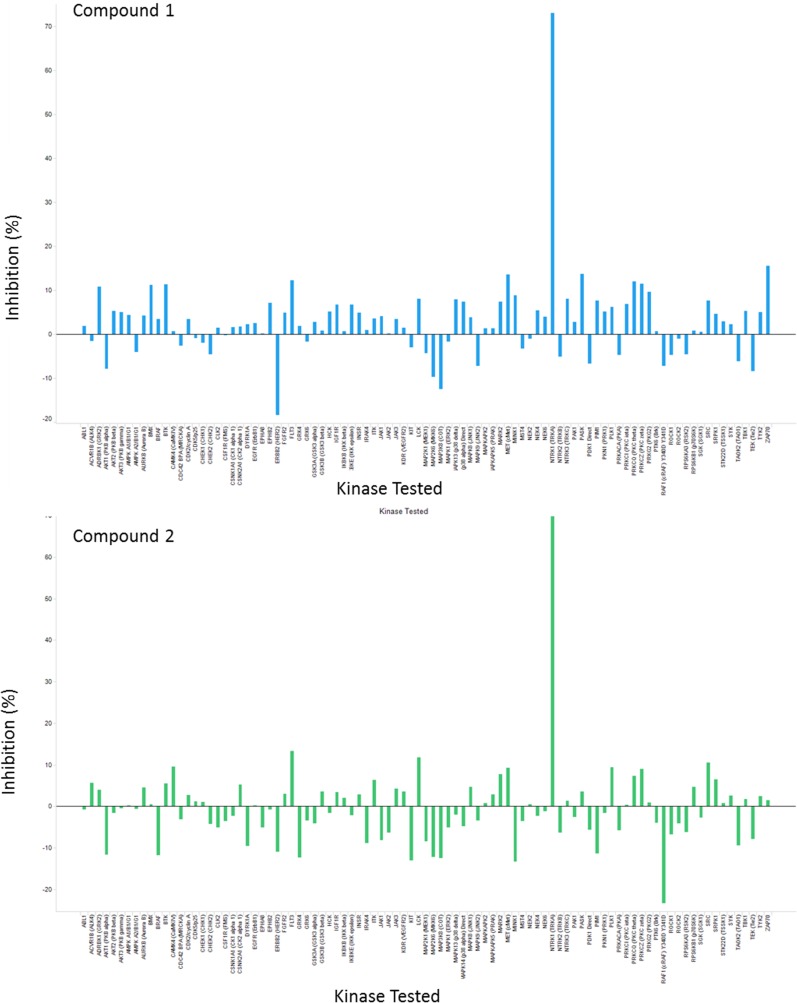
As a potential target for pain and oncology, we and others have identified small-molecule inhibitors that target the kinase active site of TrkA (5, 24–27). Although potent molecules have been identified that are highly selective for the Trk kinases, the compounds inhibited TrkB and TrkC with comparable potencies to TrkA. This finding is not surprising, as the active sites of the Trk family of kinases are highly conserved. Type 2 inhibitors, which interact with the deep pocket behind the DFG motif, were equally unable to achieve TrkA selectivity (27). A new screening campaign was initiated, searching for binding to the entire intracellular region using the affinity ligand identification system (ALIS), an affinity selection–mass spectrometry technique (28). Many hits were identified as active site binders and triaged based on cell-based kinase activity against the Trk kinases. Three hits were discovered that appeared to have selectivity for TrkA over TrkC in cell-based assays (Fig. 1A). Additionally, compounds 1 and 2 were profiled in an in vitro kinase panel (Invitrogen’s SelectScreen) against 93 kinases, which includes Trks A, B, and C. The only kinase that showed inhibition was TrkA. Fig. S1 for an illustration of percent inhibition across the panel of kinases. Because the screen is performed at 10-μM compound concentrations, compound 3 was not tested in the kinome screen because the potency was not high enough to accurately determine selectivity.
Fig. 1.
Activity and binding of compounds identified by ALIS screening. (A) The three compounds identified by ALIS screen and determined to be selective for TrkA over TrkC are shown. The IC50 values from the cell-based, PathHunter assay against TrkA and TrkC are given in nanomolars. Compound 3 did not inhibit at the highest concentration tested in the assay [limit of quantitation (LOQ) was > 27,000 nM in this assay]. (B) The schematic of full-length TrkA is shown at the top with domains indicated. Key phosphorylation sites are indicated in yellow circles. A sequence alignment is shown below for the intracellular region of TrkA, TrkB, and TrkC. Residues identical in all three sequences are colored gray. The kinase domain is shaded green and the JM region is shaded blue. The table on the right shows the residues that comprise each construct generated to test inhibition. The corresponding amino termini of the constructs are indicated on the sequence alignment in green. The constructs all extend to the natural carboxy-terminus of TrkA, followed by a hexahistidine tag for purification. Construct 1 starts immediately following the transmembrane helix. Each subsequence construct has an amino terminus closer to the kinase domain, which is indicated as construct K. (C) SPR binding traces of each compound to either the full intracellular region (construct 1) or the isolated kinase domain. (D) Lanthascreen assay of constructs with each of the three compounds. For each compound, a minimal construct can be inhibited by each inhibitor and all constructs longer than the minimal construct can also be inhibited by the compound. Construct 4 can be inhibited by all three compounds (LOQ was > 67,500 nM in this assay).
Fig. S1.
Kinase panel showing percent inhibition against 93 kinases including TrkA, TrkB, and TrkC for compounds 1 and 2.
Crystallization using the kinase domain of TrkA failed to yield structures with these compounds. Despite experience determining structures of active-site inhibitors in the tens of micromolar range, these selective compounds were recalcitrant to structural determination. In comparison with the crystallographic system, the protein used for screening contained the complete intracellular region of TrkA. Based on sequence alignment between the Trk family members, the kinase domain is well conserved compared with the JM region (Fig. 1B). Using residues 498–796 of TrkA as the kinase domain, we find that there is 75% identity between TrkA and TrkB and 72% identity between TrkA and TrkC. In contrast, for the JM region, using residues 440–497, there is 36% identity between TrkA and TrkB and 40% identity between TrkA and TrkC. To interrogate the need for a longer construct, the compounds were tested by surface plasmon resonance (SPR) for binding to the full intracellular region and a more compact kinase domain. Although there was some low-level binding of compound 1 and to a lesser extent, compounds 2 and 3, all three compounds demonstrated improved binding to the construct that contained the JM region in addition to the kinase domain (Fig. 1C).
Despite knowing that the entire intracellular region could bind the compounds, we wanted to identify a more compact construct that would be amenable to structural studies; thus, a more detailed analysis of the protein was required. A number of constructs had been generated that span the 60 residues between the transmembrane helices and the kinase domain (Fig. 1B). Construct 1 spans the entire intracellular portion of TrkA. Truncations were chosen so the starting residues are small to allow removal of the nonnative, initiating methionine during processing. To identify a minimal domain that could be used for crystallography, while minimizing disordered regions that could interfere with crystallization, these constructs were individually expressed and tested biochemically. A Lanthascreen assay was used to identify the minimal construct that could be inhibited by each compound (Fig. 1D). Whereas compound 1 could inhibit all constructs longer than the isolated kinase domain, compound 2 showed an increased ability to inhibit construct 5 but not construct 6. Compound 3 was unable to inhibit construct 5 or shorter constructs but was capable of inhibiting construct 4 and all longer constructs. SPR analysis confirmed the results of the biochemical results that construct 4 could bind all three compounds. Construct 4, the shortest construct that could be inhibited by all three compounds, encompasses residues 474–796, the native carboxy-terminus of TrkA. Construct 4 will be referred to as JM-kinase hereafter.
The Structures of Compounds 1 and 2 in Mode 1.
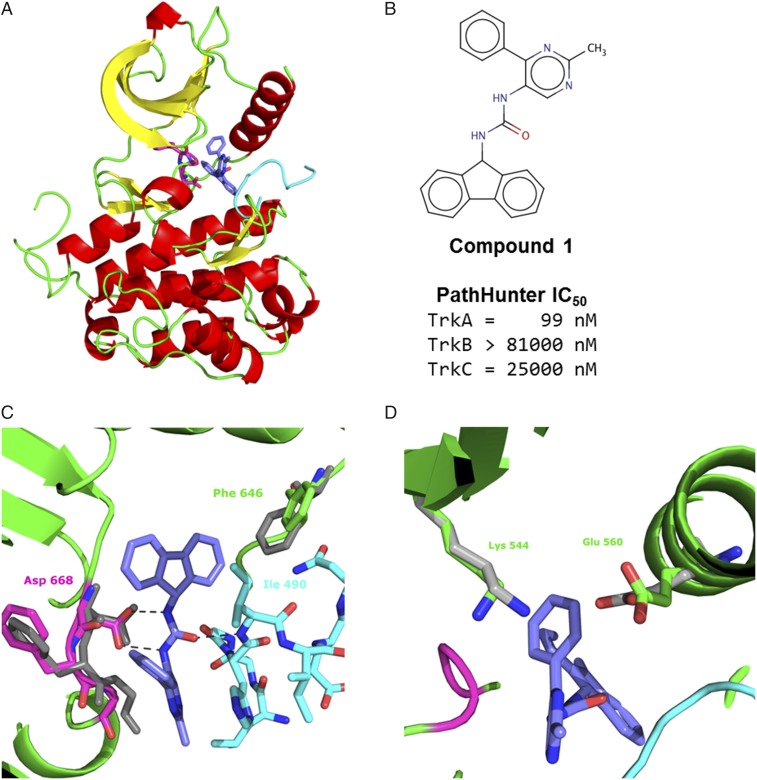
Compound 1 was the most potent compound identified in the ALIS screen. Compound 1 has an IC50 in a cell-based assay of 99 nM. In contrast, compound 1 demonstrated no inhibition against TrkB or TrkC at the upper limits of the assay (mid-micromolar range) (Fig. 2B). To determine the role the JM region in conveying efficacy of these inhibitors, we sought to determine the structures of the JM-kinase with the three inhibitors. Compound 1 was cocrystallized with the JM-kinase, revealing a binding mechanism on the surface of the kinase (Fig. 2A). (Table S1 for crystallographic statistics of the compounds described herein.) The inhibitor (shown as sticks with carbons in blue in Fig. 2A) sits behind the DFG motif (sticks with carbons in magenta in Fig. 2A) on the side opposite the kinase active site. The DFG motif is found in a DFG-out, inactive conformation, with Phe-669 pointed toward the active site. Asp-668, which would coordinate the phosphates on ATP in the kinase reaction, points away from the active site in the structure.
Fig. 2.
Structure of compound 1 in mode 1. (A) View of compound 1, in sticks with carbon atoms in blue, bound to the TrkA. The kinase domain is depicted in ribbons and colored by secondary structure with helices in red, strands in yellow, and loops in green. The DFG motif is shown in magenta as sticks. The JM is shown in cyan. (B) Cell-based, PathHunter IC50 values for compound 1 against Trks A, B, and C. (C) Zoom-in view of compound 1 binding. The central urea makes a bidentate interaction with Asp-668 of the DFG motif. Ile-490 sits above the fluorene, which is referred to as mode 1. Phe-646 of the kinase domain, which moves to accommodate the fluorene, is shown. For reference, the DFG motif and Phe-646 from the Apo kinase structure (PDB ID code 4F0I) is shown in gray. (D) The phenyl group of compound 1 sits between Lys-544 of the β3 strand and Glu-560 of helix c, causing them to move further from each other relative to their positions in the Apo structure (gray). Corresponding residues in active kinases form a salt bridge (44).
Table S1.
Crystallographic statistics
| Variables | Compound 1 | Compound 2 | Compound 3 | Compound 4 | Compound 5 | Compound 6 | Compound 7 |
| PDB ID Code | 5KMI | 5KMJ | 5KMK | 5KML | 5KMM | 5KMN | 5KMO |
| Wavelength (Å) | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Resolution range (Å) | 45.09–1.87 (1.937–1.87) | 22.55–2.04 (2.118–2.045) | 45.3–2.24 (2.325–2.245) | 22.89–2.01 (2.086–2.014) | 44.22–2.12 (2.196–2.12) | 44.3–2.14 (2.217–2.14) | 44.87–2.67 (2.768–2.672) |
| Space group | P3112 | P3112 | P3112 | P3112 | P3112 | P3112 | P3112 |
| Unit cell | a = 52.07 | a = 52.27 | a = 52.31 | a = 51.92 | a = 52.06 | a = 52.15 | a = 51.81 |
| b = 52.07 | b = 52.27 | b = 52.31 | b = 51.92 | b = 52.06 | b = 52.15 | b = 51.81 | |
| c = 227.19 | c = 225.525 | c = 224.89 | c = 230.96 | c = 226.03 | c = 228.53 | c = 229.55 | |
| α = β = 90 γ = 120 | α = β = 90 γ = 120 | α = β = 90 γ = 120 | α = β = 90 γ = 120 | α = β = 90 γ = 120 | α = β = 90 γ = 120 | α = β = 90 γ = 120 | |
| Total reflections | 294,696 (29,961) | 226,772 (22,577) | 170.646 (17,554) | 238,335 (22,879) | 196,299 (16,580) | 200,235 (20,050) | 101,105 (10,541) |
| Unique reflections | 29,752 (2,939) | 22,857 (2,257) | 17,308 (1,691) | 24,144 (2,394) | 20,453 (2,026) | 20,182 (1,987) | 10,402 (1,026) |
| Multiplicity | 9.9 (10.2) | 9.9 (10.0) | 9.9 (10.4) | 9.9 (9.6) | 9.6 (8.2) | 9.9 (10.1) | 9.7 (10.3) |
| Completeness (%) | 99.98 (100.00) | 99.98 (100.00) | 99.75 (99.59) | 100.00 (100.00) | 99.98 (100.00) | 100.00 (100.00) | 99.98 (100.00) |
| Mean I/σ(I) | 25.34 (4.19) | 19.56 (4.69) | 28.98 (4.72) | 27.06 (4.50) | 30.36 (3.83) | 11.30 (3.89) | 13.32 (4.98) |
| Wilson B-factor | 30.51 | 32.4 | 48.17 | 32.58 | 42.23 | 31.02 | 37.62 |
| Rmerge | 0.054 (0.59) | 0.077 (0.554) | 0.04716 (0.5275) | 0.05978 (0.5388) | 0.0451 (0.5801) | 0.145 (0.565) | 0.1556 (0.5287) |
| Rwork | 20.9 (27.6) | 19.2 (23.9) | 22.8 (31.4) | 20.3 (26.6) | 21.7 (29.9) | 19.4 (24.4) | 20.3 (25.6) |
| Rfree | 25.4 (30.3) | 23.1 (27.9) | 28.9 (34.3) | 24.0 (32.1) | 27.0 (35.1) | 22.6 (27.0) | 25.9 (30.0) |
| No. of nonhydrogen atoms | 2,511 | 2,488 | 2,460 | 2,431 | 2,431 | 2,449 | 2,319 |
| Macromolecules | 2,373 | 2,372 | 2,401 | 2,338 | 2,361 | 2,330 | 2,274 |
| Ligands | 30 | 25 | 28 | 30 | 27 | 31 | 23 |
| Water | 108 | 91 | 31 | 63 | 43 | 88 | 22 |
| Protein residues | 300 | 300 | 304 | 297 | 301 | 297 | 291 |
| RMS (bonds) | 0.014 | 0.014 | 0.014 | 0.013 | 0.014 | 0.014 | 0.014 |
| RMS (angles) | 1.65 | 1.67 | 1.77 | 1.69 | 1.78 | 1.69 | 1.86 |
| Ramachandran favored (%) | 98 | 97 | 94 | 99 | 96 | 99 | 94 |
| Ramachandran outliers (%) | 0 | 0 | 1 | 0.35 | 0 | 0.35 | 0.71 |
| Average B-factor | 38.2 | 39 | 57.8 | 40.2 | 51.9 | 37.7 | 40.6 |
| Macromolecules | 38.3 | 39.2 | 58 | 40.4 | 52.2 | 37.9 | 40.8 |
| Ligands | 32 | 28.9 | 52.2 | 35.5 | 44.8 | 25.9 | 27.6 |
| Solvent | 37.8 | 37.1 | 40.5 | 33.2 | 39.5 | 35.4 | 31.1 |
Numbers in parentheses represent statistics from the highest resolution bin.
The structure identifies key interactions between the inhibitor and JM-kinase. The central urea of compound 1 makes two hydrogen bonds of lengths 2.9 Å and 3.0 Å, with Asp-668 in the kinase domain (Fig. 2C). Asp-668 is part of the DFG motif, and binding to compound 1 requires a DFG-out conformation. The position of Asp-668 is more similar to the conformation in the Apo structure (in gray in Fig. 2C) than the inhibited DFG-out conformations (25). The fluorene moiety of compound 1 occupies a relatively hydrophobic pocket formed by the kinase residues, Leu-564, Leu-567, Ile-572, Val-573, Phe-589, Leu-641, Phe-646, His-648, Ile-666, and Gly-667.
The structure reveals clear interactions contributed by the JM region. Ile-490, within the JM, sits on top of the fluorene moiety, providing more of the hydrophobic pocket created by the kinase domain. Relative to other TrkA kinase structures, there is a shift of Phe-646, which allows room to accommodate the fluorene of the compound in creating the pocket. The oxygen of the central urea in compound 1 forms a 2.8-Å hydrogen bond with the amide nitrogen of Ile-490. The phenyl moiety occupies a position between Lys-544 of the β3 strand and Glu-560 of helix c. The Apo TrkA structure is in an inactive conformation, and compound binding displaces the two. In active kinases, these residues are positioned to form a salt bridge and the phenyl could further contribute to blocking the salt bridge formation (Fig. 1D).
In the structure, only a portion of the JM is resolved in electron density. The most C-terminal residue of the JM resolved is Gln-495, which leaves a gap of four residues before the first residue of the kinase domain that is resolved. Gln-495 is part of the NPQY motif that forms the docking site for the Shc phosphotyrosine binding domain. The tyrosine that becomes phosphorylated is not resolved in this structure. On the amino terminal side, Gly-485 is the first residue resolved in the structure, leaving the first 11 residues on the JM-kinase construct disordered in the structure. The resolved portion of the JM does not appear to form any discernable secondary structure.
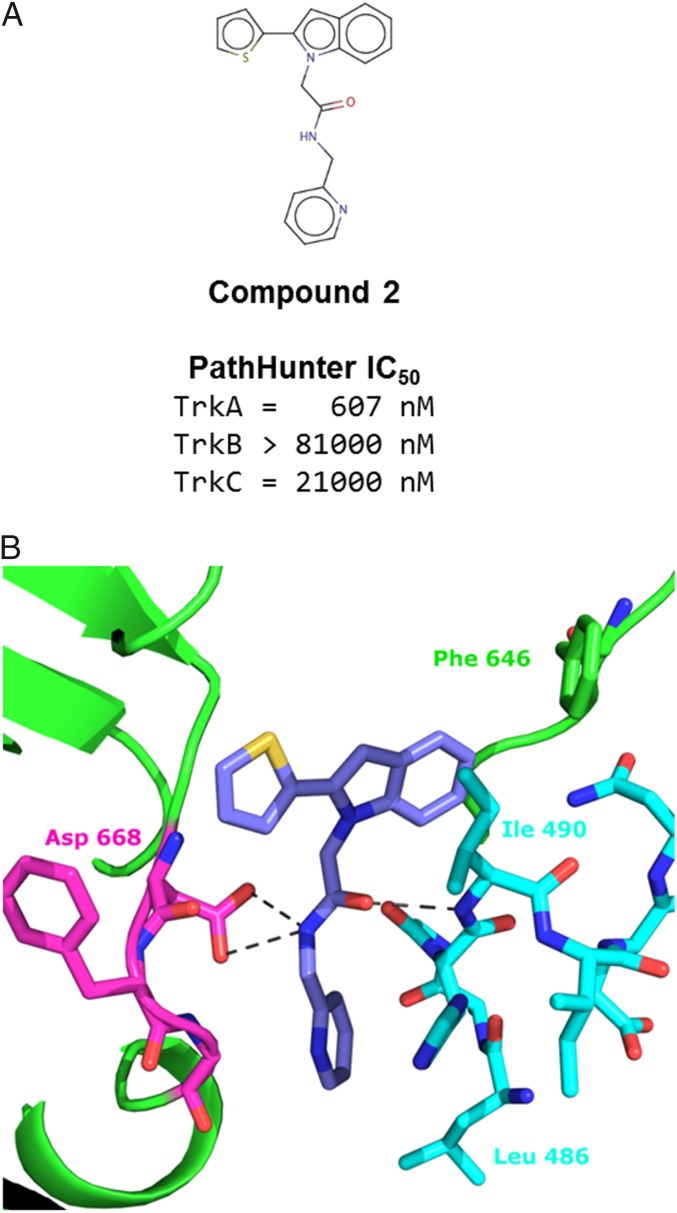
The second compound identified by the ALIS screen appeared to be less potent than compound 1. In cell-based assays, the IC50 was in the low nanomolar range against TrkA; however, compound 2 did not appear to inhibit TrkB or TrkC at concentrations tested in assays (Fig. 3A). The structure of compound 2 was found to bind with similar characteristics as compound 1 (Fig. 3B). The indole thiophene occupies the pocket that the fluorene of compound 1 occupied. The central amide oxygen makes a hydrogen bond (3.0 Å) with the amide nitrogen of Ile-490, similar to the urea oxygen of compound 1. In place of the two hydrogen bonds made by each of the urea nitrogens in compound 1, the single amide nitrogen of compound 2 sits at a distance capable of binding either oxygen of Asp-668, 3.0 Å and 3.3 Å from each. Again, this interaction requires Asp-668 to be positioned in a DFG-out orientation. In both structures, a bulky aromatic group binds in a pocket adjacent to the kinase domain. When bound, a surface is created for Ile-490 to pack on top of the aromatic group and complete the hydrophobic pocket. We refer to this as “mode 1 binding.”
Fig. 3.
Structure of compound 2 in mode 1. (A) Cell-based, PathHunter IC50 values for compound 2 against TrkA, TrkB, and TrkC. (B) Zoom-in view of compound 2 binding. The central amide nitrogen is positioned to allow interactions with both carboxyl oxygens of Asp-668. Ile-490 sits above the indole, and as such, is referred to as mode 1.
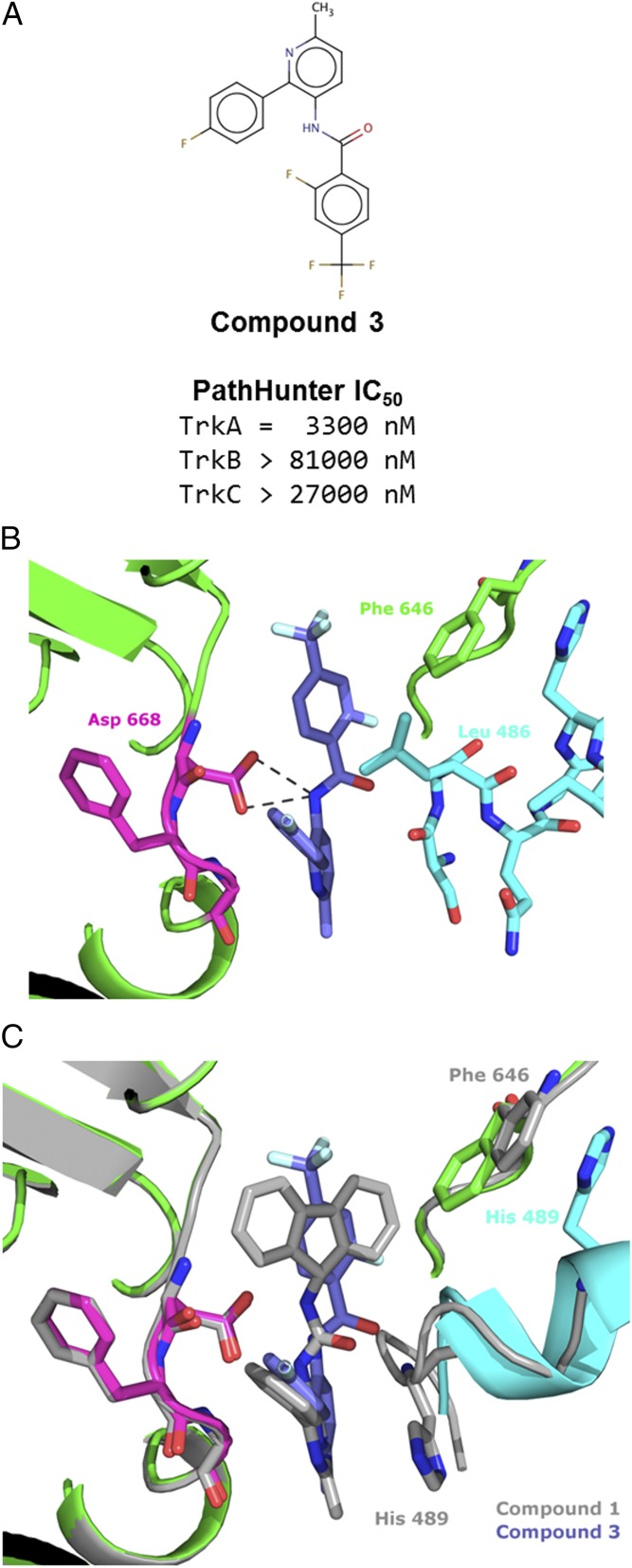
The Structure of Compound 3 in Mode 2.
Compound 3 was the weakest of the three, ALIS-identified inhibitors against TrkA. The cell-based IC50 was micromolar; however, it appeared to have at least 10-fold selectivity for TrkA vs. Trks B and C (Fig. 4A). The structure of compound 3 reveals binding to the same pocket, behind the DFG motif. The trifluorophenyl moiety sits in the pocket occupied by the fluorene of compound 1. The central amide nitrogen is positioned 3.2 Å and 3.3 Å from each of the carboxylate oxygens of Asp-668. When bound to compound 3, Phe-646 is not displaced from its Apo position as when bound to compounds 1 and 2 (Fig. 4C). Despite this difference, the kinase site is overall quite similar between the three structures.
Fig. 4.
Structure of compound 3 binding in mode 2. (A) Cell-based, PathHunter IC50 for compound 3 against Trks A, B, and C. (B) Zoom-in view of compound 3 binding. The central amide nitrogen is positioned to allow interactions with both carboxyl oxygens of Asp-668. Leu-486 sits above the fluorophenyl, which is referred to as mode 2. (C) Overlay of the structures of compounds 1 (gray) and 3, illustrating the change in the JM structure and movements of Phe-646 and His-489.
In contrast, the structure of the JM region is different in the compound 3 structure compared to those of compounds 1 and 2. Where Ile-490 packs above the aromatic rings in the first two structures, Leu-486 packs above the phenyl ring of the compound 3 structure. Mode 2 is distinguished from mode 1 (structure of compound 1 in gray for reference in Fig. 4C) by Leu-486 packing to form the hydrophobic pocket. As shown in overlay between the structures of compound 1 and compound 3 in Fig. 4C, comparison between modes 1 and 2 may best be illustrated by the shift in position of His-489, which has an 8.5-Å cα displacement between the two modes. In mode 2, the JM adopts a distinct helical, secondary structure. In mode 1, Phe-646 was displaced by the bulky moiety in the hydrophobic pocket but in mode 2, the smaller moiety accommodates Phe-646 in a position closer to its position in the Apo structure. The JM region that is resolved in the structure spans Ser-484 to Asn-493, slightly offset from those resolved in mode 1. This observation supports the biochemical data demonstrating the dependence of compound 3 on the longer construct.
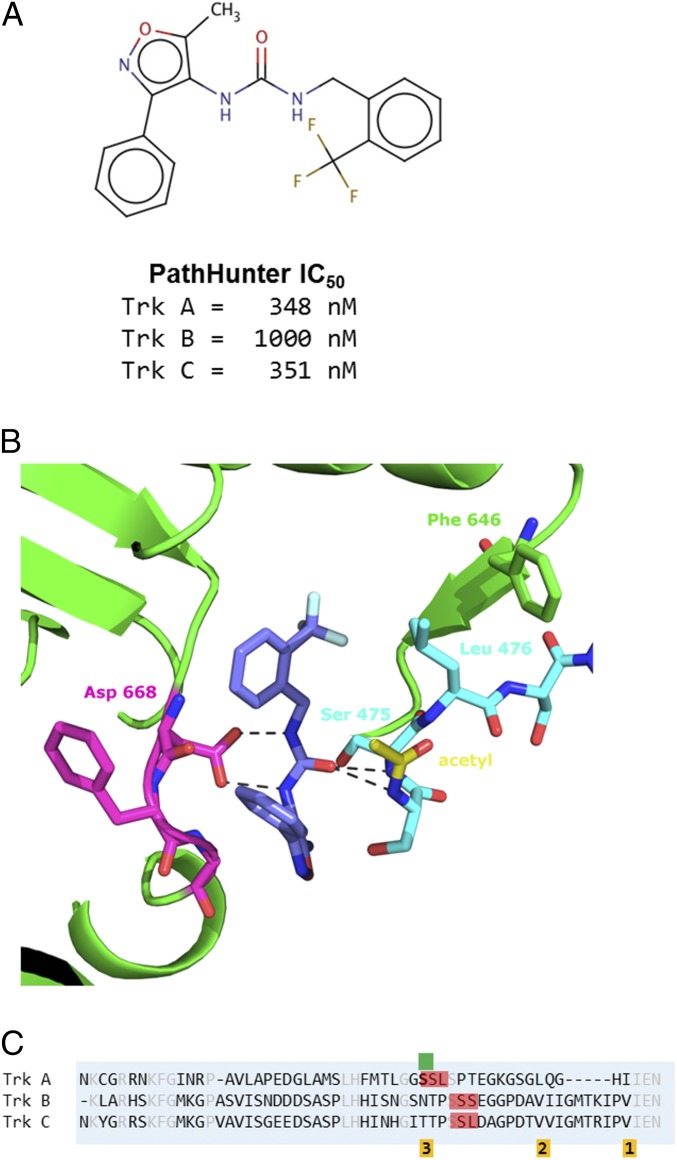
The Structure of a Non-TrkA Selective Compound in Mode 3.
With knowledge of the binding modes of the TrkA-selective compounds, it became apparent that other compounds had been identified by ALIS screening containing similar structures to these. Originally, the compounds were set aside because they did not exhibit TrkA selectivity. One such nonselective hit, compound 4, displayed submicromolar inhibition but comparable inhibition against both TrkA and TrkC (Fig. 5A). Compound 4 was cocrystallized with JM-kinase and, like the TrkA selective compounds, bound in a similar site behind the DFG motif (Fig. 5B). Asp-668 makes a pair of hydrogen bonds (2.8 Å for each) with the two urea nitrogens of compound 4, similar to compound 1. However, where Ile-490 or Leu-486 pack above the aromatic rings in modes 1 and 2 respectively, Ser-475—and to a lesser extent Leu-476—occupy this region. This conformation became mode 3 binding.
Fig. 5.
Structure of nonselective compound 4 binding in mode 3. (A) Cell-based, PathHunter IC50 values for compound 4 against Trks A and Trk C, showing that the compound is not TrkA-selective. (B) Zoom-in view of compound 4 binding. The central urea is positioned to make a bidentate interaction with the carboxyl oxygens of Asp-668. Ser-475 sits above the fluorophenyl, which is referred to as mode 3. A second JM residue, Leu-476, also occupies the pocket, forcing Phe-646 of the kinase domain to displaced further. The acetyl modification on the amino terminus of the expressed protein is shown with carbons in yellow. (C) Sequence alignment of the JM regions of Trks A, B, and C. The SSL and SSS sequences are indicated in red. The residues involved in packing against the hydrophobic pocket of modes 1, 2, and 3 are indicated below the sequence in orange. Serine 474, the first residue of the JM-kinase construct is indicated by the green marker.
The JM regions of TrkA and TrkC have low sequence identity, suggesting that addition of more nonconserved residues would lead to greater selectivity for TrkA; however, the compounds that bind to this region of the JM lose selectivity. From the structure, compound 4 interacts with residues 474-SSL-476 of the JM. As highlighted in red in the sequence alignment (Fig. 5C), a similar SSS motif is present in TrkB and an identical SSL motif is present in TrkC. Because Ser-474 is the first residue of the JM-kinase construct, the exact contribution of residues amino terminal in the native sequence cannot be determined. In the expression of JM-kinase, the initiating methionine is cleaved and the amino terminus is acetylated. The n-acetyl moiety is present in the structure, packing against the kinase. Although it is partially occluded by the JM, the position allows the backbone amine of Ser-474 to make a hydrogen bond to the urea oxygen of the compound. A bulky residue in place of the acetyl moiety would not likely be accommodated. In the native sequence, Gly-473 occupies a similar position. The comparable residue in TrkB and TrkC is proline.
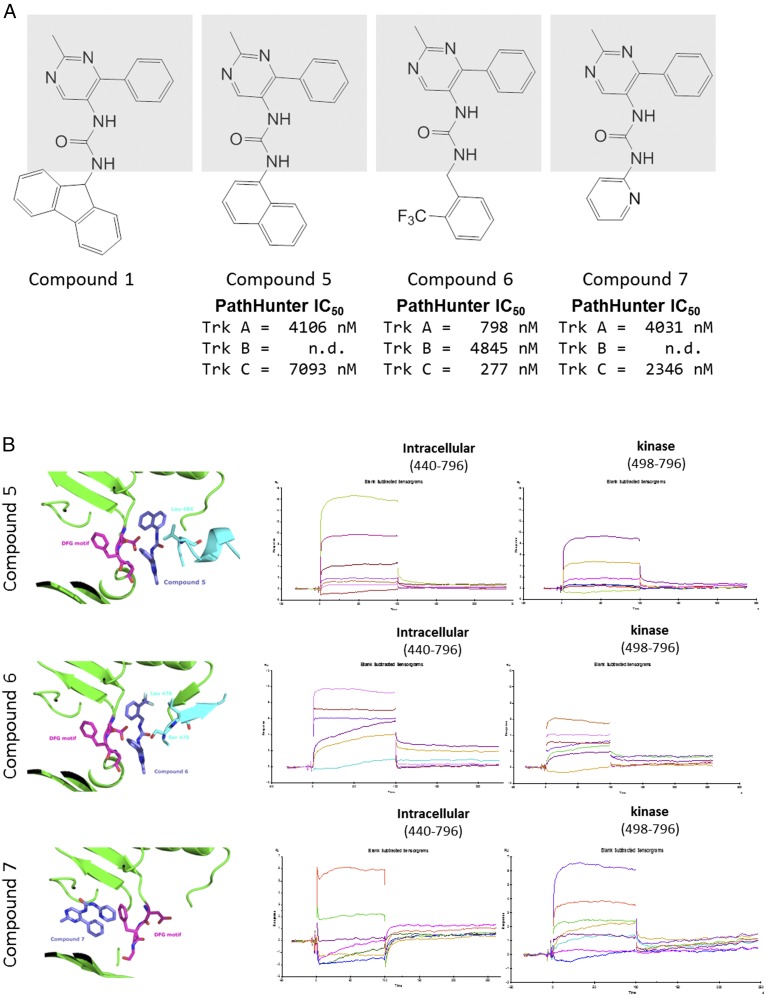
Binding Promiscuity of Similar Compounds.
From the series of structures determined in the different binding modes, we began to anticipate the binding behaviors of compounds beyond the expected structure–activity relationships one would expect from a chemical series. This can be demonstrated by the series of related compounds shown in Fig. 6A. All four compounds share the methyl pyrimidine phenyl group connected by a central urea found in compound 1 (shaded in Fig. 6A). By altering the group attached to the other side of the urea, the mode of interaction with the JM could be controlled. As shown above (Fig. 2C), compound 1 binds in mode 1 with Ile-490 packing against the fluorene. Substitution of the fluorene to a naphthalene changes the binding to mode 2 (Fig. 6B, Top), with Ile-486 packing against the naphthal moiety. Interactions with the kinase domain are similar but the JM interactions change. His-489 is shifted closer to the compound, as demonstrated previously by compound 3, another mode 2 binder. When a trifluoromethyl benzyl is used in compound 6, the compound binds as mode 3 (Fig. 6B, Middle), with Ser-474 packing above the phenyl ring. Finally, if the pyridine is added, the compound binds to the active site (Fig. 6B, Bottom), with the central urea interacting with the hinge region of the active site and the JM is disordered in the structure. Although all three compounds show some binding to the isolated kinase domain by SPR, only compound 7, which binds in the active site, appears to have comparable binding to the intracellular and isolated kinase constructs.
Fig. 6.
A series of compounds with disparate binding modes. (A) A series of compounds were chosen that share a central urea and common methylphenylpyrimidine on one end (gray box). Different substituents on the other side of the urea cause changes in binding mode. The PathHunter IC50 values are given below the compounds. For compounds 5 and 7, the TrkB value was not measured. (B) Structures of compounds 5, 6, and 7 are shown on the Left. On the Top Right is compound 5 bound in mode 2. In the Middle is compound 6 in mode 3. On the Bottom is compound 7 bound in the active site. In all three structures, the kinase is in green and the DFG motif is in magenta sticks. The JM is in cyan. On the Right are the corresponding SPR traces of the compounds with either full intracellular region (construct 1) or the isolated kinase.
Discussion
With ∼58 identified receptor tyrosine kinases (16), there is potential for obtaining selective inhibitors to other kinases with analogous JM interactions. Having a number of different assays aided the confidence to follow up on screening hits. Although the project was originally focused on active-site binders, alternative screening modalities were constantly used to identify new chemical matter. Robust cell-based assays against the different Trk kinases were needed to identify selective compounds.
Compounds with selectivity to TrkA, among the Trk family of kinases, are difficult to achieve with active-site inhibitors. The active site is well conserved among the Trk family. We have found compounds that bind outside the active site in an allosteric pocket on the distal side of the DFG motif. Despite binding to this region, interactions with the kinase domain are not unique to TrkA. Selectivity is achieved by interactions to residues of the less-conserved JM region, N terminal to the kinase. The structures described in this study illustrate three distinct modes of binding to compounds. This may appear as random ordering of the JM region; however, in determining structures in support of the project, binding appeared only in these three modes or in the active site. Additionally, when soaked into kinase crystals, most of these compounds did not bind in the absence of the JM. This observation raises the question of whether JM ordering may play a role physiologically.
JM regions have been modeled in the structures of other receptor tyrosine kinase structures. In the case of kinases such as EGFR, the JM plays a role in kinase transactivation by interacting with the carboxyl-terminal lobe of a donor kinase (29). The JM can also play a role in autoinactivation through interactions with the kinase domain. In the case of the receptor tyrosine kinase FLT3 (Fms-like tyrosine kinase 3), the aspartate of the DFG loop can make an ionic interaction with the backbone within the JM, locking it in an inactivated, DFG-out conformation (30). The TrkA binding site we demonstrate here, common to the three binding modes, also sequesters the aspartate in a DFG-out conformation; however, this sequestration is mediated through interactions with the inhibitor. In binding compounds, the JM becomes ordered and the characteristics of the inhibitor provide selectivity. We have observed that the JM does not appear to be ordered in the absence of compound binding. In other receptor tyrosine kinase structures, such as VEGFR, compounds have been involved in mediating the packing surface for the JM portion; however, these compounds all occupy space in the active site and could lead to some loss in selectivity (31).
It should be noted that the compound binding site seen in TrkA can potentially be induced in other kinases. Movements in kinase residues, like Phe-646, appear only when a compound binds. Specifically, in cases such as FLT3 and VEGFR, the compound could displace the JM in the structure and form a new mode of interaction with the DFG motif aspartate. This understanding emphasizes the importance of testing different constructs when performing screens. We believe that this could be applicable to other receptor tyrosine kinases generally.
The mode of compound binding correlates with selectivity and is largely determined by interactions with the JM region. Mode 1 binders tend to be TrkA-selective, whereas mode three binders were nonselective. Mode 2 binders could be TrkA-selective or -nonselective. In mode 1 binding, the Ile-490 of the JM region packs on the hydrophobic pocket and causes a significant shift in Phe-646. In the JM of TrkB and TrkC, the equivalent position appears to be a valine, which may not be sufficient to serve the same role. In mode 2 binding, Leu-486 packs on the hydrophobic pocket and Phe-646 is not significantly displaced relative to the Apo kinase structure. The comparable residue in TrkB and TrkC is again a valine, which may be able to partially substitute for the leucine of TrkA, necessitating cooperative interactions with other regions of the JM for selectivity.
The aim of the program is to design inhibitors of TrkA as a therapeutic for chronic pain; however, TrkA inhibitors could also be applicable for oncology. When considering TrkA as a target for cancer treatment, clinical data have emerged on resistance mutants from the pan-Trk inhibitor, Entrectinib. Russo et al. have recently identified two mutations arising from Entrectinib treatment in colorectal cancers, G595R and G667C (32). G595 is in the active site and has the greater effect of the two mutations when tested individually. However, G595 is greater than 10 Å away from the binding site identified in this report and our inhibitors should not be affected by the mutation. G667C, however, could present resistance to our compounds because the side chain could partially occlude the hydrophobic pocket that our compounds use. Although Entrectinib still appears to have submicromolar IC50 against this mutant, it could have a different and greater impact on our compounds. We would hypothesize that the mode 1 compounds would be more greatly affected by this mutation than the mode 2 compounds because of a bulkier moiety that binds to the hydrophobic pocket.
Using the Entrectinib:ALK structure (5FTO) as a model of active site inhibitors, we can also predict other residues that, when mutated, could affect our compounds within the new binding site (33). We predict four residues—Phe-589, Gly-667, Asp-668, and Phe-669—by identifying residues within 5 Å of both Entrectinib and compound 1 in TrkA. Of these residues, Gly-667 was described above. Asp-668 is critical to kinase function and should not appear as a resistance mutant. Phe-669, part of the DFG motif, is pointed away from our binding pocket and mutations may be tolerated. Phe-589 is the gatekeeper residue of TrkA. Although Phe-589 is greater than 4 Å from compound 1 and does not currently make direct interactions, it should be tracked as compounds are built for greater potency.
To evaluate the ability of the compounds to block TrkA with mutations that occur in cancer cells, we used the data curated in the Catalogue of Somatic Mutations in Cancer (COSMIC) (34). Of the 259 entries available at the time of evaluation, we focused on the 168 that were identified as missense substitutions and 1 in frame deletion. Of these 169 variants, we focused on the residues that contact the ligands from both modes 1 and 2 (Table S2). Based on this analysis, mutations of His-489, Leu-641, and Ile-66 could impact compounds within the binding site; these should be tested as an oncology program progresses in the discovery stage. Depending on the selectivity profile required, TrkA or pan-Trk selectivity can be achieved with these compounds.
Table S2.
Residues, compounds, and mutations
| Residue no. | Residue name | Compound 1 | Compound 2 | Compound 3 | Mutation(s) |
| 484 | Ser | X | |||
| 485 | Gly | X | |||
| 486 | Leu | X | X | X | |
| 487 | Gln | X | X | ||
| 488 | Gly | X | X | ||
| 489 | His | X | X | Y, Q | |
| 490 | Ile | X | X | ||
| 492 | Glu | X | |||
| 493 | Asn | X | X | ||
| 521 | Phe | X | X | X | |
| 544 | Lys | X | X | X | |
| 560 | Glu | X | X | X | |
| 564 | Leu | X | X | X | |
| 567 | Leu | X | X | X | |
| 572 | Ile | X | X | X | |
| 573 | Val | X | X | X | |
| 575 | Phe | X | |||
| 587 | Met | X | X | ||
| 589 | Phe | X | X | X | |
| 641 | Leu | X | X | X | M |
| 646 | Phe | X | X | X | |
| 647 | Val | X | X | ||
| 648 | His | X | X | X | |
| 650 | Asp | X | X | ||
| 666 | Ile | X | X | X | F |
| 667 | Gly | X | X | X | |
| 668 | Asp | X | X | X | |
| 669 | Phe | X | X | X | |
| 670 | Gly | X | X | X | |
| 673 | Arg | X | X | X |
Residues within interacting distance of compounds 1, 2, and 3 are indicated. Mutations that have been identified for those residues based on the data curated in COSMIC are indicated. If no mutations have been annotated for a residue, the cell is left blank.
Methods
Protein Expression.
Sequences of human TrkA were designed based on the sequence and numbering from Uniprot entry P04629. The carboxyl-terminal sequence, from residues 440–796, encode the entire cytoplasmic portion of the TrkA protein. Constructs were generated with amino termini starting from residues 440, 449, 461, 474, 481, 489, and 498, based on roughly even spacing and the type of residue that allow natural removal of the introduced initiating methionine. A carboxyl-terminal, 6-HIS tag was included in each construct. Proteins were expressed in Sf21 cells by baculoviral expression. Cells were pelleted by centrifugation and resuspended in lysis buffer containing 50 mM Hepes pH 7.0, 0.3N NaCl, 2 mM tris(2-carboxyethyl)phosphine (TCEP), 20 mM Imidazole, 0.3% β-octylglycoside, and 5% (vol/vol) glycerol at a ratio of 5 mL buffer per gram cell pellet. Benzonase was added at 25 U/mL and one Complete EDTA-free protease inhibitor tablet was added for each 50-mL buffer. Cells were homogenized by microfluidization and, after centrifugation, supernatant were loaded on a His Trap column. Following His purification, the protein was treated with λ-phosphatase to make a homogeneous, dephosphorylated sample. The protein was further purified by ion exchange on an SP HP column, followed by a Heparin HP column. The sample was concentrated and purified by size-exclusion chromatography (SEC) on a Superdex 75 16/60 column. The protein was concentrated to 10–12 mg/mL in a buffer containing 50 mM Mes pH 6.5, 5 mM TCEP, 150 mM NaCl, and 0.1% β-octylglycoside.
Crystallization.
Protein was concentrated to a concentration of 10 mg/mL in a buffer containing 50 mM Mes buffer pH 6.5, 5 mM TCEP, 150 mM sodium chloride, 0.1% β-octylglycoside. Compound was added to the protein from a 10 mM dimethyl sulfoxide stock to a final concentration of 250 μM and incubated at 4 °C for 1 h before crystallization.
Crystals were obtained by hanging-drop vapor diffusion over a reservoir solution containing 100 mM Mes buffer, 200 mM calcium chloride, and polyethylene glycol 3350 in using a range from 9 to 18%. To obtain crystals, streak seeding was usually performed after drops were allowed to equilibrate overnight. Crystals were harvested into a solution containing 100 mM Mes buffer, 200 mM calcium chloride, 15% (wt/vol) polyethylene glycol 3350, and 10% (vol/vol) ethylene glycol, and rapidly vitrified in liquid nitrogen.
Data were collected at the Advanced Photon Source on the Industrial Macromolecular Crystallography Associatio-Collaborative Access Team (IMCA-CAT; 17-ID) beamline. Data were processed and scaled with Global Phasing’s autoproc, running XDS (35). Phases were obtained by Fourier methods with other, in-house TrkA structures. Ligand dictionaries were generated using grade (36) and fit with Rhofit or Phenix ligandfit (37). Models were built and refined iteratively using Coot (38) and Buster or Phenix refine (37, 39). Images were rendered with Pymol (40).
Synthesis of Compounds.
The synthesis of the following compounds have been described in referenced patents: compound 1 (41), compound 2 (42), compound 4 (43), compound 5 (43), and compound 7 (43). For compound 3, the starting material 2-(4-fluorophenyl)-6-methylpyridin-3-amine (39 mg, 0.193 mmol) was dissolved in dichloromethane (2 mL); to this was added triethylamine (58.5 mg, 0.579 mmol) and 2-fluoro-4-(trifluoromethyl)benzoyl chloride (87 mg, 0.386 mmol). The resulting mixture was stirred for 1 h then purified by reverse-phase HPLC to yield the product 2-fluoro-N-(2-(4-fluorophenyl)-6-methylpyridin-3-yl)-4-(trifluoromethyl)benzamide (65 mg, 0.166 mmol). MS: m/z = 393.0 (M +1). For compound 6, a solution of 2-methyl-4-phenylpyrimidin-5-amine hydrochloride (20 mg, 0.090 mmol), 1-(isocyanatomethyl)-2-(trifluoromethyl)benzene (27.2 mg, 0.135 mmol), and triethylamine (0.025 mL, 0.180 mmol) in dichloromethane (1 mL) was stirred at 50 °C for 20 h. The dichloromethane was removed and the crude dissolved in N,N-dimethylformamide (1 mL). This material was purified by reverse-phase HPLC to yield the product 1-(2-methyl-4-phenylpyrimidin-5-yl)-3-(2-(trifluoromethyl)benzyl) (34.9 mg, 0.049 mmol). MS: m/z = 387.2 (M +1).
Lanthascreen.
To test for in vitro inhibition of kinase activity, a Lanthascreen assay (ThermoFisher Scientific) was used. For the reaction, 2 nM of the Trk kinase protein, 0.1 μM substrate peptide, and 75 μM ATP incubated for 2 h at room temperature. The reaction was quenched with 10 mM EDTA and incubated with 2-nM detection antibody Eu–anti-PY20 for 1 h at room temperature. The FRET signal was read on an Envision system (PerkinElmer). IC50 values were determined using a standard 4 parameter fit of the data.
Cell-Based PathHunter.
For cell-based activity measurements, we used a PathHunter Enzyme Fragment Complementation assay (DiscoveRx). The assay uses cell lines with the full-length TrkA, TrkB, or TrkC receptors containing a C-terminal fusion of the ProLink peptide. The peptide complements the β-galactosidase enzyme which is introduced as a fusion with the SH2 domain of SHC1. Upon activation of TrkA on the Shc-binding site and recruitment of SH2-fused, inactive β-galactosidase enzyme, the β-galactosidase is complemented by the peptide on the Trk receptors, leading to activation of the enzyme and readout by chemiluminescence. NGF, BDNF, or NT3 were used to activate TrkA, TrkB, and TrkC, respectively.
Surface Plasmon Resonance.
TrkA constructs containing the full intracellular region (containing residues 440–796 with a C-terminal 6-HIS tag) and the isolated kinase domain (containing residues 498–796 with a C-terminal 6-HIS tag) were captured on a CM5 sensor chip using amine coupling. Binding studies were conducted using a Biacore T-200 (GE Healthcare). Compounds were evaluated for binding to the proteins simultaneously and responses were normalized based on the highest response.
SI Methods
Affinity Selection–Mass Spectrometry.
ALIS, the affinity selection–mass spectrometry platform that was used here to screen TrkA, is a dual-chromatography LC-MS system that incorporates a SEC column to separate mixtures of unbound compounds from protein-binding ligands, and then separates and identifies the bound ligands from the protein target using reversed-phase LC-MS. The ALIS system and methodologies used here have been previously described (28, 45, 46).
TrkA was screened against small molecules using ALIS as follows. TrkA protein was formulated at a concentration of 1.4 mg/mL (34 μM) in 50 mM Mes pH 6.5, 5 mM TCEP, 150 mM NaCl, 0.1% octyl-glucoside. The protein was separately diluted to an enzyme concentration of 10 μM with 59 mM Hepes pH 7.5 buffer containing 150 mM NaCl, 5.9 mM MgCl2, and 0.25% n-octyl glucoside. For each ligand-binding reaction, 1.0 μL of 40× compound or compound mixtures was separately prediluted in 19 μL of 59 mM Hepes pH 7.5, 150 mM NaCl, 5.9 mM MgCl2. One-microliter of diluted protein was then mixed with an equal volume of buffer-solubilized compound mixture in a 96-well plate. Final target concentration was set at 5 μM TrkA in 50 mM Hepes, pH 7.5, 150 mM NaCl, 1 mM DTT, 5 mM MgCl2, 0.1% n-octyl glucoside, 2.5% (vol/vol) DMSO, and the final compound concentration was 1.0 μM per component for all reactions. After incubation at room temperature for 30 min, the samples, each containing a mixture of unbound compounds, protein–ligand complexes, and unliganded protein were cooled to 4 °C and then loaded onto an automated high-performance liquid chromatography system (Agilent 1100). Sample injections of 2 μL each were conducted on rapid SEC performed at 4 °C using a custom SEC column packed with proprietary gel-filtration media using a mobile phase of 0.7 M ammonium acetate pH 8 buffer running at a flow rate of 0.3 mL/min to rapidly separate protein–ligand complexes from unbound compounds. The eluent from the SEC column were directed through a UV detector (Agilent G1314A using a G1313 microflow cell) set at a UV absorbance of 230 nm to identify the protein–ligand complex. Upon detection, the complex was automatically diverted into a valve collection loop and then transferred to a reversed-phase chromatography column (Higgins Targa C18, 0.5 mm i.d × 50 mM length; Higgins Analytical Lab) running at 60 °C. Bound ligands are removed from the target protein, desalted, and eluted into the mass spectrometer using a solvent gradient of 0–95% (vol/vol) acetonitrile (0.1% formic acid) in water (0.1% formic acid) over 5 min using an Agilent G1376A capillary binary pump. Mass spectrometric detection was accomplished using a Waters LCT “Classic” or LCT “Premier” time-of-flight mass spectrometer operating in positive-ion ionization mode with a standard nebulized electrospray ionization source using a capillary voltage of 3.5 kV, desolvation temperature of 180 °C, source temperature of 100 °C, cone voltage set to 30 V, and extraction lens set to 3 V. Library mixtures that showed positive signals corresponding to calculated masses from known library components were repeated. Compounds of interest were each resynthesized as a single components, purified, and tested again against the TrkA target and also a negative control protein (Invertase) to rule out nonspecific binding. Those compounds that showed reproducible binding that were specific to the TrkA target were considered to be ALIS hits.
Synthesis of Mass-Encoded Screening Libraries.
Generation of the mass-encoded compound library mixtures used in the ALIS screening process was done using combinatorial chemistry techniques using a “core plus building block” approach that uses a template structure modified with a numerous different chemical moieties to yield a variety of compounds with broad structural diversity (28, 47). Compounds are synthesized as mixtures using software algorithms that enable multiple compounds to be generated in a fashion that minimizes mass overlap and allows each component to be identified by a unique molecular mass (47).
Acknowledgments
The authors thank Sujata Sharma and Scott Mosser for help in preparing the manuscript; Richard Hampton and Ronald Robinson for assistance with experiments; and Chris Cox for helpful discussions. Use of the IMCA-CAT beamline 17-ID (or 17-BM) at the Advanced Photon Source was supported by the companies of the Industrial Macromolecular Crystallography Association through a contract with Hauptman-Woodward Medical Research Institute. This research used resources of the Advanced Photon Source, a US Department of Energy Office of Science User Facility operated for the Department of Energy Office of Science by Argonne National Laboratory under Contract DE-AC02-06CH11357.
Footnotes
The work described in this manuscript was performed while all authors were employed at Merck and Co., Inc.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 5KMI, 5KMJ, 5KMK, 5KML, 5KMM, 5KMN, and 5KMO).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611577114/-/DCSupplemental.
References
- 1.Kalso E, Edwards JE, Moore RA, McQuay HJ. Opioids in chronic non-cancer pain: Systematic review of efficacy and safety. Pain. 2004;112(3):372–380. doi: 10.1016/j.pain.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 2.Pezet S, McMahon SB. Neurotrophins: Mediators and modulators of pain. Annu Rev Neurosci. 2006;29:507–538. doi: 10.1146/annurev.neuro.29.051605.112929. [DOI] [PubMed] [Google Scholar]
- 3.Hefti FF, et al. Novel class of pain drugs based on antagonism of NGF. Trends Pharmacol Sci. 2006;27(2):85–91. doi: 10.1016/j.tips.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Watson JJ, Allen SJ, Dawbarn D. Targeting nerve growth factor in pain: What is the therapeutic potential? BioDrugs. 2008;22(6):349–359. doi: 10.2165/0063030-200822060-00002. [DOI] [PubMed] [Google Scholar]
- 5.Wang T, Yu D, Lamb ML. Trk kinase inhibitors as new treatments for cancer and pain. Expert Opin Ther Pat. 2009;19(3):305–319. doi: 10.1517/13543770902721261. [DOI] [PubMed] [Google Scholar]
- 6.Vaishnavi A, Le AT, Doebele RC. TRKing down an old oncogene in a new era of targeted therapy. Cancer Discov. 2015;5(1):25–34. doi: 10.1158/2159-8290.CD-14-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Passiglia F, et al. The potential of neurotrophic tyrosine kinase (NTRK) inhibitors for treating lung cancer. Expert Opin Investig Drugs. 2016;25(4):385–392. doi: 10.1517/13543784.2016.1152261. [DOI] [PubMed] [Google Scholar]
- 8.Mitra G, Martin-Zanca D, Barbacid M. Identification and biochemical characterization of p70TRK, product of the human TRK oncogene. Proc Natl Acad Sci USA. 1987;84(19):6707–6711. doi: 10.1073/pnas.84.19.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Zanca D, Oskam R, Mitra G, Copeland T, Barbacid M. Molecular and biochemical characterization of the human trk proto-oncogene. Mol Cell Biol. 1989;9(1):24–33. doi: 10.1128/mcb.9.1.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parada LF, et al. The Trk family of tyrosine kinases: Receptors for NGF-related neurotrophins. Cold Spring Harb Symp Quant Biol. 1992;57:43–51. doi: 10.1101/sqb.1992.057.01.006. [DOI] [PubMed] [Google Scholar]
- 11.Kaplan DR, Hempstead BL, Martin-Zanca D, Chao MV, Parada LF. The trk proto-oncogene product: A signal transducing receptor for nerve growth factor. Science. 1991;252(5005):554–558. doi: 10.1126/science.1850549. [DOI] [PubMed] [Google Scholar]
- 12.Klein R, Jing SQ, Nanduri V, O’Rourke E, Barbacid M. The trk proto-oncogene encodes a receptor for nerve growth factor. Cell. 1991;65(1):189–197. doi: 10.1016/0092-8674(91)90419-y. [DOI] [PubMed] [Google Scholar]
- 13.Indo Y, et al. Mutations in the TRKA/NGF receptor gene in patients with congenital insensitivity to pain with anhidrosis. Nat Genet. 1996;13(4):485–488. doi: 10.1038/ng0896-485. [DOI] [PubMed] [Google Scholar]
- 14.Petty BG, et al. The effect of systemically administered recombinant human nerve growth factor in healthy human subjects. Ann Neurol. 1994;36(2):244–246. doi: 10.1002/ana.410360221. [DOI] [PubMed] [Google Scholar]
- 15.Woolf CJ, Safieh-Garabedian B, Ma Q-P, Crilly P, Winter J. Nerve growth factor contributes to the generation of inflammatory sensory hypersensitivity. Neuroscience. 1994;62(2):327–331. doi: 10.1016/0306-4522(94)90366-2. [DOI] [PubMed] [Google Scholar]
- 16.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298(5600):1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 17.Cowan-Jacob SW, Jahnke W, Knapp S. Novel approaches for targeting kinases: Allosteric inhibition, allosteric activation and pseudokinases. Future Med Chem. 2014;6(5):541–561. doi: 10.4155/fmc.13.216. [DOI] [PubMed] [Google Scholar]
- 18.Stephens RM, et al. Trk receptors use redundant signal transduction pathways involving SHC and PLC-γ 1 to mediate NGF responses. Neuron. 1994;12(3):691–705. doi: 10.1016/0896-6273(94)90223-2. [DOI] [PubMed] [Google Scholar]
- 19.Obermeier A, et al. Identification of Trk binding sites for SHC and phosphatidylinositol 3′-kinase and formation of a multimeric signaling complex. J Biol Chem. 1993;268(31):22963–22966. [PubMed] [Google Scholar]
- 20.Loeb DM, Stephens RM, Copeland T, Kaplan DR, Greene LA. A Trk nerve growth factor (NGF) receptor point mutation affecting interaction with phospholipase C-gamma 1 abolishes NGF-promoted peripherin induction but not neurite outgrowth. J Biol Chem. 1994;269(12):8901–8910. [PubMed] [Google Scholar]
- 21.McMahon SB. NGF as a mediator of inflammatory pain. Philos Trans R Soc Lond B Biol Sci. 1996;351(1338):431–440. doi: 10.1098/rstb.1996.0039. [DOI] [PubMed] [Google Scholar]
- 22.Abdiche YN, Malashock DS, Pons J. Probing the binding mechanism and affinity of tanezumab, a recombinant humanized anti-NGF monoclonal antibody, using a repertoire of biosensors. Protein Sci. 2008;17(8):1326–1335. doi: 10.1110/ps.035402.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lane NE, et al. Tanezumab for the treatment of pain from osteoarthritis of the knee. N Engl J Med. 2010;363(16):1521–1531. doi: 10.1056/NEJMoa0901510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Artim SC, Mendrola JM, Lemmon MA. Assessing the range of kinase autoinhibition mechanisms in the insulin receptor family. Biochem J. 2012;448(2):213–220. doi: 10.1042/BJ20121365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bertrand T, et al. The crystal structures of TrkA and TrkB suggest key regions for achieving selective inhibition. J Mol Biol. 2012;423(3):439–453. doi: 10.1016/j.jmb.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Choi H-S, et al. (R)-2-Phenylpyrrolidine substituted imidazopyridazines: A new class of potent and selective pan-TRK inhibitors. ACS Med Chem Lett. 2015;6(5):562–567. doi: 10.1021/acsmedchemlett.5b00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stachel SJ, et al. Maximizing diversity from a kinase screen: identification of novel and selective pan-Trk inhibitors for chronic pain. J Med Chem. 2014;57(13):5800–5816. doi: 10.1021/jm5006429. [DOI] [PubMed] [Google Scholar]
- 28.Annis DA, et al. An affinity selection–mass spectrometry method for the identification of small molecule ligands from self-encoded combinatorial libraries: Discovery of a novel antagonist of E. coli dihydrofolate reductase. Int J Mass Spectrom. 2004;238(2):77–83. [Google Scholar]
- 29.Red Brewer M, et al. The juxtamembrane region of the EGF receptor functions as an activation domain. Mol Cell. 2009;34(6):641–651. doi: 10.1016/j.molcel.2009.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffith J, et al. The structural basis for autoinhibition of FLT3 by the juxtamembrane domain. Mol Cell. 2004;13(2):169–178. doi: 10.1016/s1097-2765(03)00505-7. [DOI] [PubMed] [Google Scholar]
- 31.McTigue M, et al. Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc Natl Acad Sci USA. 2012;109(45):18281–18289. doi: 10.1073/pnas.1207759109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Russo M, et al. Acquired resistance to the TRK inhibitor Entrectinib in colorectal cancer. Cancer Discov. 2016;6(1):36–44. doi: 10.1158/2159-8290.CD-15-0940. [DOI] [PubMed] [Google Scholar]
- 33.Menichincheri M, et al. Discovery of Entrectinib: A new 3-aminoindazole as a potent anaplastic lymphoma kinase (ALK), c-ros oncogene 1 kinase (ROS1), and pan-Tropomyosin receptor kinases (Pan-TRKs) inhibitor. J Med Chem. 2016;59(7):3392–3408. doi: 10.1021/acs.jmedchem.6b00064. [DOI] [PubMed] [Google Scholar]
- 34.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue) D1:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vonrhein C, et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):293–302. doi: 10.1107/S0907444911007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smart OS, et al. 2011. Grade. (Global Phasing Ltd., Cambridge, UK), Version 1.2.9.
- 37.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 40. Schrödinger, LLC. (2014) The PyMOL Molecular Graphics System (Schrödinger, LLC., New York), Version 1.7.
- 41.Cooke AJ, Stump CA, Zhang X-F, Li CS, Mao Q. 2015 WO2015042088A1: TrkA Kinase Inhibitors, Compositions and Methods thereof. Available at www.google.com/patents/WO2015042088A1, Accessed April 29, 2016.
- 42.Cooke AJ, et al. 2016 WO2016054807: TrkA Kinase Inhibitors, Compositions and Methods Thereof. Available at https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2016054807&recNum=2&docAn=CN2014088321&queryString=EN_ALL:nmr%20AND%20PA:merck&maxRec=12704, Accessed April 29, 2016.
- 43.Cooke AJ, Stump CA, Zhang X-F, Li CS, Mao Q. 2015 WO2015042085A3: TrkA Kinase Inhibitors, Compositions and Methods Thereof. Available at www.google.com/patents/WO2015042085A3, Accessed April 29, 2016.
- 44.Cowan-Jacob SW. Structural biology of protein tyrosine kinases. Cell Mol Life Sci. 2006;63(22):2608–2625. doi: 10.1007/s00018-006-6202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Annis A, Chuang C-C, Nazef N. ALIS: An affinity selection–mass spectrometry system for the discovery and characterization of protein–ligand interactions. In: Wanner KT, Höfner G, editors. Mass Spectrometry in Medicinal Chemistry. Wiley-VCH; Hoboken, NJ: 2007. pp. 121–156. [Google Scholar]
- 46.Sheth PR, et al. Novel benzimidazole inhibitors bind to a unique site in the kinesin spindle protein motor domain. Biochemistry. 2010;49(38):8350–8358. doi: 10.1021/bi1005283. [DOI] [PubMed] [Google Scholar]
- 47.Nash HM, et al. 1999 Method for producing mass-coded combinatorial libraries. Available at www.google.com/patents/WO1999035109A1, Accessed April 15, 2016.