Significance
CD74 has been associated with tumor progression and metastasis. Its expression has been suggested to serve as a prognostic factor in many cancers, with higher relative expression behaving as a marker of tumor progression. Our previous studies showed that stimulation of CD74 expressed on chronic lymphocytic cells initiates a signaling cascade leading to chronic lymphocytic survival. The present study demonstrates that CD74’s cytoplasmic domain binds chromatin and regulates transcription and expression of genes involved in immune regulation, cell survival, and hematopoietic cancers. Further analysis of these processes will identify new targets that regulate tumor cell maintenance.
Keywords: CD74, transcription, CLL, NF-κB, RUNX
Abstract
CD74 is a cell-surface receptor for the cytokine macrophage migration inhibitory factor. Macrophage migration inhibitory factor binding to CD74 induces its intramembrane cleavage and the release of its cytosolic intracellular domain (CD74–ICD), which regulates cell survival. In the present study, we characterized the transcriptional activity of CD74–ICD in chronic lymphocytic B cells. We show that following CD74 activation, CD74–ICD interacts with the transcription factors RUNX (Runt related transcription factor) and NF-κB and binds to proximal and distal regulatory sites enriched for genes involved in apoptosis, immune response, and cell migration. This process leads to regulation of expression of these genes. Our results suggest that identifying targets of CD74 will help in understanding of essential pathways regulating B-cell survival in health and disease.
The type II transmembrane protein CD74 is expressed on antigen-presenting cells and was initially demonstrated to function as an MHC class II chaperone (1). At steady state, a small fraction of CD74 is expressed on the cell surface. Some of this cell-surface pool of CD74 is modified by the addition of chondroitin sulfate (2). Cell surface CD74 serves as a receptor for the cytokine, macrophage migration inhibitory factor (MIF) on many cell types (3). In immune cells, MIF binding to CD74 induces a signaling cascade that results in regulation of cell proliferation and survival (3).
In B cells, CD74 expression is directly involved in shaping the B-cell repertoire by regulating mature B-cell survival (4). MIF binding to CD74 induces a signaling pathway that involves the Syk tyrosine kinase and the PI3K/Akt pathway, induction of CD74 intramembrane cleavage, and the release of the CD74 intracellular domain (CD74–ICD) (5, 6). CD74–ICD translocates to the nucleus, where it induces activation of transcription mediated by the NF-κB RelA protein and its coactivator, TAFII105, resulting in regulation of transcription of genes that control B-cell proliferation and survival (5–9). Thus, MIF binding to CD74 initiates a cascade that results in B-cell proliferation and the rescue of these cells from apoptotic death.
Interestingly, both MIF and CD74 are associated with tumor progression and metastasis. CD74 expression has been suggested to serve as a prognostic factor in many cancers, with higher relative expression behaving as a marker of tumor progression (10). Our previous studies showed that stimulation of CD74 expressed on chronic lymphocytic leukemia (CLL) cells with the MIF ligand, as well as with anti-CD74 agonistic antibody, initiates a signaling cascade leading to CD74 intramembrane cleavage, NF-κB activation, and elevation of TAp63 expression (11, 12). TAp63 regulates transcriptional elevation of several genes in CLL, among them IL-8, which promotes CLL cell survival regardless of the clinical status of the patients (11, 12). In addition, we showed that stimulation of CD74 with MIF induces the expression and secretion of the cytokine, midkine. Midkine suppresses CLL apoptosis by elevating the expression of Bcl-2 and inhibiting caspase 3 and 7 activity (13). Thus, CD74 functions as a survival receptor on CLL cells. Furthermore, a positive correlation between CD74 and ZAP70 expression was detected in CLL patients, and high expression of CD74 was positively correlated with more advanced stage of the disease (14).
Quite a few regulatory proteins, including transcription factors, are kept in a dormant state to be activated in response to internal or environmental cues. Previously, a novel strategy, requiring proteolytic cleavage, was described for the mobilization of dormant transcription factors. These transcription factors are initially synthesized in an inactive form, while “nesting” in integral membrane precursor proteins. Following a cleavage event, these new active factors are released from the membrane and can migrate into the nucleus to drive regulated gene transcription. This mechanism, “regulated intramembrane proteolysis” (RIP), is known for its conservation in several systems, and for its ability to control diverse biological processes in prokaryotes and eukaryotes in response to a variety of signals (15). Examples include membrane proteins such as Notch, a plasma membrane receptor, whose cytosolic domain is released throughout development (16).
Because CD74 is cleaved in the plane of the membrane in a manner similar to other RIP proteins, in the present study we wished to characterize the transcription regulation activity of CD74–ICD and map the specific DNA binding sites and identify the genes it activates. Our findings demonstrate that CD74–ICD binds chromatin and regulates transcription and expression of genes involved in cell survival and the immune response in health and disease.
Results
CD74 Enters the Nucleus and Binds Chromatin in Proximity to Genes Related to Immune Regulation and Immune Diseases.
To characterize CD74–ICD function as a transcriptional regulator that interacts with the nuclear chromatin, chromatin-binding CD74–ICD was immunoprecipitated and sequenced (ChIP-seq). To calibrate the procedure, we performed ChIP-seq of CD74 in CD74–ICD-transfected 293T cells (5). This experiment yielded fewer than 50 reproducible peaks compared with control IgG. One example is presented in Fig. S1A.
Fig. S1.
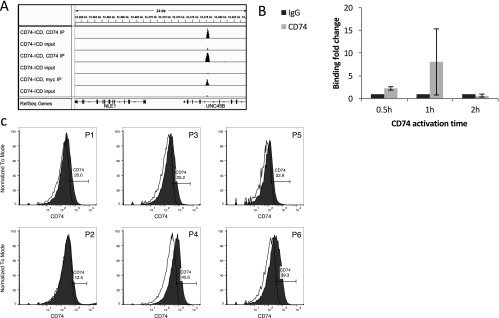
CD74 binding to the chromatin. (A) 293T cells were transfected with CD74–ICD–myc plasmid. ChIP-seq was done using CD74 or myc antibodies. Binding of CD74 in the proximity of the NLE1 gene was detected with both antibodies. (B) CLL cells were activated with anti-CD74 Ab for 0.5 h, 1 and 2 h. Following activation, the cells were fixed, and Chip-RT was conducted using CD74 Ab. qRT-PCR of a binding area in the proximity of the NLE1 gene was used to determine binding levels of CD74 to the chromatin in CLL cells. Binding of CD74–ICD to the chromatin is detected 0.5 and 1 h following activation (n = 2). (C) CD74 cell-surface expression on CLL cells was determined by FACS analysis.
Next, we wished to determine CD74–ICD binding to the chromatin in B cells. Because the B-cell leukemia cells, CLL, overexpress CD74 compared with healthy mature B cells, and CD74–ICD is released at higher levels in these cells (11), we followed CD74–ICD binding to the chromatin in CLL cells. CLL cells were activated with anti-CD74 activating Ab for 0.5, 1, and 2 h. The cells were then fixed with formaldehyde and di(N-succinimidyl). Binding to the chromatin was followed by quantitative RT-PCR (qRT-PCR) analysis of the binding regions that were identified in the 293T-transfected cells (Fig. S1B). The highest binding was detected 1 h postactivation; therefore, this time-point was used in our further studies.
Next, we tested CD74 binding to the chromatin in human cells derived from three CLL patients with progressive disease. One patient was analyzed before (P1) and after (P2) Rituximab-treatment, with samples taken 1-y apart (Table S1). All samples expressed cell-surface CD74 (Fig. S1C). The cells were activated for 1 h with IgG or anti-CD74 antibodies. The cells were then fixed with formaldehyde and di(N-succinimidyl) glutarate (DSG), to improve cross-linking of protein–protein interactions (17), and CD74 was immunoprecipitated. There was almost no binding of CD74–ICD to the chromatin in nonactivated cells, and ChIP-seq could not be performed because of the low amount of DNA obtained, demonstrating that activation of CD74 induces its intramembrane cleavage and binding to the chromatin. Clear peaks were detected in CD74-activated cells compared with the input background (Fig. 1A). Significant binding of CD74 to the chromatin was detected in all samples, at several hundred to several thousand binding sites, with no correlation to cell-surface CD74 levels (Table S2), and major overlap between the peaks in different samples (Table S3). As expected, the highest overlap was observed in the P1 and P2 peaks from the same patient, although the vast majority (89%) of these shared peaks was shared by the other samples as well. The high overlap between the peaks in the different samples implies that CD74 has a specific binding pattern in CLL cells, supporting a role for this fragment as a transcription regulator in CLL cells. Analysis of the total peaks shared by different samples revealed 3,575 peaks that were detected in at least two of the four samples. These peaks were treated as high-confidence peaks, because they appeared in more than one sample, and were chosen for further analysis. As many as 766 peaks were shared in all four samples (total peaks shared by the samples: two and up, 3,575; three and up, 1,697; all four, 766).
Table S1.
Patient list
| Patient | Age (y) | Sex | Binet (37) | WBC* | Disease state | Previous therapy | CD38 | ZAP70 | Experiments |
| P1 | 70 | M | B | 138.8 | PD | No | 5 | 26 | siRNA, ChIP-Seq (CD74) |
| P2 | 71 | M | B | 51.8 | PD | FCR | 1 | 26 | ChIP-Seq (CD74) |
| P3 | 57 | F | A | 68.6 | PD | No | N/D | N/D | RNA-Seq, ChIP-Seq (Me2, Me3, CD74) |
| P4 | 75 | M | B | 45.5 | PD | BR | 79 | 40 | ChIP-Seq (Ac, CD74) |
| P5 | 62 | M | B | 309.10 | PD | FCR | 8 | N/D | RNA-Seq |
| P6 | 81 | M | B | 108.41 | PD | No | 10 | 1 | RNA-Seq, ChIP-Seq (Me2, Me3) |
| P7 | 61 | M | B | 173.0 | PD | No | 11 | 11 | ChIP-Seq (Ac) |
| P8 | 72 | M | A | 50 | SD | No | N/D | N/D | siRNA |
| P9 | 64 | F | C | 150.2 | SD | No | 4 | 0 | siRNA |
| P10 | 73 | F | A | 145.0 | PD | No | 0 | pos | siRNA |
| P11 | 58 | M | B | 174.0 | PD | No | 0 | N/D | siRNA |
| P12 | 72 | M | B | 55.4 | PD | No | 2 | 1 | siRNA |
| P13 | 60 | M | C | 100.0 | PD | No | 44 | 0 | siRNA, CD74 stim |
| P14 | 60 | F | A | 101.8 | PD | No | 8 | N/D | siRNA |
| P15 | 61 | M | A | 113.5 | SD | No | 0 | 0 | CD74 stim |
| P16 | 69 | M | C | 47.0 | SD | FCM | 2 | N/D | CD74 stim |
The fourth column indicates the stage of the disease as defined by Binet (37). BR, bendamustine/rituximab; F, female; FCM, fludarabine/cyclophosphamide/mitoxantrone; FCR, fludarabine/cyclophosphamide/rituximab; M, male; ND, not determined; PD, progressive disease; pos, positive; SD, stable disease; WBC, white blood cells.
×103/µL.
Fig. 1.
CD74–ICD binds the chromatin in regulatory areas. CLL cells from four different samples were stimulated for 1 h with anti-CD74 Ab. ChIP-seq was then performed using anti-CD74 or isotype control antibodies. Binding of CD74 to the chromatin was compared with the input. (A) A representative analysis of binding of CD74–ICD to a promoter region. ChIP-Seq peaks are shown for anti-CD74 and input DNA of bigWig files of two representative experiments. (B) Binding was analyzed in all four samples and overlaps were examined. Peaks that were found in two or more samples were considered as high-confidence peaks, and their distances (kilobases) from the nearest TSS were analyzed using GREAT. (C) The distribution of important genomic features in the whole genome (Left) and in CD74 ChIP-seq peaks (Right) was obtained from CEAS. A representative analysis is shown. (D) CLL cells from two different patients were stimulated for 1 h with anti-CD74 Ab. ChIP-seq was performed with anti-H3K27Ac, anti-H3K4Me3, or anti-H3K4Me2. The diagram shows the overlap between CD74 peaks and the acetylation and methylation peaks detected by these antibodies. (E) CLL cells from two different patients were stimulated for 1 h with either anti-CD74 Ab or IgG control Ab. ChIP-seq was done with anti-H3K4Me2. The ngs.plot tool was used to plot average reads per million mapped reads (RPKM) over high-confidence CD74 peaks. The center on the x axis relates to the summits of the peaks. The SE is shaded. The diagram shows the correlation between CD74 peaks and H3K4Me2, with (orange) and without (green) CD74 activation. WCE, whole cell extract.
Table S2.
Correlation between CD74 cell-surface expression and the number of peaks binding the chromatin in each patient
| Patient | No. of peaks | CD74 experiment level |
| P1 | 5,520 | 20 |
| P2 | 1,694 | 12.5 |
| P3 | 1,774 | 25.2 |
| P4 | 6,530 | 45.8 |
Table S3.
The overlap between the peaks in the different samples
| Patient | P1 (%) | P2 (%) | P3 (%) | P4 (%) |
| P1 | 100.0 | 83.8 | 70.4 | 52.0 |
| P2 | 83.8 | 100.0 | 51.7 | 81.1 |
| P3 | 70.4 | 51.7 | 100.0 | 80.8 |
| P4 | 52.0 | 81.1 | 80.8 | 100.0 |
Binding patterns of transcriptional regulators in the genome are typically enriched in enhancer and promoter regions, where they regulate the histone structure and transcription (18). To mediate this function, the regulators interact with the transcription start site (TSS). This interaction can occur via trans binding, distal to the TSS, and looping of the chromatin, or by cis binding in proximity to the TSS. cis binding is more frequent and can be easily recognized in a ChIP-seq experiment (19). To determine the binding pattern of CD74, GREAT (Genomic Regions Enrichment of Annotations Tool) analysis was used (20). This analysis attributes each peak to its adjacent genes. As shown in Fig. 1B, the majority of high confidence peaks were located within 50 kb of a known TSS, and almost one-third of the peaks were detected within 5 kb of a known TSS.
The CD74 binding pattern was further analyzed by the cis Regulatory Element Annotation System (CEAS) analysis of the binding regions (21). The analysis identified enhanced binding in several genomic regions: promoters, downstream of genes (defined as up to 3,000 bp 3′ to the transcription termination sites), and at the 5′ UTR (Fig. 1C). Because these are regions of transcription factor binding, this binding pattern further strengthens the notion that CD74 exhibits transcription regulation function.
GREAT and CEAS analyses are based solely on the pattern of binding compared with known features of the genome. However, different cell types exhibit different activity patterns of enhancers and promoters. Histone acetylation and methylation patterns have been shown to mark regulatory areas (22). To determine whether CD74 binds active regulatory regions, we performed ChIP-seq on cells activated with an agonistic anti-CD74 Ab for 1 h and detected various chromatin regions: H3K27Ac, a marker for active enhancers and promoters (23); H3K4me2, a marker for active promoters, and active and poised enhancers; and H3K4Me3, a marker for promoters (24). As can be seen in Fig. 1D, as many as 23%, 70%, and 92.6% of the CD74 peaks were bound by H3K4Me3, H3K27Ac, and H3K4me2, respectively. Furthermore, CD74 activation led to up-regulation of H3K4me2 in areas of CD74 binding, which is linked to active transcription (Fig. 1E). Thus, CD74 activation leads to changes in the landscape of the genome methylation pattern. The released CD74–ICD binds the genome in these regulatory areas.
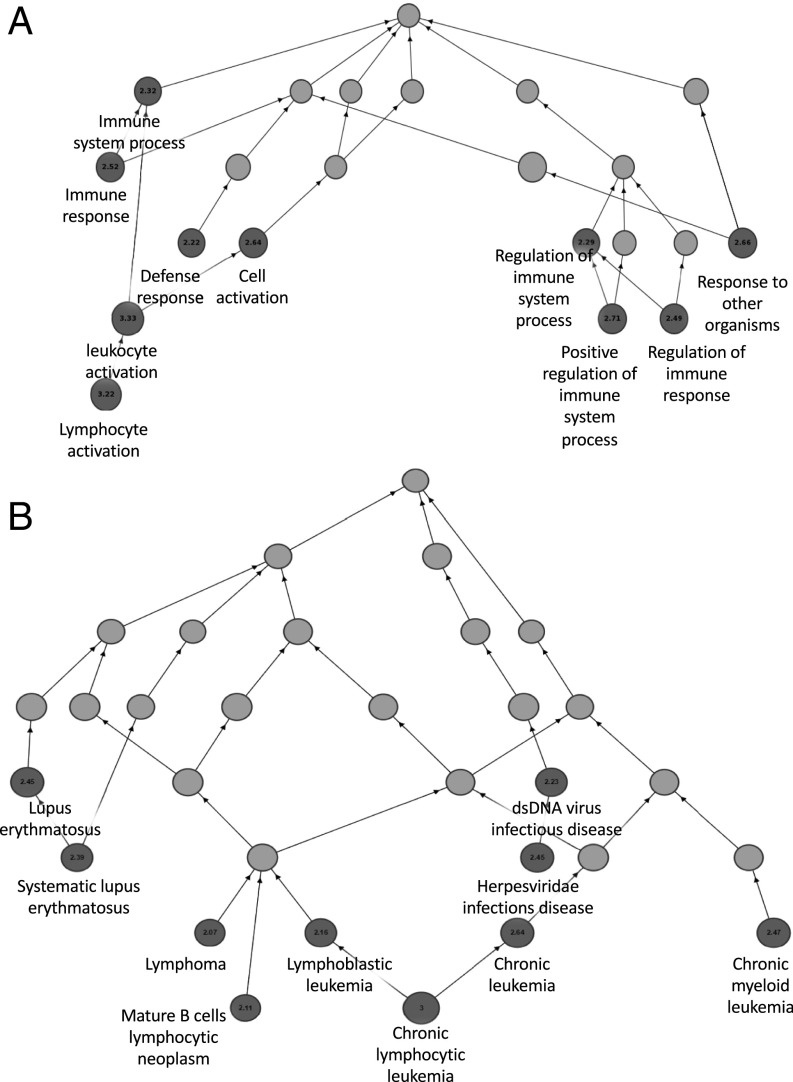
ChIP-seq analysis of CD74 binding to the chromatin in activated CLL cells yielded several hundred to several thousand peaks for each patient. This relatively small number of peaks indicated that CD74 binding to the chromatin is limited to genes controlling specific cellular processes. We next aimed to identify the processes regulated by CD74. GREAT analysis revealed that CD74 binding to the genome in CLL activated cells was specific to pathways related to the immune system. The predominant biological processes found were related to immune activation and response (Fig. 2A). In addition, analysis of diseases related to the genes associated with the CD74–ICD binding peaks revealed association to hematopoietic cancers, including CLL, acute lymphoblastic leukemia, and other immune disorders (Fig. 2B).
Fig. 2.
CD74 has a specific immune oriented role as a transcription factor. CLL cells from four samples were activated with anti-CD74 activating Ab for 1 h. Following activation, the cells were fixed, and ChIP-seq was conducted using CD74 Ab. GREAT analysis of the high confidence peaks was performed. The local directed acyclic graph is shown. The numbers relates to the binomial fold-change. (A) The biological processes controlled by CD74 identified by Gene Ontology (GO) analysis. (B) Enriched terms in disease ontology showing that CD74 specifically binds in the proximity of genes that are involved in diseases relating to lymphocytes, specifically in different types of lymphomas. dsDNA, double-stranded DNA.
CD74 Activation Leads to Regulation of Transcription.
We next wished to correlate the regions bound by CD74–ICD to genes whose expression is regulated by CD74. To this end, an RNA-seq analysis of mRNA levels in CD74 activated cells was performed. CLL cells from three different patients were stimulated with anti-CD74 or isotype control IgG antibodies. After 2- or 8-h stimulation, mRNA was extracted and sequenced. mRNA-seq analysis demonstrated a significant change (absolute change >2; false-discovery rate < 0.05) in 106 genes 2 h postactivation, and in 584 genes 8 h postactivation.
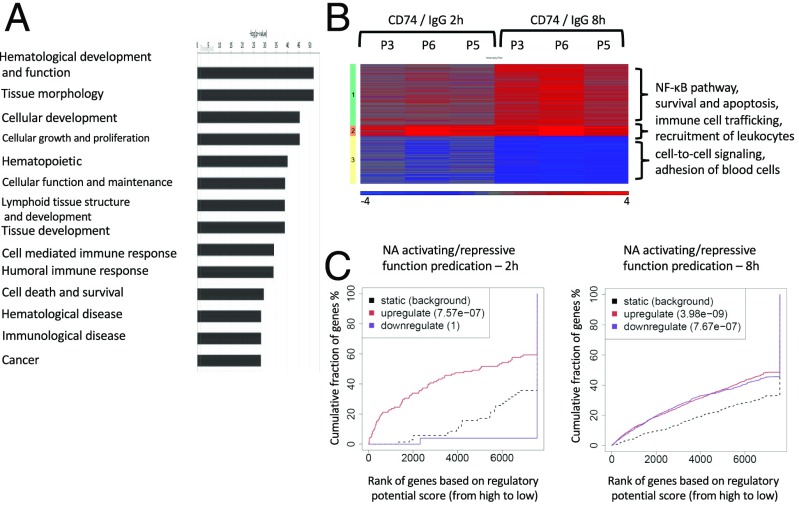
Functional annotation using Qiagen’s Ingenuity Pathway Analysis (IPA; www.ingenuity.com) revealed that the CD74-regulated genes were enriched with genes involved in the inflammatory responses and hematological system development and function (Fig. 3A). The differentially expressed genes were clustered to three clusters (Fig. 3B). In cluster 1, genes were moderately up-regulated 2 h following activation, and strongly up-regulated 8 h postactivation. This cluster was enriched in genes involved in the NF-κB cell death and survival of immune cells (P = 7.93 × 10−22), consistent with the role of CD74 in B-cell survival. Cluster 2 was characterized by genes that were up-regulated shortly after activation, and remained at high levels at the later time point. Those genes were mainly related to immune cell trafficking and recruitment of leukocytes (P = 9.47 × 10−17). Cluster 3, on the other hand, was characterized by a slight down-regulation in the 2-h postactivation time-point, and strong down-regulation 8 h postactivation. This cluster was enriched with genes related to cell–cell signaling and adhesion of blood cells (P = 4.34 × 10−09). Thus, at both the mRNA level, as well as at the DNA-binding level, CD74 had a significant effect on immune response and survival pathways.
Fig. 3.
CD74 activation leads to transcription regulation. Samples from three patients were activated with either anti-CD74 or control IgG antibodies for 2 and 8 h. mRNA was extracted and subjected to mRNA sequencing. mRNA levels were compared between CD74- and IgG-activated cells. (A) Differentially expressed genes at the 8-h time point (Padj < 0.05, absolute fold-change > 2, reads count of at least 20 in at least one sample) were analyzed using the Ingenuity system. Log2 ratios of CD74 activation versus IgG are depicted in a color scale. (B) Differentially expressed genes in response to CD74 activation after 2 or 8 h (total of 603 genes) were clustered to three clusters using Euclidean distance measure. For each patient and time point, log2 ratios of CD74 activation versus IgG is shown. Cluster 1 includes genes that are up-regulated at 2 h, and more strongly up-regulated at 8 h; cluster 2 is characterized by genes that were up-regulated shortly after activation, and remained at high levels at the later time point; and cluster 3, in which the genes are down-regulated at 2 h, with stronger down-regulation seen at 8 h. Enriched pathways are indicated near each cluster. (C) BETA software was used to correlate between the genes that were bound in their proximity by CD74, and genes that were regulated at the mRNA level.
Our next aim was to correlate between the genes that were bound in their proximity by CD74–ICD and were regulated at the mRNA level. To perform this analysis, BETA software was used (25). BETA (binding and expression target analysis) is a software package that integrates ChIP-seq of transcription factors or chromatin regulators with differential gene-expression data to infer direct target genes. At 2 h following CD74 activation, the regulatory potential of the up-regulated genes was significantly higher relative to the nonregulated genes (P = 7.57 × 10−7). At 8 h following CD74 activation, both the up- and down-regulated genes had a significantly higher regulatory potential relative to the nonregulated genes (P = 3.98 × 10−09 and P = 7.67 × 10−07, respectively) (Fig. 3C), indicating that CD74 association with the chromatin directly affects gene transcription, and that CD74 functions both as an inhibitor and an activator of gene transcription.
We next wished to examine the function of the regulated genes. The output of BETA was used to extrapolate genes that have regulatory potential based on both the ChIP-seq and the mRNA-seq. All genes that were up-regulated 2 h postactivation were also elevated at the 8-h time point; thus, function analysis was performed for genes regulated at 8 h. Ingenuity analysis revealed that within the set of genes that were bound by CD74 and regulated at the mRNA level, genes whose mRNA levels were down-regulated 8 h following CD74 activation did not have a clear pattern of functions, whereas genes whose mRNA was up-regulated 8 h after CD74 activation were greatly enriched for apoptosis-regulating genes, which also correlates with CD74 function in cell survival.
CD74 Forms a Complex with Known Transcription Factors.
Transcription regulators often function as part of a transcription complex (26). The complex formed by a transcription factor determines its binding and function. Because CD74–ICD is a short peptide of 42 amino acids, it is plausible that it forms complexes with additional proteins. We therefore searched for common binding motifs for CD74 and additional transcription factors using the Genomatix overrepresented transcription factor binding site tool (27).
As shown in Table S4, CD74 was predicted to bind the same binding sites used by additional transcription factors. Although AP1F is the most highly represented transcription factor family, it has been shown that the binding-motif profile of AP1 is overrepresented in many ChIP-seq assays in a nonspecific manner (28). Taking into account the mRNA levels found in CLL cells in the mRNA-seq assay, we focused on two families of transcription factors that are predicted to interact with CD74, HAML [the Runt related transcription factor (RUNX) family], and the NF-κB family, both with established function in CLL. CD74 activates the p65 member of the NF-κB family in B cells (5, 7) and CLL cells (11), and RUNX1/acute myeloid leukemia (AML)1 is the most frequently implicated transcription factor in human leukemias (29). The binding motifs of CD74 that were shared with HAML and NF-κB are presented in Table S5.
Table S4.
Transcription factor’s families that are predicted to bind in the same sites as CD74
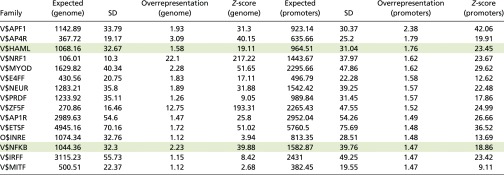 |
The shaded rows indicate the families of transcription factors selected for further study.
Table S5.
The binding motifs of CD74 that were shared with HAML and NF-κB
| Matrix name | International Union of Pure and Applied Chemistry DNA sequences |
| HAML | |
| V$AML1.01 | CTGYGGTYW |
| V$AML1.02 | NSTGTGGTNNG |
| V$AML2.01 | NNTGTGGTTWNN |
| V$AML3.01 | NNSTGTGGTTTGTG |
| V$AML3.02 | NYTGTGGTTTNN |
| NF-κB | |
| V$CREL.01 | BGGGNTTTCC |
| V$HIVEP1.01 | WDGGGAMTTTCCN |
| V$NFKAPPAB.01 | GGGANTYYCC |
| V$NFKAPPAB.02 | NGGGACTTTCCN |
| V$NFKAPPAB50.01 | GGGGATYCCC |
| V$NFKAPPAB65.01 | BGGRRTTTCC |
Shows a multiple alignment of the consensus sequences of all matrices belonging to the family. The anchor position used for marking the position in MatInspector is printed in underlie.
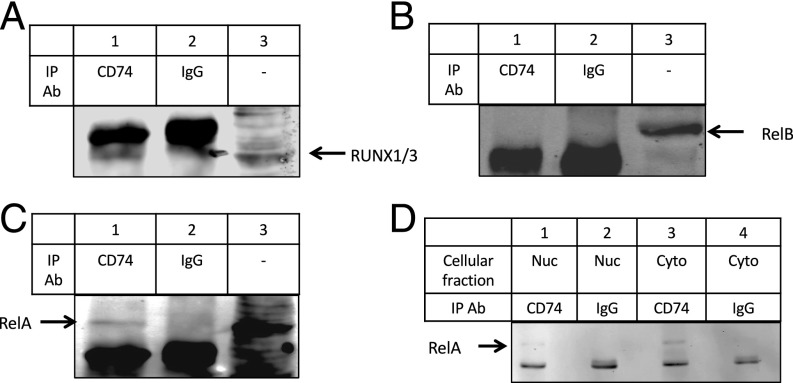
First, mRNA levels of RUNX family members were followed in CLL cells. mRNA sequencing revealed that RUNX1 and RUNX3 were both highly expressed in CLL cells [reads per million mapped reads (RPKM) RUNX1 > 7, RPKM RUNX3 > 13]. Moreover, RUNX3 levels were significantly up-regulated (fold-change at 8 h = 2.03, Padj = 4.4 × 10−10) following CD74 activation (GEO accession no. GSE88957). We next wished to determine whether CD74 and RUNX form a protein complex. CLL cells were activated for 1 h with CD74 Ab. Total proteins were extracted, and CD74–ICD was immunoprecipitated. Immunoprecipitates were analyzed for RUNX1 and RUNX3. As can be seen in Fig. 4A, CD74 pulled down RUNX1/3, suggesting complex formation of these two factors in activated CLL cells.
Fig. 4.
CD74 forms a complex with other transcription factors to regulate transcription. CLL cells were activated for 1 h with CD74 Ab. Total proteins were extracted, and immunoprecipitation was performed with anti-CD74 or isotype control antibodies. Proteins were separated on a 10% gel, and immunoblots were analyzed for RUNX1 and RUNX3 (A), RelB (B), and RelA (C). (D) CLL cells were activated for 1 h with anti-CD74 Ab. Nuclear and cytoplasmic fractions were separated using the Nucbuster extraction kit. Immunoprecipitation was performed using CD74 Ab (lanes 1 and 3) or IgG control Ab (lanes 2 and 4). Immunoprecipitates were separated on 10% SDS/PAGE, and analyzed for RelA.
The other family of proteins predicted to work in a complex with CD74 is the NF-κB family. We previously showed that CD74–ICD activates the p65 (RelA) member of NF-κB (5, 7). In addition, activation of NF-κB was detected in CD74-activated CLL cells (11). To determine whether CD74 forms a physical interaction with NF-κB, a coimmunoprecipitation experiment was performed. CD74 was immunoprecipitated from lysates of CD74 activated CLL cells and the presence of RelB (Fig. 4B) and RelA (Fig. 4C) in the pulled-down proteins was analyzed. RelA, but not RelB, was pulled down with CD74, suggesting a specific complex formation between CD74 and RelA. CD74–ICD translocates to the nucleus and induces the activation of the p65 member of NF-κB within the nuclear compartment (6). As shown in Fig. 4D, the CD74/RelA complex was detected in both cytoplasmic and nuclear fractions, suggesting that the complex of these two proteins is formed in the cytoplasm and translocates to the nucleus to regulate transcription.
We next analyzed the mRNA levels of genes that are NF-κB or Runx1/3 targets following CD74 activation. A significant up-regulation of NF-κB family members and of genes that are part of the NF-κB survival pathways (RNA-seq results and Fig. 5A) and Runx1/3 (RNA-seq results and Fig. 5B) was detected in CLL cells following CD74 activation. To verify our results, the regulation of the expression of two NF-κB target genes, TRAF1 and BIRC3, bound in their proximity by CD74–ICD and involved in cell survival, was analyzed. As shown in Fig. 5C, protein levels of TRAF1 and BIRC3 were significantly up-regulated 48 h following CD74 activation. Furthermore, the message levels of these proteins were significantly reduced when CD74 expression levels were down-regulated by small-interfering RNA (siRNA) (Fig. 5D).
Fig. 5.
CD74 up-regulates expression of genes that are part of the NF-κB–induced survival cascade. (A and B) CLL cells from three patients were activated with anti-CD74 Ab for 2 and 8 h. (A) mRNA-seq showing elevation in genes that take part in the NF-κB survival pathway. For B-cell activating factor (BAFF)-R level determination, CLL cells were activated with CD74 Ab for 18 h. mRNA was extracted, and BAFF-R levels were determined using qRT-PCR (n = 4, P < 0.05). (B) mRNA-seq showing elevation in genes that are Runx1/3 targets. (C) TRAF1 and BIRC3 protein levels 48 h following activation were analyzed by FACS. Graph presents fold-change relative to unactivated cells (n = 4, P < 0.01; n = 6, P < 0.05). (D) CLL cells (5 × 106) were transfected with siRNA against CD74 or control siRNA. After 24 h, cells were harvested, RNA was purified and mRNA levels of TRAF and BIRC3 were analyzed by qRT-PCR (n = 7, *P = 0.047 for TRAF, P = 0.0258 for BIRC3). *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001.
These results imply an intricate connection between CD74 and additional transcription factors, and show that CD74 and NF-κB or RUNX1/3 bind chromatin in a complex to regulate gene transcription.
Discussion
Transcription regulators are characterized by their ability to enter the nucleus, bind the chromatin, and affect gene transcription. In the present study, using ChIP-seq, we demonstrated that activation of CD74 expressed on CLL cells induces binding of CD74–ICD to chromatin. This fragment binds to transcription binding sites and regulatory areas of genes that control the immune response, and are overexpressed in leukemia and other immune diseases, indicating that CD74 has a specific role in regulating genes controlling those diseases. To further understand the effect of CD74 on gene regulation, we used mRNA-seq and analyzed the levels of mRNA following CD74 activation. Genes with a gain in gene expression after 2 h of CD74 activation tended to also have an enrichment of CD74 binding sites. After 8 h of CD74 activation, both up- and down-regulated genes were enriched in CD74 binding sites. This finding further strengthens our hypothesis of a role for CD74 as a transcription factor.
Our results suggest that CD74–ICD binding to chromatin induces the expression of various processes, including cell survival and cell–cell signaling. The prosurvival genes are part of several survival pathways, including the TNF-, B-cell receptor-, CD40-, and NF-κB–induced signaling cascades. These results indicate that the role of CD74 in survival is achieved by CD74–ICD binding to survival gene regulatory areas in chromatin following its activation, and causing up-regulation at the mRNA and at the protein levels.
CD74–ICD is a small 42-amino acid peptide, with no distinctive DNA binding region, supporting the notion that it exerts its function in a complex with other transcription factors. Based on bioinformatics predictions of potential coregulators, we focused on the RUNX family and NF-κB. RUNX1 and RUNX3 are both expressed at various hematopoietic stages (25). Additionally, RUNX3 was found to bind the promoter of RUNX1 and suppress its expression (30), and thus the expression of these proteins is linked. Our results suggest that CD74 and the RUNX proteins form a complex. Although the exact function of this complex is, as yet, unknown, these results suggest a joint function for CD74 and the RUNX proteins.
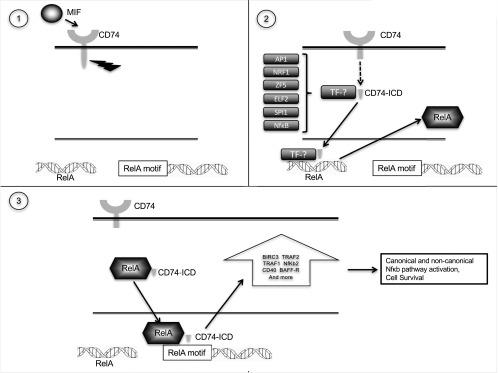
It has been known for some time that there is cross-talk between the CD74 and NF-κB pathways (5, 7). Based on the current results, we suggest that CD74 regulates NF-κB activity at several levels (Fig. S2). First, CD74 activation leads to its binding to the RelA promoter and up-regulating its levels in the cell, conceivably in a complex with other transcription factors. Examination of the transcription factors that share a binding motif with CD74 and are expressed in CLL cells, leads us to propose that this transcription factor might be AP1, NRF1, ZF5, ELF2, or SPI1. RelA levels are then up-regulated and form a complex with CD74 in the cytoplasm, which is translocated to the nucleus to bind chromatin and regulate transcription. Binding of the complex to the chromatin in proximity to genes of NF-κB family members, such as RelB and NFKB2, promotes their transcription. In addition, binding of this complex to the chromatin leads to up-regulation of genes involved in the NF-κB–induced cascade, such as TRAF1, TRAF2, BIRC3, CD40, and others. Although CD74 activation leads to up-regulation of both RelA and RelB, a complex of CD74 and RelB was not detected, indicating that CD74 specifically binds RelA.
Fig. S2.
A summarizing model. TF, transcription factor.
The nuclear localization sequence of CD74–ICD is not known. Small peptides like CD74–ICD generally migrate into the nuclear pore by diffusion (31). However, migration by diffusion can only occur until equilibrium is achieved. It appears that an active translocation process induces entry of CD74–ICD to the nucleus. Our results suggest that binding of CD74–ICD to additional transcription factors facilitates its translocation to the nucleus. It is also possible that the intracellular domain of CD74 is phosphorylated before the RIP process, which may lead to binding to a carrier protein that transports the CD74–ICD to the nucleus.
MIF and the newly characterized MIF2 are the natural ligands of CD74 (32, 33). Depending on the cell type, this pathway activation requires the involvement of the coreceptors CD44, CXCR2, or CXCR4 (34–36). The role of MIF2 in B-cell function and B-cell–dependent diseases has not been addressed. It will be essential to compare the binding region and target genes activated by MIF and MIF2, and whether the various CD74 coreceptors influence the transcription regulatory role of CD74.
Our results indicate a crucial role for CD74–ICD as a transcription regulator, controlling pathways involved in immune regulation and immune diseases. Further analysis of these processes will identify specific DNA sequences that are involved in this interaction, and essential pathways regulating B-cell survival in health and disease.
Materials and Methods
Cells.
B cells obtained from the peripheral blood of CLL patients at varying disease stages were provided in compliance with the Institutional Review Board of Kaplan Medical Center in Rehovot, Sourasky Medical Center in Tel Aviv, and Laniado Hospital–Sanz Medical Centers in Netanya, Israel, as previously described (11). Patients were staged according to Binet et al. (37). A list of patients appears in Table S1. CLL lymphocytes were purified as described previously (11). Informed consent was obtained from all participants.
RNA Isolation and Reverse Transcription.
mRNA from primary human CLL cells was isolated using PerfectPure RNA Cultured Cell Kit (5 Prime), according to the manufacturer’s instructions. mRNA levels were analyzed by quantitative real-time PCR, as previously described (11). Primer sequences (Sigma-Aldrich) are listed in Table S6.
Table S6.
Primers list
| Primer | Forward | Reverse |
| TRAF1 | TTACGGAGCTGTGTGGACTG | GACTGTGGGCTTCCCTTGAA |
| BIRC3 | GGCAGCAGGTTTACAAAGGAG | CTCCCGAGATTAGACTAAGTCCCTT |
| NLE1 promoter | TTTCCGTACGCCACATTTC | GTACGGAAGACCCACTCC |
| BAFF-R | ACCTTGTCCAGGGGCTCT | CTGGTCCTGGTGGGTCTG |
RNA-Seq.
Total RNA was extracted from 5 × 106 cells from three patients, with CD74 or IgG activation. Illumina libraries were constructed from total RNA using the Illumina TruSeq RNA Sample Preparation v2 (Cat. no. RS-122–2002, Illumina) according to the manufacturer's instructions. Indexed samples were sequenced in an Illumina HiSEq. 2500 machine in a single-read mode. The sequence yield was between 19.3 and 23.8 million reads per sample.
RNA-Seq Analysis.
TopHat (v2.0.10) was used to align the reads to the human genome (hg19) (38). Of the reads, 92% were uniquely aligned to the genome. Counting reads on hg19 RefSeq genes (downloaded from igenomes) was done with HTSeq-count (v0.6.1p1) (39). Differential expression analysis was performed using DESeq2 (40). A two-factor model including CD74 activation (CD74 antibody or IgG) and the patient was built using DESeq2 (1.6.3). Raw P values were adjusted for multiple testing using the procedure of Benjamini and Hochberg.
Immunofluorescence and Flow Cytometry.
Isolated CLL cells were stained using specific antibodies, as previously described (11). The antibodies list can be found in SI Materials and Methods, by the Genomic unit, Weizmann Institute of Science.
CD74 Stimulation.
CD74 was activated, as previously described (11).
Calcium Phosphate Transfection.
The 293T HEK cells were cultured and transfected as previously described (6).
ChIP-seq and Its Analysis.
ChIP-seq of 293T HEK cells: ChIP-seq was performed as previously described (26), with some modifications described in SI Materials and Methods, by the Genomic unit, Weizmann Institute of Science.
ChIP-seq for CLL: CD74 activated CLL cells were fixed with 1 mL of 2 mM DSG solution and shaken at room temperature. After 35 min, 66.7 µL of formaldehyde was added. After 8 min at room temperature, 52 µL 2.5 M glycine was added to the culture to quench the fixation reaction. ChIP DNA was processed as previously described (41). Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow multiplexing of different samples on one lane. Between 14 and 40 million single-end 60-bp reads were sequenced per sample on an Illumina HiSEq. 2500 v4 instrument.
Adapters were trimmed using the Cutadapt tool (42). Following adapter removal, reads that were shorter than 40 nucleotides were discarded (Cutadapt option –m 40). The reads were aligned uniquely to the human genome (hg19) using Bowtie (v1.0.0) (43). CD74 bound regions, H3K4Me2 and H3K4Me3 peaks were detected using MACS2 (v2.0.10.20131216) (44). H3K27Ac peaks were also detected with MACS2 using the broad option. CEAS (v1.0.2) was used to obtain statistics on ChIP enrichment of genomic features (21). MACS2 bedGraph files were converted to bigWig files for visualization.
To define “high-confidence peaks,” 3,575 peaks that were shared by at least two samples were obtained using the Bedtools suite (45). The summit of each shared peak was extracted from one of the samples.
The ngs.plot package was used for plotting H3K4Me2 profiles over the high-confidence CD74 peak summits (46). Overlap between the CD74 peaks and the histone modification peaks was identified using the Bedtools suite.
GREAT analysis was done using the 3,575 high confidence peaks with the two nearest gene-association rule settings (20).
ChIP-Seq and RNA-Seq Integration.
The BETA tool was used for the integration of the RNA-Seq and ChIP-SEq. (25). Detailed descriptions can be found in SI Materials and Methods.
Coimmunoprecipitation.
For coimmunoprecipitation, 1–5 µg Ab was added to a total of 500 µL lysate and incubated overnight at 4 °C. Dynabead Protein G Magnetic Beads (12–25 μL; Thermofisher) were washed with binding/blocking buffer (0.5% BSA, 0.5% Tween 20 in PBS), and then added to each tube and incubated for 1 h at 4 °C on a rotator. Beads were pelleted, washed and resuspended in 20 μL of 1× Reducing Loading Buffer and separated on 10% (wt/vol) acrylamide gel.
Cell Fractionation.
Cell fractionation was preformed using the NucBuster protocol, as described in detail in SI Materials and Methods.
CD74 Down-Regulation by siRNA.
CD74 siRNA was introduced by electroporation using a Nepagene (Ichikawa) as previously described (47). Sequences are listed in SI Materials and Methods.
SI Materials and Methods
ChIP-seq.
Transfected 293T HEK cells were fixed using formaldehyde. Samples were sonicated in 1% (wt/vol) SDS lysis buffer to obtain fragments of 200–1,000 bp. Anti-CD74 (BD Bioscience) was used for CD74 immunoprecipitation.
ChIP-Seq and RNA-Seq Integration.
The summits of the shared peaks and the differential expression dataset were used as input for the BETA tool with the options: –da 1–df 0.05. For the RNA-Seq, 12,781 genes that had a count of at least 20 in at least one of the samples were included in the analysis. Clustering of the differently expressed genes was generated using Partek Genomic Suite 6.6 (Partek).
Immunofluorescence and Flow Cytometry.
The following antibodies were used: ab129279 anti-TRAF1, ab23223 anti-BIRC3, ab6881 anti-goat (Abcam), 406414 anti-rabbit (Biolegend), and 12-0748-42 antiCD74 (eBioscience).
Cell Fractionation.
Following brief centrifugation (500 × g, 3 min at 4 °C), the cells were resuspended in NucBuster (Merck Millipore) reagent containing protease inhibitors at 150 μL NucBuster Reagent-1 per 50 μL packed cell volume. Intact nuclei were separated from cytoplasmic proteins by centrifugation, and nuclear proteins were collected following addition of 75 μL of Nucbuster Reagent-2. Nuclear proteins were stored at −70 °C.
siRNA was purchased from GE Lifescience; human CD74: Dharmacon ON-TARGETplus SMARTpool human CD74 (Cat# L-012667-00); nonspecific control siRNA: Dharmacon ON-TARGETplus Nontargeting Pool (D-001810-10).
Acknowledgments
We thank Prof. Rivka Dikstein for advice, support, and critical reading of the manuscript. This research was supported by Binational Science Foundation Grant 711979; the Rising Tides Foundation; and the Quinquin Foundation. I.S. is the incumbent of the Dr. Morton and Ann Kleiman Professorial Chair. G.F. is the incumbent of the David and Stacey Cynamon Research fellow Chair in Genetics and Personalized Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE88957).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612195114/-/DCSupplemental.
References
- 1.Stumptner-Cuvelette P, Benaroch P. Multiple roles of the invariant chain in MHC class II function. Biochim Biophys Acta. 2002;1542(1-3):1–13. doi: 10.1016/s0167-4889(01)00166-5. [DOI] [PubMed] [Google Scholar]
- 2.Arneson LS, Miller J. The chondroitin sulfate form of invariant chain trimerizes with conventional invariant chain and these complexes are rapidly transported from the trans-Golgi network to the cell surface. Biochem J. 2007;406(1):97–103. doi: 10.1042/BJ20070446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bucala R, Shachar I. The integral role of CD74 in antigen presentation, MIF signal transduction, and B cell survival and homeostasis. Mini Rev Med Chem. 2014;14(14):1132–1138. doi: 10.2174/1389557515666150203144111. [DOI] [PubMed] [Google Scholar]
- 4.Cohen S, Shachar I. Cytokines as regulators of proliferation and survival of healthy and malignant peripheral B cells. Cytokine. 2012;60(1):13–22. doi: 10.1016/j.cyto.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 5.Matza D, Kerem A, Medvedovsky H, Lantner F, Shachar I. Invariant chain-induced B cell differentiation requires intramembrane proteolytic release of the cytosolic domain. Immunity. 2002;17(5):549–560. doi: 10.1016/s1074-7613(02)00455-7. [DOI] [PubMed] [Google Scholar]
- 6.Becker-Herman S, Arie G, Medvedovsky H, Kerem A, Shachar I. CD74 is a member of the regulated intramembrane proteolysis-processed protein family. Mol Biol Cell. 2005;16(11):5061–5069. doi: 10.1091/mbc.E05-04-0327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matza D, Wolstein O, Dikstein R, Shachar I. Invariant chain induces B cell maturation by activating a TAF(II)105-NF-kappaB-dependent transcription program. J Biol Chem. 2001;276(29):27203–27206. doi: 10.1074/jbc.M104684200. [DOI] [PubMed] [Google Scholar]
- 8.Starlets D, et al. Cell-surface CD74 initiates a signaling cascade leading to cell proliferation and survival. Blood. 2006;107(12):4807–4816. doi: 10.1182/blood-2005-11-4334. [DOI] [PubMed] [Google Scholar]
- 9.Gore Y, et al. Macrophage migration inhibitory factor induces B cell survival by activation of a CD74-CD44 receptor complex. J Biol Chem. 2008;283(5):2784–2792. doi: 10.1074/jbc.M703265200. [DOI] [PubMed] [Google Scholar]
- 10.Mizue Y, et al. Quantitation of macrophage migration inhibitory factor (MIF) using the one-step sandwich enzyme immunosorbent assay: Elevated serum MIF concentrations in patients with autoimmune diseases and identification of MIF in erythrocytes. Int J Mol Med. 2000;5(4):397–403. doi: 10.3892/ijmm.5.4.397. [DOI] [PubMed] [Google Scholar]
- 11.Binsky I, et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci USA. 2007;104(33):13408–13413. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Binsky I, et al. TAp63 regulates VLA-4 expression and chronic lymphocytic leukemia cell migration to the bone marrow in a CD74-dependent manner. J Immunol. 2010;184(9):4761–4769. doi: 10.4049/jimmunol.0904149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen S, et al. The cytokine midkine and its receptor RPTPζ regulate B cell survival in a pathway induced by CD74. J Immunol. 2012;188(1):259–269. doi: 10.4049/jimmunol.1101468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butrym A, Majewski M, Dzietczenia J, Kuliczkowski K, Mazur G. High CD74 expression correlates with ZAP70 expression in B cell chronic lymphocytic leukemia patients. Med Oncol. 2013;30(2):560. doi: 10.1007/s12032-013-0560-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schröder B, Saftig P. Intramembrane proteolysis within lysosomes. Ageing Res Rev. 2016;32:51–64. doi: 10.1016/j.arr.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 16.Weinmaster G. Notch signal transduction: A real rip and more. Curr Opin Genet Dev. 2000;10(4):363–369. doi: 10.1016/s0959-437x(00)00097-6. [DOI] [PubMed] [Google Scholar]
- 17.Nowak DE, Tian B, Brasier AR. Two-step cross-linking method for identification of NF-kappaB gene network by chromatin immunoprecipitation. Biotechniques. 2005;39(5):715–725. doi: 10.2144/000112014. [DOI] [PubMed] [Google Scholar]
- 18.Plank JL, Dean A. Enhancer function: Mechanistic and genome-wide insights come together. Mol Cell. 2014;55(1):5–14. doi: 10.1016/j.molcel.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanyal A, Lajoie BR, Jain G, Dekker J. The long-range interaction landscape of gene promoters. Nature. 2012;489(7414):109–113. doi: 10.1038/nature11279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLean CY, et al. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28(5):495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25(19):2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- 22.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev. 2002;12(2):142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 23.Creyghton MP, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107(50):21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang DX, Glass CK. Towards an understanding of cell-specific functions of signal-dependent transcription factors. J Mol Endocrinol. 2013;51(3):T37–T50. doi: 10.1530/JME-13-0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, et al. Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat Protoc. 2013;8(12):2502–2515. doi: 10.1038/nprot.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garber M, et al. A high-throughput chromatin immunoprecipitation approach reveals principles of dynamic gene regulation in mammals. Mol Cell. 2012;47(5):810–822. doi: 10.1016/j.molcel.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cartharius K, et al. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21(13):2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 28.Worsley Hunt R, Wasserman WW. Non-targeted transcription factors motifs are a systemic component of ChIP-seq datasets. Genome Biol. 2014;15(7):412. doi: 10.1186/s13059-014-0412-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Osato M, Ito Y. Increased dosage of the RUNX1/AML1 gene: A third mode of RUNX leukemia? Crit Rev Eukaryot Gene Expr. 2005;15(3):217–228. doi: 10.1615/critreveukargeneexpr.v15.i3.40. [DOI] [PubMed] [Google Scholar]
- 30.Spender LC, Whiteman HJ, Karstegl CE, Farrell PJ. Transcriptional cross-regulation of RUNX1 by RUNX3 in human B cells. Oncogene. 2005;24(11):1873–1881. doi: 10.1038/sj.onc.1208404. [DOI] [PubMed] [Google Scholar]
- 31.Wang R, Brattain MG. The maximal size of protein to diffuse through the nuclear pore is larger than 60kDa. FEBS Lett. 2007;581(17):3164–3170. doi: 10.1016/j.febslet.2007.05.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leng L, et al. MIF signal transduction initiated by binding to CD74. J Exp Med. 2003;197(11):1467–1476. doi: 10.1084/jem.20030286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merk M, et al. The D-dopachrome tautomerase (DDT) gene product is a cytokine and functional homolog of macrophage migration inhibitory factor (MIF) Proc Natl Acad Sci USA. 2011;108(34):E577–E585. doi: 10.1073/pnas.1102941108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi X, et al. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 2006;25(4):595–606. doi: 10.1016/j.immuni.2006.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bernhagen J, et al. MIF is a noncognate ligand of CXC chemokine receptors in inflammatory and atherogenic cell recruitment. Nat Med. 2007;13(5):587–596. doi: 10.1038/nm1567. [DOI] [PubMed] [Google Scholar]
- 36.Schwartz V, et al. A functional heteromeric MIF receptor formed by CD74 and CXCR4. FEBS Lett. 2009;583(17):2749–2757. doi: 10.1016/j.febslet.2009.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Binet JL, et al. A clinical staging system for chronic lymphocytic leukemia: Prognostic significance. Cancer. 1977;40(2):855–864. doi: 10.1002/1097-0142(197708)40:2<855::aid-cncr2820400239>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 38.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Pyl PT, Huber W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anders S, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–1786. doi: 10.1038/nprot.2013.099. [DOI] [PubMed] [Google Scholar]
- 41.Blecher-Gonen R, et al. High-throughput chromatin immunoprecipitation for genome-wide mapping of in vivo protein-DNA interactions and epigenomic states. Nat Protoc. 2013;8(3):539–554. doi: 10.1038/nprot.2013.023. [DOI] [PubMed] [Google Scholar]
- 42.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet Journal. 2011;17(1):10–12. [Google Scholar]
- 43.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9(9):R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Quinlan AR. BEDTools: The Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marom A, et al. CD84 mediates CLL-microenvironment interactions. Oncogene. July 25, 2016 doi: 10.1038/onc.2016.238. [DOI] [PubMed] [Google Scholar]