Significance
The Zika virus (ZIKV) causes various neurologic defects including microcephaly and the Guillain-Barré syndrome. However, little is known about how ZIKV causes those diseases or which viral protein(s) is responsible for the observed cytopathic effects involved in restricted neuronal cellular growth, dysregulation of the cell cycle, and induction of cell hypertrophy or cell death. A genome-wide analysis of ZIKV proteins and peptides was conducted using fission yeast as a surrogate host. Seven ZIKV proteins conferred various cytopathic effects in which NS4A-induced cellular hypertrophy and growth restriction were mediated through the target of rapamycin (TOR) cellular stress-response pathway. These findings provide a foundation for identifying viral pathogenicity factors associated with the ZIKV diseases.
Keywords: Zika genome, Zika proteins, fission yeast, Schizosaccharomyces pombe, cytopathic factors
Abstract
The Zika virus (ZIKV) causes microcephaly and the Guillain-Barré syndrome. Little is known about how ZIKV causes these conditions or which ZIKV viral protein(s) is responsible for the associated ZIKV-induced cytopathic effects, including cell hypertrophy, growth restriction, cell-cycle dysregulation, and cell death. We used fission yeast for the rapid, global functional analysis of the ZIKV genome. All 14 proteins or small peptides were produced under an inducible promoter, and we measured the intracellular localization and the specific effects on ZIKV-associated cytopathic activities of each protein. The subcellular localization of each ZIKV protein was in overall agreement with its predicted protein structure. Five structural and two nonstructural ZIKV proteins showed various levels of cytopathic effects. The expression of these ZIKV proteins restricted cell proliferation, induced hypertrophy, or triggered cellular oxidative stress leading to cell death. The expression of premembrane protein (prM) resulted in cell-cycle G1 accumulation, whereas membrane-anchored capsid (anaC), membrane protein (M), envelope protein (E), and nonstructural protein 4A (NS4A) caused cell-cycle G2/M accumulation. A mechanistic study revealed that NS4A-induced cellular hypertrophy and growth restriction were mediated specifically through the target of rapamycin (TOR) cellular stress pathway involving Tor1 and type 2A phosphatase activator Tip41. These findings should provide a reference for future research on the prevention and treatment of ZIKV diseases.
The recent Zika virus (ZIKV) outbreak was surprising in its rapidity and alarming in its association with microcephaly in newborns (1–3) and the Guillain-Barré syndrome in adults (4, 5). Currently, little is known about ZIKV pathogenicity. ZIKV infects various neuronal cells and affects brain neurogenesis by reducing cellular proliferation and inducing cell-cycle abnormalities, cell death/apoptosis, and autophagy (2, 3, 6–8). However, it is not known which viral determinant(s) cause these cytopathic effects. The objective of this study was to identify ZIKV factors responsible for the ZIKV-mediated cytopathic effects that underlie the diseases associated with ZIKV.
The Zika virus is a member of the flavivirus family, which is comprised of highly pathogenic viruses, including dengue virus (DENV), yellow fever virus (YFV), West Nile virus (WNV), and Japanese encephalitis virus (JEV). ZIKV is a positive-sense, single-stranded RNA virus that has a viral genome of ∼10.7 kb (Fig. 1A) (9). The ZIKV genome encodes a single polyprotein that is processed by viral and host proteases into six structural proteins [membrane-anchored capsid protein (anaC)→mature capsid protein (C), premembrane protein (prM)→membrane protein (M) + protein pr (Pr), and envelope protein (E)], and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) and the nonstructural peptide 2K. (Fig. 1A) (10). The cleavage of anaC by a viral protease generates mature C protein, which in turn triggers the cleavage of prM to produce M and Pr proteins by a host signalase, Furin (11, 12). The 2K signal peptide, which lies between NS4A and NS4B, regulates the translocation of NS4B into the endoplasmic reticulum (ER) lumen and RNA synthesis (13, 14). The overall architecture of ZIKV is similar to that of other flaviviruses (15–17).
Fig. 1.
Genome-wide analysis of intracellular localizations of ZIKV proteins. (A) Schematic structure of the ZIKV genome with the predicted association of each ZIKV protein with the intracellular membrane. The ZIKV genome shown is based on the MR766 viral genome (GenBank accession number: Ay632535). Each of the viral proteins is drawn based on the relative orientation in the entire RNA genome. The ZIKV viral protease, host protease, and Furin protease are indicated by arrows. Each arrow points to the specific protease cleavage site. The numbers shown above each protein product indicate the start/end position. NS5 encodes methyltransferase at its N-terminal end and RNA-dependent RNA polymerase at its C-terminal end. ZIKV protein products information based on Kuno et al. (16). The protein membrane-associated structure and processing are information from Assenberg et al. (17). (B) The primary intracellular localizations of ZIKV proteins. All GFP-ZIKV proteins were produced at low gene-expression levels and observed within 20 h of GI. (a) An empty vector control (Vec). (b) Structural ZIKV proteins. (c) Nonstructural ZIKV proteins. (Scale bars, 10 μm.) (C, a) Comparison of ZIKV protein localization with cellular proteins that are known to localize in ER (Gpi1-YFP), Golgi (Ynd1-YFP), and cytoplasmic puncta (Atg1-YFP) (1). (b) Colocalization of GFP-NS1 with Atg1-YFP under nitrogen starvation. Note that both GFP and YFP were detectable under the GFP/YFP filter. However, only the GFP signal was detectable under the CFP filter. Arrows indicate where Atg1-YFP signals are missing from the CFP filter. (Scale bar, 10 μm.) The interpretation of the specific subcellular localization of each ZIKV protein is summarized in Table 1. A comparison of subcellular localization patterns of each ZIKV protein at low and high expression levels is presented in Fig. S2.
Similar to other flaviviruses, ZIKV viral replication is believed to occur on virus-associated intracellular membranes, including the ER network and the trans-Golgi network (TGN) (18–20). The ER is a complex network of continuous sheets and tubules extending from the nuclear envelope to the plasma membrane (20). The TGN is a secretory sorting apparatus that transports newly synthesized ZIKV proteins. One goal of this study was to identify the intracellular localization of each of the ZIKV proteins to provide better understanding of ZIKV’s activities within cells.
During the ZIKV viral life cycle, ZIKV presumably enters host cells via endocytosis through the phosphatidylserine receptor, AXL (6, 21). The viral particle is disassembled in the endosome, and the E protein undergoes trimerization leading to fusion of the viral and cell membranes (22). After virus–host cell fusion, the viral genome is released into the cytoplasm where viral RNA is translated into a single polyprotein, which is subsequently processed by viral and host proteases into 14 proteins or short peptides. Viral genome replication typically occurs on virus-associated intracellular membranes, primarily in the ER network (19). Thus, based on the protein structures, most of the ZIKV proteins are expected to be membrane-associated proteins, with the possible exception of mature Pr, NS1, NS3, 2K, and NS5 (Fig. 1A) (17). Virus assembly takes place on the surface of the ER where the structural proteins (C, prM, and E) and newly synthesized viral RNA bud into the lumen of the ER and form immature viral particles (23, 24). In DENV, E protein forms a heterodimer with prM and affects viral particle formation and secretion (25). The resultant noninfectious and immature viral particles are transported through the TGN, where prM is cleaved by a host protease Furin, resulting in mature infectious particles (26, 27). The transition from prM to M via the cleavage of host protease Furin is required for viral infectivity (11, 12). Mature virions are subsequently released from the host cell by exocytosis.
One of the major challenges in understanding ZIKV activities and viral pathogenicity is that ZIKV infects a wide range of neural and other cell types, including skin fibroblasts, astroglia, neurons, and neuronal progenitor cells (7, 18). Also, a wide variety of geographically and genetically distinct viral variants have been associated with the known ZIKV diseases. Given the number of host cells and viral variants, it has been difficult to identify pathogenic factors and to generate a consensus on ZIKV protein functionality. To meet this challenge, in the present study, we used fission yeast (Schizosaccharomyces pombe) as a surrogate system to characterize the activities of the ZIKV genome. Fission yeast is a single-cell eukaryote that has been used extensively as a model organism to study human cell biology (28–31) and virology (32–35). Importantly, fission yeast is a well-tested model for studying highly conserved cellular activities (36, 37) such as those affected by ZIKV, including cellular growth, cell-cycle regulation, cell death, and autophagy (6–8, 18, 38). The study of ZIKV effects on these vital cellular activities in fission yeast is expected to be functionally and clinically relevant for understanding the ZIKV-associated diseases. Here, using this approach, we report on the results of cloning and functional characterization of all 14 proteins and small peptides of the ZIKV genome.
Results
Subcellular Localizations of ZIKV Proteins.
The ER is the primary subcellular reservoir for ZIKV replication and propagation (18). However, little is known about the subcellular localization of ZIKV proteins. The presumed associations of ZIKV proteins with specific intracellular membranes are based largely on protein sequence structures and on known associations in other flaviviruses (Fig. 1A) (17). In our initial experiment, we examined the subcellular localization of each of the ZIKV proteins.
Each ZIKV protein sequence was fused with N-terminal GFP on a gene-expression vector pYZ3N with an inducible nmt1 (no message in the thiamine) promoter (Fig. S1) (39). GFP-ZIKV proteins were expressed in a wild-type fission yeast strain, SP223 (40, 41). The subcellular localization of each GFP-ZIKV protein was determined using fluorescence microscopy. Fission yeast cells that expressed GFP from an otherwise empty pYZ3N vector were used as a control (39). The results of this experiment are summarized in Table 1. The primary subcellular localization of each ZIKV viral protein is shown in Fig. 1B.
Fig. S1.
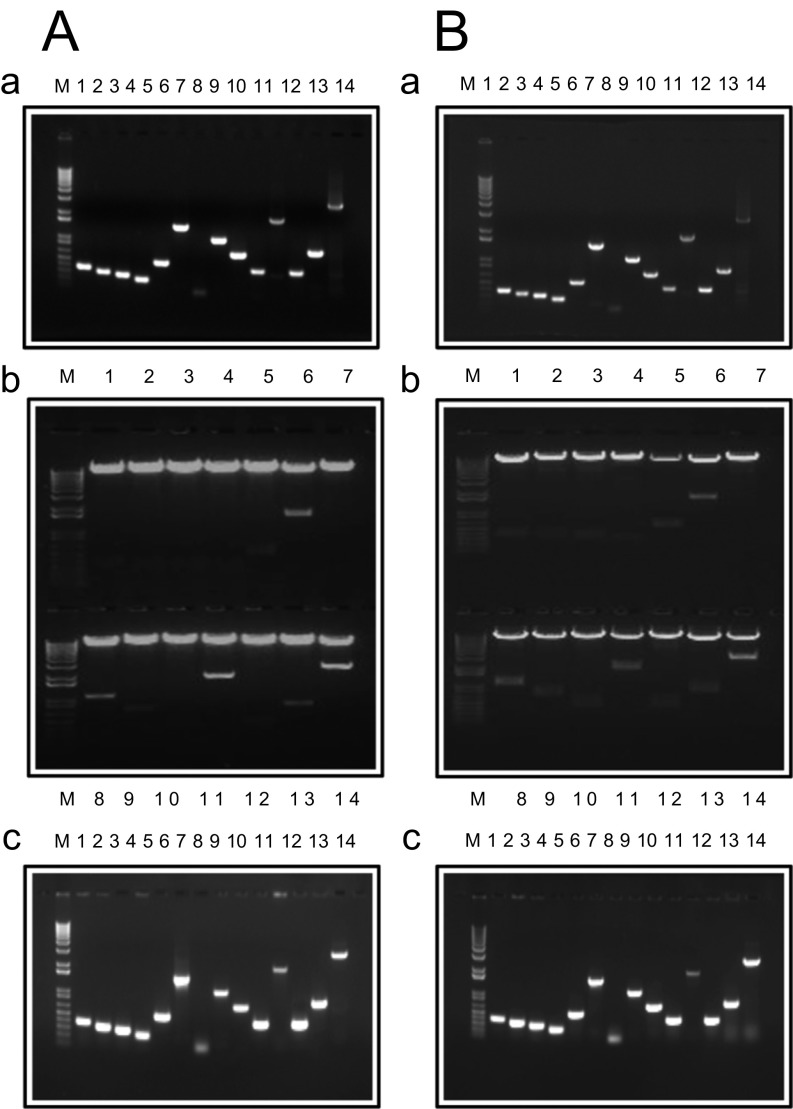
Confirmation of molecular cloning of the ZIKV MR766 viral genome into the fission yeast gene-expression pYZ1N and pYZ3N system. (A) Molecular cloning into the pYZ1N gene expression vector. (a) Results of PCR-amplified products of each ZIKV ORF-encoding DNA sequence from ZIKV viral cDNA. (b) Endonuclease restriction digestions of the ZIKV ORF-encoding DNA inserts that were cloned into the pYZ1N. Plasmid DNAs were propagated and isolated from E. coli. (c) Results of yeast colony PCR showing correct inserts of respective ZIKV ORF-carrying plasmids from transformed fission yeast SP223 cells. (B, a–c) Gels for molecular cloning of the ZIKV genome into the pYZ3N vector are shown as in A. The order of ORFs from 1–14: anaC, C, PR, M, prM, E, 2K, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. M, molecular marker.
Table 1.
Summary of primary subcellular localizations of ZIKV proteins
| ZIKV proteins | cDNA, bp | No. amino acids | Predicted intracellular membrane association* | Intracellular membrane association | Predominant subcellular location | Additional protein features* |
| Structural proteins | ||||||
| anaC | 366 | 122 | Yes | Yes | Nuclear membrane | Nuclear localization signals |
| C | 312 | 104 | Yes | Yes | Nuclear membrane | Nuclear localization signals |
| prM | 504 | 168 | Yes | Yes | ER with speck appearance on cytoplasmic membrane | N-glycosylation |
| M | 225 | 75 | Yes | Yes | ER | |
| Pr | 279 | 93 | No | No | Uniformly distributed with stronger nuclear presentation | |
| E | 1,512 | 504 | Yes | Yes | ER with cytoplasmic specks | Transmembrane helix |
| Nonstructural proteins | ||||||
| NS1 | 1,056 | 352 | No | No | Cytoplasmic puncta | |
| NS2A | 678 | 226 | Yes | Yes | Golgi and low in ER | |
| NS2B | 390 | 130 | Yes | Yes | ER | Cofactor of protease |
| NS3 | 1,851 | 617 | No | No | Uniformly distributed | Protease |
| NS4A | 381 | 127 | Yes | Yes | ER | |
| 2K | 69 | 23 | No | No | Uniformly distributed | |
| NS4B | 753 | 251 | Yes | Yes | ER | |
| NS5 | 2,709 | 903 | No | No | Uniformly distributed | Methyl transferase; RNA-directed RNA polymerase |
In fission yeast cells that expressed a vector control plasmid, the expression of GFP protein alone showed no preference for subcellular localization, and GFP dispersed uniformly throughout the cells (Fig. 1 B, a) (39). In contrast, individual ZIKV proteins appeared to localize primarily to a particular subcellular location. Specific subcellular locations of structural and nonstructural proteins are shown in Fig. 1 B, b and c. ZIKV proteins could be divided into membrane-associated and non–membrane-associated proteins. Membrane-associated proteins associated primarily with the ER network, including the nuclear membrane, ER, and Golgi. Both anaC and C localized predominantly to the nuclear membrane, although both also were observed in the cytoplasm. prM, M, E, NS2B, NS4A, and NS4B were associated primarily with the ER network, which was easily recognized by its association with the nuclear membrane and the cytoplasmic membrane (the “double circles” phenotype) (42). NS2A was localized primarily in the Golgi compartment, with a low level of association with the ER. In contrast to the membrane-associated proteins, five proteins, Pr, NS1, NS3, 2K, and NS5, showed no intracellular membrane association. NS1 formed unique and compact cytoplasmic specks that mimicked cytoplasmic puncta (42). The other four non–membrane-associated ZIKV proteins showed no apparent structure within cells. The ER, Golgi, or cytoplasmic puncta-associated subcellular location of ZIKV proteins was confirmed by comparing each ZIKV protein localization with that of cellular proteins that are known to localize to ER [glycosylphosphatidylinositol 16 (Gpi16)], Golgi [yeast nucleosidediphosphatase 1 (Ynd1)], and cytoplasmic puncta [autophagy-related gene 1 (Atg1)] (Fig. 1 C, a) (42). Induction of cytoplasmic puncta by NS1 under nitrogen starvation was further verified by its colocalization with Atg1 (Fig. 1 C, b) (42).
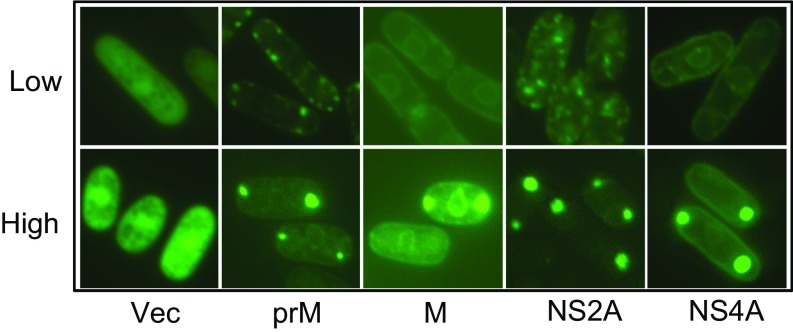
The listings in Table 1 represent the dominant intracellular localization of ZIKV proteins under conditions of low to moderate expression. For some proteins, localization was more complex and depended upon the time and the amount of protein expressed. At low expression levels, both M and NS4A were associated exclusively with the ER, whereas at high expression levels these two proteins accumulated and formed protein pouches in specific cytoplasmic locations, appearing as cytoplasmic puncta (Fig. S2). Similarly, at low expression levels NS2A localized to Golgi-like protein webs, whereas at high expression levels puncta-like protein specks were formed. One of the common features of ZIKV proteins was that, at high expression levels, they all formed cytoplasmic puncta-like structures, as is often seen with cellular autophagy linked to cellular or oxidative stress (43, 44). These structures were not artifacts of protein overproduction, because a high level of GFP alone did not show a similar effect.
Fig. S2.
Comparison of intracellular localization patterns of ZIKV proteins under high and low levels of protein expression. Only ZIKV proteins that showed differences in localization patterns and possible cytoplasmic puncta are shown here. All GFP-ZIKV proteins were expressed from the pYZ3N gene-expression vector through the inducible nmt1 promoter. The high level of ZIKV protein was produced with full induction of the nmt1 promoter by complete removal of thiamine from the growth medium. The subcellular localizations of each ZIKV protein were observed between 24 and 48 h after GI. The low level of ZIKV protein was produced with partial induction of the nmt1 promoter using 10 nM thiamine in the growth medium (14, 51). The subcellular localizations of each ZIKV protein were observed within 20 h after GI.
Overall, the results of these experiments indicated that the subcellular localizations of ZIKV proteins, as observed in fission yeast cells, were in agreement with the predicted membrane associations based on their protein structures (Fig. 1 A and B) (17).
Seven ZIKV Proteins Affect Cellular Proliferation.
One of the hallmarks of microcephaly is reduced brain development. ZIKV-induced microcephaly may be caused by some combination of intrauterine growth restriction, reduced cell proliferation, reduced neuronal cell layer volume, or cell death/apoptosis (2, 7, 45). Here we used the colony-forming assay to identify viral factors that affect cell proliferation (46, 47).
The 14 ZIKV proteins or small peptides, cloned into the fission yeast pYZ1N gene expression vector with an inducible nmt1 transcriptional promoter, were expressed in wild-type fission yeast, the SP223 strain, grown on selective agar plates (Fig. S1) (39). Fission yeast cells expressing an empty pYZ1N plasmid were used as a control. The ability of ZIKV protein-expressing cells to form yeast colonies on agar plates was used as an indication of cellular growth and of potential ZIKV-induced cytotoxicity. ZIKV protein-specific effects were identified by comparing the yeast colony-forming ability of transformed cells expressing the same plasmid grown on agar plates under conditions of gene induction (GI) (gene-on) vs. gene repression (gene-off). Because there are no specific antibodies against ZIKV proteins, expression of all 14 ZIKV genes in fission yeast cells was confirmed by measuring mRNA transcripts with RT-PCR analysis (Fig. 2 A, a).
Fig. 2.
The cytopathic effects of ZIKV proteins. (A) The effects on cell proliferation. (a) Expression of ZIKV mRNA transcripts measured 24 h after GI by RT-PCR. The order of proteins from 1–14: anaC, C, PR, M, prM, E, 2K, NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5. M, molecular marker. (b) Effect of ZIKV expression on fission yeast colony formation. Agar plates are numbered as in a. An empty pYZ1N vector was used as a control and designated as zero. ZIKV-carrying cells grown on selective EMM agar plates were incubated at 30 °C for 4–5 d before the images were captured. Off, gene-suppressed; On, gene-induced. (c) Growth curve analysis was used to quantify growth inhibition by ZIKV proteins. Only the effect of ZIKV proteins (anaC, C, E, prM, M, NS2B, and NS4A) that affected yeast colony-forming abilities as demonstrated in b are shown. Cell growth was measured spectrophotometrically at OD650 over the indicated time period. The experiment was repeated at least three times, and the SEs of each time point were calculated. ●, gene-on, the ZIKV gene was induced; ○, gene-off, the ZIKV gene was suppressed. (B) The effects on cell morphology. Only ZIKV proteins that affected cell proliferation as demonstrated in A, c are shown. The effects of all ZIKV proteins on fission yeast nuclear morphology are included in Fig. S3. (a) Individual cell morphology. Each image was taken 45 h after GI using bright-field microscopy. Note cell hypertrophy (arrow) in the NS4A-expressing cells. Vec, empty vector. (b) Overall cell morphology as shown by the FSC analysis. Ten thousand cells were measured 48 h after GI. The FSC measures the distribution of all cell sizes. The SSC determines intracellular complexity. (C) Effects on nuclear morphology and cell-cycle regulation. (a) Nuclear morphology. Cells were stained with Hoechst blue fluorescent DNA dye. All cell and nuclear morphologies were examined 45 h after GI. (Scale bar, 10 μm.) Effects of all ZIKV proteins on fission yeast nuclear morphology are included in Fig. S4. (b) The effect of ZIKV protein on cell-cycle G1 regulation. Cells were grown in regular EMM medium in which cells normally reside in the G2/M phase of the cell cycle. (c) Effect of ZIKV proteins on cell-cycle G2/M regulation. Cells were grown in a low-nitrogen EMM medium that enriches cells in the G1 phase of the cell cycle, as described previously (1, 2). Cell-cycle profiles were measured by DNA content using flow cytometric analysis 48 h after GI. Average and SD values were calculated based on the results of three independent experiments. A pairwise Student t test was conducted to compare the DNA content values of each ZIKV protein with and without GI. *P < 0.01. Arrows indicate the location of differences (C, b and c) or where the abnormal cells reside (B, b and C, a).
As shown in Fig. 2 A, b, all cells in which the ZIKV viral gene was repressed formed normal-sized colonies on the agar plates. Also, on the gene-inducing plates, yeast cells carrying an empty pYZ1N plasmid formed normal-sized colonies. However, fission yeast strains expressing seven ZIKV proteins (anaC, C, prM, M, E, NS2B, and NS4A) showed various levels of inhibitory effects on yeast colony formations. In each case, either no colony or very small colonies were seen under ZIKV gene-inducing conditions.
The expression of these seven ZIKV proteins in the fission yeast cells may have prevented colony formation by inhibiting cellular growth or otherwise affecting cell viability. To test whether these seven ZIKV proteins affected cell proliferation, we measured the growth kinetics of these ZIKV-carrying yeast cells. Fission yeast cells were grown under the gene-off and gene-on conditions in liquid minimal and selective EMM medium. Cellular growth was measured as cell concentration by spectrophotometry (optical density OD650) as a function of time at 16–48 h after ZIKV GI. In control cultures, two indistinguishable growth curves with typical logarithmic kinetics were observed for both the gene-inducing and gene-suppressing pYZ1N-carrying yeast cells. In contrast, cells expressing the seven selected ZIKV proteins (anaC, C, prM, M, E, NS2B, and NS4A) showed various degrees of delayed proliferation (Fig. 2 A, c). During the first 24 h after GI, the ZIKV gene-off and gene-on cells grew at about the same rate. When the ZIKV genes were fully expressed 24 h after GI (41, 48), the growth of cells producing the seven ZIKV proteins became slower than that of cells without gene expression. Among the cells expressing ZIKV proteins, those expressing prM and NS2B showed nearly complete inhibition of cellular growth, whereas those expressing anaC, C, M, E, and NS4A showed reduced growth (Fig. 2 A, c).
ZIKV Proteins Induce Cell Elongation and Hypertrophy.
An early study reported that ZIKV infection induces hypertrophy (gross cell enlargement) of mouse astroglial cells (18). To determine which ZIKV protein produces this effect, we examined the morphology of cells producing ZIKV proteins using bright-field microscopy (Fig. S3). Fission yeast cells carrying the control vector pYZ1N appeared normal (36), with a typical rod-shaped morphology measuring 3–4 µM in diameter and 7–14 µM in length (Fig. 2 B, a) (36). Interestingly, the same seven ZIKV proteins (anaC, C, prM, M, E, NS2B, and NS4A) that reduced the yeast colony-forming ability (Fig. 2 A, b) and cellular growth (Fig. 2 A, c and Fig. S2) also affected cell morphology (Fig. 2 B, a). Some of the cells producing ZIKV proteins were clearly longer than cells without ZIKV gene expression. About 5–20% of the cells expressing prM, NS2B, or NS4A appeared to be grossly enlarged (Fig. 2 B, a and b). Some of the cells expressing NS4A appeared balloon-like, suggesting induction of cell hypertrophy (Fig. 2 B, a, arrow).
Fig. S3.
Effect of ZIKV protein production on fission yeast cell morphology. (A) Individual cell morphology. (a) Structural ZIKV proteins. (b) Nonstructural ZIKV proteins. Each image was obtained 45 h after GI using bright-field microscopy. (B) Overall cell morphology as shown by FSC analysis. Ten thousands cells were measured 48 h after GI. The FSC measures the distribution of all cell sizes. The SSC determines intracellular complexity. (a) Structural ZIKV proteins. (b) Nonstructural ZIKV proteins.
To measure the overall changes in cell morphology induced by ZIKV proteins, cells expressing ZIKV proteins were analyzed by forward-scatter analysis using flow cytometry. Both the forward-scattered light (FSC) and side-scattered light (SSC) were measured for each cell population (Fig. S3). The FSC is proportional to the cell-surface area and thus measures cell size. The SSC is proportional to cell granularity and thus determines intracellular complexity. Correlated measurements of FSC and SSC allow the differentiation of cell shapes in a heterogeneous cell population. As shown in Fig. 2 B, b, no significant difference of overall cell architecture was found between control (vector-carrying) yeast cells grown in the gene-inducing and gene-repressing conditions. In contrast, marked differences were observed in the cells expressing prM, E, NS2B, and NS4A (Fig. 2 B, b, arrows).
Together, our findings showed that seven of the ZIKV proteins (anaC, C, prM, M, E, NS2B, and NS4A) caused elongation of cells. The prM, NS2B, and NS4A proteins also induced cell hypertrophy.
ZIKV Proteins Cause Nuclear Fragmentation and Cell-Cycle Dysregulation.
Early studies showed that ZIKV infection induces hyperchromatic nuclear debris and cell-cycle dysregulation in mouse astrocytes and human progenitor stem cells (7, 8, 18). To test whether the hyperchromatic debris resulted from chromosomal aberrations, we examined the nuclear morphology of all ZIKV proteins using Hoechst DNA-staining fluorescent dye (41). Results of the seven ZIKV proteins tested above are shown in Fig. 2 C, a, and data on all the ZIKV proteins are provided in Fig. S4. A normal, round, compact nuclear morphology was seen in vector-carrying control cells. As in controls, no nuclear morphological abnormality was observed in M-expressing cells. In contrast, fragmented nuclei were observed in cells expressing anaC, prM, E, NS2B, and NS4A. These abnormal nuclear morphologies were particularly evident in elongated cells with abnormal cellular morphology (Fig. 2 C, a, arrows). In C-expressing cells, although most of the nuclei appeared to be normal, some of the dividing cells were either missing a nucleus or had two nuclei (Fig. 2 C, a, arrows).
Fig. S4.
Effects of ZIKV proteins on nuclear morphology and cell-cycle regulation. (A) Nuclear morphology. Cells were stained with Hoechst blue fluorescent DNA dye. All cell and nuclear morphologies were examined 45 h after GI. (a) An empty pYZ1N vector control. (b) Structural ZIKV proteins. (c) Nonstructural ZIKV proteins. (B) Cell-cycle profiling with starting cells predominantly in the G2 phase of the cell cycle. All cells were grown in regular EMM medium. The effect of ZIKV protein on the cell cycle was measured by flow cytometry 48 h after GI. (a) Structural ZIKV proteins. (b) Nonstructural ZIKV proteins. (C) Cell-cycle profiling with starting cells predominantly in the G1 phase of the cell cycle. All cells were grown in the low-nitrogen EMM medium to enrich G1 cells (6). The effect of ZIKV protein on cell cycle was measured by flow cytometry 48 h after GI. (a) Structural ZIKV proteins. (b) Nonstructural ZIKV proteins.
Elongated fission yeast cells are indicative of cell-cycle dysfunction. Next, we tested whether the expression of the ZIKV proteins affected cell-cycle phase distributions by measuring the overall cellular DNA content using flow cytometric analysis. In the standard growth EMM medium, fission yeast cells reside predominantly in the G2 phase of the cell cycle (Fig. 2 C, b, Upper) (49). If a ZIKV protein induces cell-cycle G1 delay, we would expect a shift from a predominantly G2 cell population to a G1 population after GI. Similarly, ZIKV-induced cell-cycle G2 delay would be detected by first synchronizing cells in the G1 phase of the cell cycle (Fig. 2 C, c, Top) and then turning on the ZIKV gene expression to measure the ZIKV effect (Fig. 2 C, c).
As shown in Fig. 2 C, b, the cell-cycle profile was essentially the same in control (vector-carrying) cells in the gene-inducing and gene-suppressing conditions. However, expression of prM resulted in an ∼20% increase in the G1 cell population (Fig. 2 C, b). Similarly, in the G1-synchronized cells (Fig. 2 C, c), the percent of G2 cells was essentially the same in control cells in the gene-inducing and gene-suppressing conditions. However, 23.3, 37.8, 52.3, and 40.3% increases in the G2 cell populations were observed in the cells expressing anaC, M, E, and NS4A, respectively. A pairwise statistical t test suggested that the observed differences were significant (P < 0.01). Therefore, the seven ZIKV proteins, with the exception of the C protein, affected cell-cycle regulation.
ZIKV Proteins Induce Cell Death and Cellular Oxidative Stress.
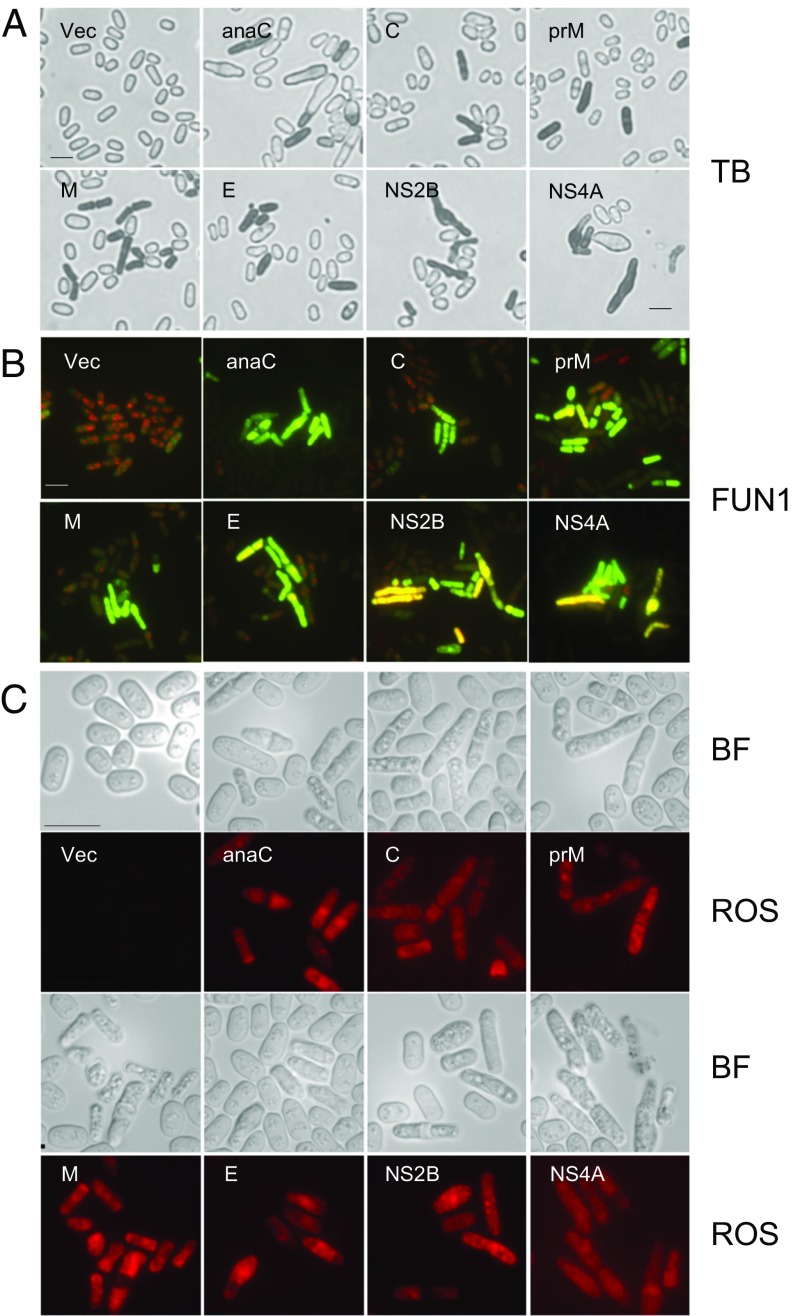
ZIKV infection leads to cell death and apoptosis in some neuronal cells (7, 8). The cytopathic effects described above could have potentially lethal consequences. We tested whether any of the seven ZIKV proteins tested above would cause cell death. Cell death was first evaluated by staining cells expressing ZIKV protein with a vital diazo dye, Trypan blue (50). As shown in Fig. 3A, live fission yeast cells exclude Trypan blue. However, Trypan blue staining was observed in all seven ZIKV protein-expressing cells, indicating the presence of dead cells.
Fig. 3.
ZIKV proteins induce cell death and cellular oxidative stress. ZIKV-induced cell death was measured 48 h after GI by Trypan blue (TB) staining (A) and the yeast live/dead assay (B), which was measured by FUN-1 staining 40–45 h after GI. (C) ZIKV induces oxidative stress, as indicated by DHE staining showing ROS expression. Images were taken 48 h after GI. (Scale bar, 10 μm.) BF, bright-field.
To further assess the viability of cells expressing ZIKV protein, we used an assay that determines the intracellular metabolic status as a measure of cell viability by FUN-1 staining (47, 51). As shown in Fig. 3B, yeast cells without ZIKV gene expression (vector-expressing cells) stained orange-red, but the cells expressing ZIKV protein remained yellow-green, indicating they were metabolically inactive.
Induction of oxidative stress leads to cell death in other flaviviruses, including DENV and JEV (52–54). To explore the possible molecular mechanism underlying ZIKV-induced cell death, we evaluated ZIKV-induced intracellular stress by measuring reactive oxygen species (ROS). A ROS-specific dye, Dihydroethidium (DHE), which produces red fluorescence in the presence of ROS, was used to measure cellular oxidative stress 48 h after ZIKV GI (Fig. 3C). Strong red fluorescence was detected in all cells expressing the seven ZIKV proteins, whereas no red fluorescence was observed in control cells. These data suggested that the observed cell death induced by the seven ZIKV proteins was caused at least in part by the induction of intracellular oxidative stress.
NS4A-Induced Cell Hypertrophy and Growth Delay Are Mediated Through the Target of Rapamycin-Mediated Cellular-Stress Response Pathway.
Because NS4A induced cellular oxidative stress (Fig. 3 C, c), we hypothesized that the ROS-induced Target of rapamycin (TOR) cellular-stress response would be affected (55). NS4A protein was expressed via the nmt1 inducible promoter in wild-type, Tor1-deletion (∆Tor1), and type 2A phosphatase activator Tip41-deletion (∆Tip41) mutant strains (Fig. 4A). Tor1 protein is a key protein in the TOR pathway; it is required for cellular responses to external stresses such as nitrogen starvation (55), and it regulates cellular growth and the control of cell size (56). Indeed, deletion of the Tor1 gene affected cell size and growth (Fig. 4 A, a, Left and 4 D, Middle). The Tip41 protein also is involved in the cellular response to nitrogen starvation, and it negatively regulates the TOR signaling pathway in budding yeast (57, 58). Deletion of the Tip41 gene had no clear effect on cell size (Fig. 4 A, a) and cellular growth (Fig. 4 D, Bottom). However, when TIP41 was produced in fission yeast cells, it induced short, spherical cells (Fig. 4 A, b, Right), suggesting that it also is involved in the control of cell size (58).
Fig. 4.
NS4A impacts the TOR pathway. (A) The effect of TOR pathway-related gene deletions on NS4A-induced hypertrophy and cellular growth. (a) Cell morphologic changes when NS4A protein was expressed 48 h after GI in wild-type, ∆Tor1, and ∆Tip41 mutant strains. Note that NS4A induced hypertrophy in wild-type cells. However, no apparent gross cell enlargement was seen in the NS4A-expressing ∆Tor1 cells. Conversely, NS4A induced a spherical cell phenotype in the ∆Tip41 mutant strain. (b) Overexpression of the Tip41 gene in the ∆Tip41 mutant strain produced a spherical cell phenotype similar to that shown in a. (Scale bar, 10 μm.) (B) ZIKV NS4A gene transcription by RT-PCR 24 h after GI. (C) The ∆Tor1 deletion suppressed the effect of NS4A on yeast colony formation. (D) ∆Tor1 deletion restored (↑) cellular growth to nearly the normal level in the NS4A-expressing cells; ∆Tip41 worsened NS4A-induced growth inhibition (↓). ●, NS4A gene expressed; ○, NS4A gene suppressed.
We showed above that the expression of NS4A protein in the fission yeast cells induced gross enlargement and growth delay (Fig. 2 B). When the NS4A-carrying plasmid was expressed in the ∆Tor1 fission yeast cells, NS4A-induced cell hypertrophy was largely abolished (Fig. 4 A, a, Right). Moreover, cell growth was restored to nearly normal levels, as indicated by the colony-forming assay (Fig. 4 C, Center) and growth kinetics analysis (Fig. 4 D, Middle). In contrast, the expression of NS4A protein in the ∆Tip41 cells worsened NS4A-induced growth delay (Fig. 4 D, Bottom). Moreover, it intensified NS4A-induced cell hypertrophy (Fig. 4 A, a, Bottom Right) and showed a phenotype that mimicked the effect of Tip41 overexpression. Indeed, as shown in Fig. 4 A, b, Right, overexpression of the Tip41 gene in the same ∆Tip41 mutant cells generated a similar spherical morphology.
The results of these experiments suggested that NS4A-induced hypertrophy and growth delay are likely mediated by the TOR cellular stress-response pathway, specifically via Tor1 and Tip41.
Discussion
In this study we characterized the ZIKV genome in a fission yeast cell system. We demonstrated that the ZIKV proteins that contain membrane-associated domains localized along the ER-associated network, including the nuclear membrane, ER to Golgi (Fig. 1 B and C and Table 1). All the structural proteins, with the exception of Pr, and two nonstructural ZIKV proteins (NS2B and NS4A) conferred cytopathic effects that included inhibition of growth/proliferation (Fig. 2A), cell hypertrophy (Fig. 2B), cell-cycle dysregulation (Fig. 2C), and cell death (Fig. 3). prM promoted a G1 shift of the cell cycle, whereas anaC, M, E, and NS4A caused cell-cycle G2/M phase accumulation. Our study further showed that these seven ZIKV proteins induced cellular oxidative-stress responses (Fig. 3C) that were associated with cell death. Genetic study of the molecular actions underlying the NS4A-induced cytopathic effects suggested that cellular hypertrophy and growth restriction were mediated through the Tor1 and Tip41 proteins of the TOR cellular stress-response pathway (Fig. 4). Whether the other six ZIKV proteins exert the same effect as NS4A on the TOR pathway is yet to be determined.
Two types of ZIKV proteins were identified in this study, membrane-associated and non–membrane-associated proteins (Table 1). Consistent with the idea that the ER is the major ZIKV subcellular “viral factory” in which ZIKV proliferates (18, 20, 59), membrane-associated ZIKV proteins were found to associate mostly with the ER network (Fig. 1B). The mature C protein is derived from anaC, and both were found predominantly on nuclear membranes. Notably, in DENV, three nuclear localization signals are embedded in these two proteins (60). The prM and M proteins also are related structurally. The transition of prM to M by Furin cleavage is crucial for viral maturation and infectivity in flaviviruses (26, 27). It thus was of interest to note the subtle differences in the localization of these three proteins: Both prM and M localized primarily on the ER, but M localized almost exclusively on the ER, whereas prM also accumulated along cytoplasmic membranes (Fig. 1B). Moreover, prM appeared to enhanced the G1 phase of the cell cycle (Fig. 3 B, a), whereas M enriched the G2/M phase of the cell cycle (Fig. 3 B, b). The Pr peptide, which is generated after the cleavage of prM to form M, showed relatively strong presence in nuclei but also was dispersed throughout the cell.
Interestingly, even at low levels of expression, the expression of NS1 formed unique, compact cytoplasmic protein specks within cells (Fig. 1 B, c). Similar protein specks and patterns also were observed in a number of highly expressed ZIKV proteins, including prM-, M-, NS2A-, and NS4A (Fig. S2). Because viral infection of cells could produce both low and high levels of proteins, depending on the degree of viremia, the formation of these accumulated protein specks potentially could be relevant to the severity of ZIKV infection. The nature of these ZIKV protein accumulations in such a unique pattern is unclear at present. However, some of these cytoplasmic protein specks mimic the appearance of cytoplasmic puncta, which often are an indication of autophagy. Autophagy is part of the cellular response to cellular stress such as nitrogen starvation or oxidative stress (43, 44). Consistent with this role, activation of cellular autophagy, which is part of the host innate immune response to eliminate invading intracellular pathogens, is a hallmark of flavivirus infection (59). Autophagy is activated in part following its interruptive association with intracellular membranes and the accumulation of viral factors within ER (59). However, which viral protein triggers cellular autophagy is not fully understood. In fission yeast, the activation of autophagy can be recognized by the formation of random single or multiple cytoplasmic puncta caused by the accumulation of Atg proteins (61). As with cytoplasmic puncta of autophagy (61), the ZIKV-induced puncta-like structures also were seen as either single random protein specks or multiple specks, suggesting that these ZIKV proteins might mimic the actions of the Atg proteins (Fig. 1 C, a). Indeed, under nitrogen-starvation conditions, specific protein specks appeared in cells and showed a distribution pattern similar to that of Atg1 (Fig. 1 C, b). Further detailed study of ZIKV proteins is needed to determine whether they activate autophagy.
Several studies have demonstrated that a number of ZIKV-induced cytopathic effects may contribute to microcephaly, including the inhibition of neuronal cell proliferation, disturbance of host cell-cycle regulation, and the induction of cell death or apoptosis (3, 7, 8, 62). However, the ZIKV protein(s) responsible for these cytopathic effects was unknown. We demonstrate here that five structural ZIKV proteins (anaC, C, prM, M, and E) and two nonstructural ZIKV proteins (NS2B and NS4A) contribute, at least in part, to ZIKV-induced cytopathic effects, including growth restriction (Fig. 2A), cell hypertrophy (Fig. 2B), cell-cycle dysfunction (Fig. 2C), and the induction of cell death (Fig. 3). Interestingly, all the structural ZIKV proteins, with the exception of the Pr signal peptide, conferred cytopathic activities. Because anaC/C or prM/M are essentially the same protein present in different forms at different phases of the viral life cycle, it would be interesting to examine their differences and delineate their potential roles in interacting with host cells.
The E protein is a major viral surface protein involved in various steps of the viral life cycle (63). Early sequence and structural comparisons of the ZIKV E protein with that of other flaviviruses suggest that overall the ZIKV E protein is unique among flaviviruses, although parts of it resemble its homologs in WNV, JEV, or DENV (64, 65). During flaviviral assembly, E interacts with prM to form the prM–E heterodimers that protrude from the viral surface in the noninfectious and immature viral particles (25). It also is involved in fusing the viral membrane with the host endosome membrane. Therefore, it potentially could be a useful antigen for the development of vaccines (66, 67). In addition, we demonstrated here that the ZIKV E protein has strong cytotoxic effects, including cell death. Thus, it may be an attractive viral target for weakening ZIKV-induced cytopathic effects.
Two nonstructural proteins, NS2B and NS4A, also displayed cytopathic effects similar to those of the structural proteins. Like the structural proteins, both NS2B and NS4A are membrane-associated proteins that localized primarily to the ER (Fig. 1 B, c). The lethal effect of NS2B is interesting because it encodes a cofactor of viral protease that is required for enhancing enzymatic activity and substrate specificity (68, 69). Coexpression of the NS2B-NS3 protease induces cell death and apoptosis in DENV, WNV, and JEV (70–72). It is not known whether the ZIKV NS2B-NS3 protease also induces apoptosis. Conceivably, the observed ZIKV-induced cytopathic effects also could be caused by combinations of different ZIKV proteins. As a result, it is possible that proteins that are nontoxic by themselves may become toxic when they form a complex, and vice versa.
The ER is the virus factory in ZIKV-infected neuronal cells in the newborn mouse brain (18). Examination of ZIKV-infected neurons, astrocytes, and pyriform cells by electron microscopy suggested that the most striking changes were hypertrophy of astrocytes and hyperchromatic nuclear debris in pyriform cells (18). Here we demonstrated that the NS4A protein localized specifically to the ER of fission yeast cells, where it induced hypertrophy and nuclear fragmentation (Figs. 1 B, c and 2 B and C, a).
Our genetic and mechanistic studies of the molecular actions of NS4A revealed that NS4A-induced growth retardation and cytopathic effects were mediated specifically through Tor1 and Tip41 of the TOR cellular stress-response pathway (Fig. 4). In particular, we showed that deletion of Tor1 essentially blocked NS4A-induced cell hypertrophy, inhibition of colony formation, and growth delay (Fig. 4), suggesting that Tor1 was primarily responsible for these effects. These findings are consistent with the role of Tor1 in regulating cellular growth and cell size control (55, 56). Our data further suggest that the NS4A effects also were mediated, at least in part, via inhibition of Tip41 activity. The expression of NS4A protein in the ∆Tip41 cells worsened the NS4A-induced growth delay (Fig. 4 D, Bottom). Moreover, it intensified NS4A-induced cell hypertrophy (Fig. 4 A, a, Bottom Right) by producing a spherical phenotype (Fig. 4 A, a, Bottom Right). Most interestingly, this spherical phenotype mimicked the results of Tip41 (Fig. 4 A, b, Right), suggesting the action of NS4A behaved at least in part like that of Tip41 protein. This premise is supported by the fact that Tip41 is a negative regulator of Tor1 (57, 58). Additional experiments are needed to verify this possibility.
The TOR signaling pathway is a highly conserved regulatory system across the evolutionary span from yeast to humans (73, 74). It is a central cellular sensor system that is responsible for the reaction and adjustment to cellular stress such as oxidative stress, nitrogen starvation, and flaviviral infections (75, 76). The fine balance between autophagy and programmed cell death following viral infection is controlled in part by the TOR pathway (77). Our findings on the actions of NS4A indicate that NS4A plays a functional role in manipulating this regulatory signaling network. Notably, during the preparation of this manuscript, a report was published showing that NS4A and NS4B proteins induce autophagy and inhibit neurogenesis through deregulation of Akt–mTOR signaling in human fetal neural stem cells (38). Our results described here are largely in agreement with that report. In addition, we showed that NS4A confers its cytopathic effects specifically through Tor1 and Tip41, which are the human equivalents of Tsc1 and Tip41 proteins (58, 78). It would be interesting to examine further whether the other six ZIKV proteins exert an effect similar to that of NS4A.
In summary, we characterized all the ZIKV proteins and small peptides in the fission yeast model system. In searching for the viral factors that are responsible for inhibiting cell proliferation, disturbance of cell-cycle, and induction of cell death during ZIKV infection, we discovered that five structural ZIKV proteins and two nonstructural ZIKV proteins conferred effects similar to those observed in ZIKV-infected mouse or human neuronal cells (1, 3, 7, 8). The results of our mechanistic studies suggest that those cytopathic effects likely resulted from cellular oxidative stress and the activation of cellular autophagy. In particular, we showed that NS4A causes cytopathic effects by modulating the TOR signaling pathway through coordinated and counterbalancing actions on Tor1 and Tip41. The results presented here provide a primary reference for future studies validating these effects in mammalian cells.
Materials and Methods
Cell and Growth Media.
A wild-type fission yeast SP223 strain (h-, ade6-216, leu1-32, ura4-294) was used in this study (36, 41). The Tor1-deletion mutant strain MA99 (h-, tor1::ura4+ leu1-32, ura4-∆18, ade6-M216) and the Tip41-deletion mutant strain I3-24 (h−, vsp24::ura4+; leu1-32, ura4-294, ade6-M216) were described previously (55, 58). Standard yeast extract with supplements (YES) complete medium or minimal EMM selective medium supplemented with adenine, uracil, leucine, or thiamine (20 µM) was used to grow fission yeast cells or to select for plasmid-carrying cells, respectively. LB medium supplemented with Ampicillin (100 µg/mL) was used for growing Escherichia coli Top 10 or DH5α cells and for DNA transformation. All reagents used to prepare for the yeast and bacterial culture media, including thiamine, were purchased from Sigma-Aldrich.
Plasmids.
A fission yeast gene-expression vector system, which includes pYZ1N and pYZ3N gene-expression vectors, was used as previously described (39). The pYZ1N vector was used to test ZIKV gene functions. The pYZ3N vector was used to visualize the intracellular location of each viral protein, with a GFP tag at the 5′ end of the protein. These plasmids carry an nmt1 promoter. Under this inducible gene-expression system, viral gene expression can be either repressed or induced in the presence or absence of thiamine, respectively (48, 79). These vectors carry a LEU2 gene as a selection marker.
Molecular Cloning of ZIKV Proteins in Fission Yeast.
ZIKV MR766 viral genomic cDNA was generated by reverse transcription of ZIKV genomic viral RNA. It was derived from a viral extract that was isolated from ZIKV-infected Vero cells (a gift from H. Tang, Florida State University, Tallahassee, FL). The ZIKV reference strain MR766 was used for this study because it was the first ZIKV strain isolated from the Zika forest of Uganda (80). It is highly neurotropic and is virulent in young mice, and its replication in mouse brain causes death (81). It also causes a number of cytopathic effects including cell-cycle abnormality and cell death in human neuronal progenitor cells (7), and it induces a microcephaly-like phenotype in a human BRSOs model (2, 7).
A total of 14 known MR766 ZIKV viral protein and peptide products were shotgun-cloned into a fission yeast gene-expression vector system that we developed for large-scale cloning (39, 82). All gene cloning was done in a unidirectional fashion with positive identification of the gene insertions, based on α-complementation of X-Gal (Sigma) in E. coli. An inducible gene transcriptional nmt1 promoter (39, 48) was used to allow the measurement of gene-specific effects. Molecular cloning of the ZIKV proteins into the pYZ3N GFP-carrying vector generated 5′ GFP-tagged ZIKV proteins; ZIKV genes cloned in the pYZ1N gene-expression vector are without tags. All the primers that contained specific restriction enzymes for molecular cloning and were used to clone the 14 ZIKV viral protein-encoding nucleotides into pYZ1N and pYZ3N vectors are listed in Tables S1 and S2. The PCR DNA fragments that were amplified with specific primers for cloning into the pYZ1N and pYZ3N vectors are shown on agarose gels (Fig. S1 A, a and B, a). The ZIKV viral gene inserts cloned into the pYZ1N and pYZ3N vectors were verified by restriction digestions (all restriction endonucleases were purchased from New England Biolabs) (Fig. S1 A, b and B, b) and by Sanger sequencing.
Table S1.
Primers used to clone the Zika genes into the pYZ1N fission yeast expression vector
| Gene name | Size, bp | 5′ primer | 3′ primer | Cloning sites |
| anaC | 366 | CTCG CATATG AAAAACCCAAAGAAGAAATCC | CTCG GGATCC TTA TGCCATGGCTGTAGTCAGCAG | NdeI/BamHI |
| C | 312 | CTCG CATATG AAAAACCCAAAGAAGAAATCC | CTCG GGATCC TTA ACGTCTCTTCCTCTCTTTCCTAGC | NdeI/BamHI |
| Pr | 279 | CTCG CATATG GCAGAGATCACTAGACGCGGG | CTCG GGATCC TTA TCTTCTAGATCGCCGTGCCTC | NdeI/BamHI |
| M | 225 | CTCG CATATG GCCGTGACGCTCCCTTCTC | CTCG GGATCC TTA ACTGTATGCCGGGGCAATC | NdeI/BamHI |
| prM | 504 | CTCG CATATG GCAGAGATCACTAGACGCGGG | CTCG GGATCC TTA ACTGTATGCCGGGGCAATC | NdeI/BamHI |
| E | 1,512 | CTCG CATATG ATCAGGTGCATTGGAGTCAGC | CTCG GGATCC TTA AGCAGAAACAGCCGTGGAGAG | NdeI/BamHI |
| 2K | 69 | CTCG CATATG TCTCCCCAAGATAACCAGATGG | CTCG GGATCC TTA TGCAGTTATCAAACCTAGAAGGC | NdeI/BamHI |
| NS1 | 1,056 | CTCG CATATG GACGTGGGGTGCTCAGTGGAC | CTCG GGATCC TTA CGCTGTCACCATTGACCTCAC | NdeI/BamHI |
| NS2A | 678 | CTCG GGATCC ATG GGGTCAACCGATCATATGGAC | CTGTC GCGGCCGC TTA CCGCTTCCCACTCCTTGTG | BamHI/NotI |
| NS2B | 390 | CTCG CATATG AGCTGGCCCCCTAGTGAAG | CTCG GGATCC TTA CCTTTTCCCAGTCTTCACATACAC | NdeI/BamHI |
| NS3 | 1,851 | CTCG CATATG AGTGGCGCCCTCTGGGAC | CTCG GGATCC TTA TCTTTTTCCAGCGGCGAATT | NdeI/BamHI |
| NS4A | 381 | CTCG CATATG GGAGCGGCTTTGGGAGTAAT | CTCG GGATCC TTA TCTTTGCTTCTCTGGCTCGG | NdeI/BamHI |
| NS4B | 753 | CTCG CATATG AACGAACTTGGATGGCTGG | CTCG GGATCC TTA ACGTCTCTTAACCAGGCCAG | NdeI/BamHI |
| NS5 | 2,709 | CTCG GTCGAC ATG GGAGGTGGGACGGGAGAGAC | CTGTC GCGGCCGC TTA CAACACTCCGGGTGTGGACC | SalI/NotI |
Restriction enzymatic sites are underlined.
Table S2.
Primers used to clone the Zika genes into the pYZ3N fission yeast expression vector for the construction of GFP-fusion proteins
| Gene name | Size, bp | 5′ primer | 3′ primer | Cloning sites |
| anaC | 366 | CTGTC GCGGCCGC T AAAAACCCAAAGAAGAAATCCG | CTCG GGATCC TTA TGCCATGGCTGTAGTCAGCAG | NotI/BamHI |
| C | 312 | CTGTC GCGGCCGC T AAAAACCCAAAGAAGAAATCCG | CTCG GGATCC TTA ACGTCTCTTCCTCTCTTTCCTAGC | NotI/BamHI |
| Pr | 279 | CTGTC GCGGCCGC A GCAGAGATCACTAGACGCGGG | CTCG GGATCC TTA TCTTCTAGATCGCCGTGCCTC | NotI/BamHI |
| M | 225 | CTGTC GCGGCCGC A GCCGTGACGCTCCCTTCTC | CTCG GGATCC TTA ACTGTATGCCGGGGCAATC | NotI/BamHI |
| prM | 504 | CTGTC GCGGCCGC A GCAGAGATCACTAGACGCGGG | CTCG GGATCC TTA ACTGTATGCCGGGGCAATC | NotI/BamHI |
| E | 1,512 | CTGTC GCGGCCGC A ATCAGGTGCATTGGAGTCAGC | CTCG GGATCC TTA AGCAGAAACAGCCGTGGAGAG | NotI/BamHI |
| 2K | 69 | CTGTC GCGGCCGC A TCTCCCCAAGATAACCAGATGG | CTCG GGATCC TTA TGCAGTTATCAAACCTAGAAGGC | NotI/BamHI |
| NS1 | 1,056 | CTGTC GCGGCCGC A GACGTGGGGTGCTCAGTGGAC | CTCG GGATCC TTA CGCTGTCACCATTGACCTCAC | NotI/BamHI |
| NS2A | 678 | CTGTC GCGGCCGC A GGGTCAACCGATCATATGGAC | CTCG GGATCC TTA CCGCTTCCCACTCCTTGTG | NotI/BamHI |
| NS2B | 390 | CTGTC GCGGCCGC A AGCTGGCCCCCTAGTGAAG | CTCG GGATCC TTA CCTTTTCCCAGTCTTCACATACAC | NotI/BamHI |
| NS3 | 1,851 | CTGTC GCGGCCGC A AGTGGCGCCCTCTGGGAC | CTCG GGATCC TTA TCTTTTTCCAGCGGCGAATT | NotI/BamHI |
| NS4A | 381 | CTGTC GCGGCCGC A GGAGCGGCTTTGGGAGTAAT | CTCG GGATCC TTA TCTTTGCTTCTCTGGCTCGG | NotI/BamHI |
| NS4B | 753 | CTGTC GCGGCCGC T AACGAACTTGGATGGCTGG | CTCG GGATCC TTA ACGTCTCTTAACCAGGCCAG | NotI/BamHI |
| NS5 | 2,709 | CTGTC GCGGCCGC A GGAGGTGGGACGGGAGAGAC | CTCG GTCGAC TTA CAACACTCCGGGTGTGGACC | NotI/SalI |
Restriction enzymatic sites are underlined.
Recombinant DNA Transformation and Inducible ZIKV Gene Expression.
The ZIKV gene-carrying pYZ plasmids were transformed into the wild-type fission yeast SP223 strain by electroporation (39, 82). Successful transformation of the respective ZIKV-containing plasmids was verified by single-colony PCR (Fig. S1 A, c and B, c) as described previously (82). The transformants were selected for leucine on a minimal selective medium. To measure ZIKV gene-specific activities, a single yeast colony, which carries a specific ZIKV gene-containing plasmid, was grown to log phase on the special EMM liquid medium supplemented with 20 μM of thiamine. Cells then were harvested and washed three times with distilled water to remove thiamine. Finally, 2 × 105 cells/mL were reinoculated into fresh specific EMM liquid medium without thiamine to induce gene expression (gene-on condition) or with thiamine to suppress gene expression (gene-off condition, used as controls). The cell suspensions were incubated at 30 °C with constant shaking before observation (48, 79). Successful expression of the respective ZIKV mRNA transcripts was verified by RT-PCR using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity (catalog no. 12574-030; Invitrogen).
Determination of Subcellular Localization.
The ZIKV viral proteins fused to GFP at their 5′ ends were produced in fission yeast cells from the pYZ3N gene-expression vector. To avoid artifacts caused by high-level expression of the ZIKV proteins, 10 nM of thiamine was added to the EMM medium to reduce the level of ZIKV protein expression (42, 82). The subcellular localization of each ZIKV protein was visualized, typically within 20 h after GI, by fluorescent microscopy. To verify the specific subcellular location of ZIKV protein, fission yeast cellular proteins that were known to locate specifically in ER (Gpi16), Golgi (Ynd1), and cytoplasmic puncta (Atg1) were used for comparison. These proteins were tagged at the C-terminal ends with YFP and were previously described by Matsuyama et al. (42). A Leica DMR fluorescence microscope equipped with a high-performance charge-coupled device camera (Hamamatsu) and Openlab software (Improvision, Inc.) were used for all image analyses. For the observation of GFP alone, a Leica L5 filter, which has an excitation of 480/40 nm and emission of 527/30 nm, was used. For the colocalization of GFP- and YFP-tagged proteins, a Leica S Green (YFP/GFP) filter, which has an excitation of 500/20 nm and emission of 535/30 nm, was used for the observation of both GFP and YFP. Because YFP cannot be excited under the excitation spectrum of the Leica A Aqua (CFP) filter that has an excitation of 436/20 nm and emission of 480/30 nm, no YFP signal is detected under the CFP filter. Thus, that filter was used to distinguish the signal of YFP from that of GFP.
Because ZIKV infection could have high viremia resulting in high levels of ZIKV proteins, each ZIKV protein also was expressed over time with the full strength of the nmt1 promoter without thiamine. The effect of high-level ZIKV protein expression on the subcellular localization of each protein also was documented with the GFP alone as a control and was compared with the localization of each ZIKV protein at low levels of expression.
Measurement of Cell Proliferation and Cell-Cycle Profiling.
To identify which ZIKV protein is responsible for ZIKV-associated growth inhibition, we used the colony-forming assay to measure cell proliferation and viability (46, 47) and the cellular growth kinetics to quantify cellular growth, as described previously (41, 83). Briefly, the fission yeast culture was prepared as described above. About 100 µL of liquid culture was spread onto the selective EMM agar plates with and without thiamine. The agar plates were incubated at 30 °C for 4–6 d to observe the presence or absence and, when present, the size of the forming colonies. The absence of colonies on the agar plates indicated a possible cell-killing effect. Colony sizes smaller than the control typically suggest reduced cellular growth.
For quantitative measurements of growth kinetics, liquid cell cultures were grown in a 96-well microtiter plate containing the selective EMM medium. Cell cultures were grown at 30 °C in an incubator with moisture. Cellular growth was measured at OD650 over the indicated time period using a spectrophotometer.
Cell-cycle profiles of the cells expressing ZIKV proteins were obtained by measuring DNA content using flow cytometry, as described previously (41, 82). To measure the effect of ZIKV protein on the G1 phase of the cell cycle, the fission yeast cells were cultured in the regular EMM medium, in which fission yeast cells are mostly in the G2/M phase of the cell cycle. To measure the effect on the G2/M phase of the cell cycle, fission yeast cells were first prepared in low-nitrogen medium, in which fission yeast cells are enriched in the G1 phase of the cell cycle (41). The DNA content was analyzed on a FACSCanto II flow cytometer (Becton Dickinson) using FACS DIVA 6.3 software (Becton Dickinson). Ten thousand events were collected, and the DNA content corresponding to cells in G1 and G2 phases was determined as the FL-2 parameter. FL-2 measures the amount of propidium iodide fluorescence emitted through a 585-nm band-pass filter.
Measurement of Cell Hypertrophy, Nuclear Morphology, and Cell Death.
The effects of ZIKV protein expression on fission yeast cell and nuclear morphology were examined 45 h after GI. Cell morphology was observed using bright-field microscopy. The overall cell morphology was evaluated by flow cytometer using FSC analysis (41). Ten thousand cells were collected, and the FSC and SSC were measured for each cell population. The FSC is proportional to the cell surface area and thus measures cell size. The SSC determines intracellular complexity because it is proportional to cell granularity. Correlated measurements of FSC and SSC allow the differentiation of cell shapes in a heterogeneous cell population.
The nuclear morphology was observed with Hoechst stain, a blue fluorescent dye that specifically stains DNA. Stained cells were observed with a Leica A4 filter with an excitation of 360/40 nm and emission of 470/40 nm.
Cell viability was evaluated by Trypan blue staining (50) and by a commercial live/dead yeast viability assay (Invitrogen) (47, 51). Trypan blue is a diazo dye that stains only dead cells; live cells with intact cell membranes are not stained. The live/dead yeast viability assay measures cell viability by monitoring intracellular metabolic activities with FUN-1 staining (Molecular Probes) using a Leica DM fluorescent microscope with an 11001v2 long-path Chroma filter cube. Metabolically active cells convert yellow-green fluorescent intracellular FUN-1 into red-orange intravacuolar structures, which emit at 590 nm. Metabolically inert or dead cells exhibit bright, diffuse, green-yellow fluorescence at ∼540 nm. FUN-1–stained cell images were collected using an S Green filter for green and an N2.1 filter (emission: LP590/excitation: 515–560 nm) for red fluorescence. Final images were generated by fluorescence merging.
The induction of cellular oxidative stress by ZIKV proteins was measured as the expression of ROS, which was detected by a ROS-specific dye, DHE (Sigma), that produces red fluorescence in the presence of ROS, as described previously (82, 84, 85). Cells were grown as described above; 48 h after GI, DHE was added to a final concentration of 5 μg/mL, and ROS were detected by fluorescent microscopy.
Statistical Analysis.
We used a pairwise Student t test (Microsoft Excel 2010) for the described statistical analysis.
Acknowledgments
We thank Dr. Hongli Tang, Florida State University, for providing the ZIKV MR766 viral extract and Dr. Volodymyr Gerzanich, University of Maryland, for assistance with fluorescence microscopy. This work was supported in part by funding from the University of Maryland Medical Center (R.Y.Z.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1619735114/-/DCSupplemental.
References
- 1.Dang J, et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 2016;19(2):258–265. doi: 10.1016/j.stem.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qian X, et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165(5):1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cugola FR, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534(7606):267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao-Lormeau VM, et al. Guillain-Barré syndrome outbreak associated with Zika virus infection in French Polynesia: A case-control study. Lancet. 2016;387(10027):1531–1539. doi: 10.1016/S0140-6736(16)00562-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith DW, Mackenzie J. Zika virus and Guillain-Barré syndrome: Another viral cause to add to the list. Lancet. 2016;387(10027):1486–1488. doi: 10.1016/S0140-6736(16)00564-X. [DOI] [PubMed] [Google Scholar]
- 6.Hamel R, et al. Biology of Zika virus infection in human skin cells. J Virol. 2015;89(17):8880–8896. doi: 10.1128/JVI.00354-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tang H, et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell. 2016;18(5):587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li C, et al. Zika virus disrupts neural progenitor development and leads to microcephaly in mice. Cell Stem Cell. 2016;19(1):120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 9.Faye O, et al. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS Negl Trop Dis. 2014;8(1):e2636. doi: 10.1371/journal.pntd.0002636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harris E, Holden KL, Edgil D, Polacek C, Clyde K. Molecular biology of flaviviruses. Novartis Found Symp. 2006;277:23–39. discussion 40–71–23. [PubMed] [Google Scholar]
- 11.Amberg SM, Nestorowicz A, McCourt DW, Rice CM. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: In vitro and in vivo studies. J Virol. 1994;68(6):3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lobigs M, Lee E, Ng ML, Pavy M, Lobigs P. A flavivirus signal peptide balances the catalytic activity of two proteases and thereby facilitates virus morphogenesis. Virology. 2010;401(1):80–89. doi: 10.1016/j.virol.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 13.Miller S, Kastner S, Krijnse-Locker J, Bühler S, Bartenschlager R. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J Biol Chem. 2007;282(12):8873–8882. doi: 10.1074/jbc.M609919200. [DOI] [PubMed] [Google Scholar]
- 14.Zou G, et al. A single-amino acid substitution in West Nile virus 2K peptide between NS4A and NS4B confers resistance to lycorine, a flavivirus inhibitor. Virology. 2009;384(1):242–252. doi: 10.1016/j.virol.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mukhopadhyay S, Kuhn RJ, Rossmann MG. A structural perspective of the flavivirus life cycle. Nat Rev Microbiol. 2005;3(1):13–22. doi: 10.1038/nrmicro1067. [DOI] [PubMed] [Google Scholar]
- 16.Kuno G, Chang GJ. Full-length sequencing and genomic characterization of Bagaza, Kedougou, and Zika viruses. Arch Virol. 2007;152(4):687–696. doi: 10.1007/s00705-006-0903-z. [DOI] [PubMed] [Google Scholar]
- 17.Assenberg R, et al. Crystal structure of a novel conformational state of the flavivirus NS3 protein: Implications for polyprotein processing and viral replication. J Virol. 2009;83(24):12895–12906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bell TM, Field EJ, Narang HK. Zika virus infection of the central nervous system of mice. Arch Gesamte Virusforsch. 1971;35(2):183–193. doi: 10.1007/BF01249709. [DOI] [PubMed] [Google Scholar]
- 19.Kaufusi PH, Kelley JF, Yanagihara R, Nerurkar VR. Induction of endoplasmic reticulum-derived replication-competent membrane structures by West Nile virus non-structural protein 4B. PLoS One. 2014;9(1):e84040. doi: 10.1371/journal.pone.0084040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero-Brey I, Bartenschlager R. Endoplasmic reticulum: The favorite intracellular niche for viral replication and assembly. Viruses. 2016;8(6):E160. doi: 10.3390/v8060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nowakowski TJ, et al. Expression analysis highlights AXL as a candidate Zika virus entry receptor in neural stem cells. Cell Stem Cell. 2016;18(5):591–596. doi: 10.1016/j.stem.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allison SL, et al. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J Virol. 1995;69(2):695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guirakhoo F, Heinz FX, Mandl CW, Holzmann H, Kunz C. Fusion activity of flaviviruses: Comparison of mature and immature (prM-containing) tick-borne encephalitis virions. J Gen Virol. 1991;72(Pt 6):1323–1329. doi: 10.1099/0022-1317-72-6-1323. [DOI] [PubMed] [Google Scholar]
- 24.Guirakhoo F, Bolin RA, Roehrig JT. The Murray Valley encephalitis virus prM protein confers acid resistance to virus particles and alters the expression of epitopes within the R2 domain of E glycoprotein. Virology. 1992;191(2):921–931. doi: 10.1016/0042-6822(92)90267-S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin JC, et al. Dengue viral protease interaction with NF-κB inhibitor α/β results in endothelial cell apoptosis and hemorrhage development. J Immunol. 2014;193(3):1258–1267. doi: 10.4049/jimmunol.1302675. [DOI] [PubMed] [Google Scholar]
- 26.Stadler K, Allison SL, Schalich J, Heinz FX. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71(11):8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elshuber S, Allison SL, Heinz FX, Mandl CW. Cleavage of protein prM is necessary for infection of BHK-21 cells by tick-borne encephalitis virus. J Gen Virol. 2003;84(Pt 1):183–191. doi: 10.1099/vir.0.18723-0. [DOI] [PubMed] [Google Scholar]
- 28.Hartwell LH. Yeast and cancer. Biosci Rep. 2004;24(4-5):523–544. doi: 10.1007/s10540-005-2743-6. [DOI] [PubMed] [Google Scholar]
- 29.Nasmyth K. A prize for proliferation. Cell. 2001;107(6):689–701. doi: 10.1016/s0092-8674(01)00604-3. [DOI] [PubMed] [Google Scholar]
- 30.Nurse PM. Nobel Lecture. Cyclin dependent kinases and cell cycle control. Biosci Rep. 2002;22(5-6):487–499. doi: 10.1023/a:1022017701871. [DOI] [PubMed] [Google Scholar]
- 31.Ray K. From fission to fusion: A perspective on the research that won the Nobel Prize in Physiology or Medicine, 2013. J Biosci. 2014;39(1):3–12. doi: 10.1007/s12038-014-9416-1. [DOI] [PubMed] [Google Scholar]
- 32.Zhao Y, Elder RT. Yeast perspectives on HIV-1 VPR. Front Biosci. 2000;5:D905–D916. doi: 10.2741/zhao. [DOI] [PubMed] [Google Scholar]
- 33.Zhao RY, Elder RT. Viral infections and cell cycle G2/M regulation. Cell Res. 2005;15(3):143–149. doi: 10.1038/sj.cr.7290279. [DOI] [PubMed] [Google Scholar]
- 34.Andréola ML, Litvak S. Yeast and the AIDS virus: The odd couple. J Biomed Biotechnol. 2012;2012:549020. doi: 10.1155/2012/549020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lista MJ, et al. The long-lasting love affair between the budding yeast Saccharomyces cerevisiae and the Epstein-Barr virus. Biotechnol J. 2015;10(11):1670–1681. doi: 10.1002/biot.201500161. [DOI] [PubMed] [Google Scholar]
- 36.Zhao Y, Lieberman HB. Schizosaccharomyces pombe: A model for molecular studies of eukaryotic genes. DNA Cell Biol. 1995;14(5):359–371. doi: 10.1089/dna.1995.14.359. [DOI] [PubMed] [Google Scholar]
- 37.Olsson I, Bjerling P. Advancing our understanding of functional genome organisation through studies in the fission yeast. Curr Genet. 2011;57(1):1–12. doi: 10.1007/s00294-010-0327-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Q, et al. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell. 2016;19(5):663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Elder RT, Chen M, Cao J. Fission yeast expression vectors adapted for positive identification of gene insertion and green fluorescent protein fusion. Biotechniques. 1998;25(3):438–440, 442, 444. doi: 10.2144/98253st06. [DOI] [PubMed] [Google Scholar]
- 40.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123(1):127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 41.Zhao Y, Cao J, O’Gorman MR, Yu M, Yogev R. Effect of human immunodeficiency virus type 1 protein R (vpr) gene expression on basic cellular function of fission yeast Schizosaccharomyces pombe. J Virol. 1996;70(9):5821–5826. doi: 10.1128/jvi.70.9.5821-5826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matsuyama A, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24(7):841–847. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 43.Mikawa T, Kanoh J, Ishikawa F. Fission yeast Vps1 and Atg8 contribute to oxidative stress resistance. Genes Cells. 2010;15(3):229–242. doi: 10.1111/j.1365-2443.2009.01376.x. [DOI] [PubMed] [Google Scholar]
- 44.Yiang GT, et al. The NS3 protease and helicase domains of Japanese encephalitis virus trigger cell death via caspase-dependent and -independent pathways. Mol Med Rep. 2013;7(3):826–830. doi: 10.3892/mmr.2013.1261. [DOI] [PubMed] [Google Scholar]
- 45.Zhang R, et al. A CRISPR screen defines a signal peptide processing pathway required by flaviviruses. Nature. 2016;535(7610):164–168. doi: 10.1038/nature18625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen M, et al. Mutational analysis of Vpr-induced G2 arrest, nuclear localization, and cell death in fission yeast. J Virol. 1999;73(4):3236–3245. doi: 10.1128/jvi.73.4.3236-3245.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao Y, et al. Pleiotropic effects of HIV-1 protein R (Vpr) on morphogenesis and cell survival in fission yeast and antagonism by pentoxifylline. Virology. 1998;246(2):266–276. doi: 10.1006/viro.1998.9208. [DOI] [PubMed] [Google Scholar]
- 48.Maundrell K. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265(19):10857–10864. [PubMed] [Google Scholar]
- 49.Nurse P, Bissett Y. Gene required in G1 for commitment to cell cycle and in G2 for control of mitosis in fission yeast. Nature. 1981;292(5823):558–560. doi: 10.1038/292558a0. [DOI] [PubMed] [Google Scholar]
- 50.Kucsera J, Yarita K, Takeo K. Simple detection method for distinguishing dead and living yeast colonies. J Microbiol Methods. 2000;41(1):19–21. doi: 10.1016/s0167-7012(00)00136-6. [DOI] [PubMed] [Google Scholar]
- 51.Benko Z, Elder RT, Li G, Liang D, Zhao RY. HIV-1 Protease in the Fission Yeast Schizosaccharomyces pombe. PLoS One. 2016;11(3):e0151286. doi: 10.1371/journal.pone.0151286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin RJ, Liao CL, Lin YL. Replication-incompetent virions of Japanese encephalitis virus trigger neuronal cell death by oxidative stress in a culture system. J Gen Virol. 2004;85(Pt 2):521–533. doi: 10.1099/vir.0.19496-0. [DOI] [PubMed] [Google Scholar]
- 53.Yang TC, et al. Japanese encephalitis virus down-regulates thioredoxin and induces ROS-mediated ASK1-ERK/p38 MAPK activation in human promonocyte cells. Microbes Infect. 2010;12(8-9):643–651. doi: 10.1016/j.micinf.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 54.Olagnier D, et al. Cellular oxidative stress response controls the antiviral and apoptotic programs in dengue virus-infected dendritic cells. PLoS Pathog. 2014;10(12):e1004566. doi: 10.1371/journal.ppat.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Weisman R, Choder M. The fission yeast TOR homolog, tor1+, is required for the response to starvation and other stresses via a conserved serine. J Biol Chem. 2001;276(10):7027–7032. doi: 10.1074/jbc.M010446200. [DOI] [PubMed] [Google Scholar]
- 56.Ikai N, Nakazawa N, Hayashi T, Yanagida M. The reverse, but coordinated, roles of Tor2 (TORC1) and Tor1 (TORC2) kinases for growth, cell cycle and separase-mediated mitosis in Schizosaccharomyces pombe. Open Biol. 2011;1(3):110007. doi: 10.1098/rsob.110007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jacinto E, Guo B, Arndt KT, Schmelzle T, Hall MN. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol Cell. 2001;8(5):1017–1026. doi: 10.1016/s1097-2765(01)00386-0. [DOI] [PubMed] [Google Scholar]
- 58.Fenyvuesvolgyi C, Elder RT, Benko Z, Liang D, Zhao RY. Fission yeast homologue of Tip41-like proteins regulates type 2A phosphatases and responses to nitrogen sources. Biochim Biophys Acta. 2005;1746(2):155–162. doi: 10.1016/j.bbamcr.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 59.Morán M, et al. Bulk autophagy, but not mitophagy, is increased in cellular model of mitochondrial disease. Biochim Biophys Acta. 2014;1842(7):1059–1070. doi: 10.1016/j.bbadis.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 60.Netsawang J, et al. Nuclear localization of dengue virus capsid protein is required for DAXX interaction and apoptosis. Virus Res. 2010;147(2):275–283. doi: 10.1016/j.virusres.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 61.Sun LL, et al. Global analysis of fission yeast mating genes reveals new autophagy factors. PLoS Genet. 2013;9(8):e1003715. doi: 10.1371/journal.pgen.1003715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Garcez PP, et al. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352(6287):816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- 63.Lindenbach BD, Rice CM. Molecular biology of flaviviruses. Adv Virus Res. 2003;59:23–61. doi: 10.1016/s0065-3527(03)59002-9. [DOI] [PubMed] [Google Scholar]
- 64.Cox BD, Stanton RA, Schinazi RF. Predicting Zika virus structural biology: Challenges and opportunities for intervention. Antivir Chem Chemother. 2015;24(3-4):118–126. doi: 10.1177/2040206616653873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kostyuchenko VA, et al. Structure of the thermally stable Zika virus. Nature. 2016;533(7603):425–428. doi: 10.1038/nature17994. [DOI] [PubMed] [Google Scholar]
- 66.Coller BA, Clements DE, Bett AJ, Sagar SL, Ter Meulen JH. The development of recombinant subunit envelope-based vaccines to protect against dengue virus induced disease. Vaccine. 2011;29(42):7267–7275. doi: 10.1016/j.vaccine.2011.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miller N. Recent progress in dengue vaccine research and development. Curr Opin Mol Ther. 2010;12(1):31–38. [PubMed] [Google Scholar]
- 68.Li L, Li HS, Pauza CD, Bukrinsky M, Zhao RY. Roles of HIV-1 auxiliary proteins in viral pathogenesis and host-pathogen interactions. Cell Res. 2005;15(11-12):923–934. doi: 10.1038/sj.cr.7290370. [DOI] [PubMed] [Google Scholar]
- 69.Melino S, et al. The active essential CFNS3d protein complex. FEBS J. 2006;273(16):3650–3662. doi: 10.1111/j.1742-4658.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 70.Shafee N, AbuBakar S. Dengue virus type 2 NS3 protease and NS2B-NS3 protease precursor induce apoptosis. J Gen Virol. 2003;84(Pt 8):2191–2195. doi: 10.1099/vir.0.19022-0. [DOI] [PubMed] [Google Scholar]
- 71.Ramanathan MP, et al. Host cell killing by the West Nile Virus NS2B-NS3 proteolytic complex: NS3 alone is sufficient to recruit caspase-8-based apoptotic pathway. Virology. 2006;345(1):56–72. doi: 10.1016/j.virol.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 72.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr Top Microbiol Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cutler NS, Heitman J, Cardenas ME. TOR kinase homologs function in a signal transduction pathway that is conserved from yeast to mammals. Mol Cell Endocrinol. 1999;155(1-2):135–142. doi: 10.1016/s0303-7207(99)00121-5. [DOI] [PubMed] [Google Scholar]
- 74.Jacinto E, Hall MN. Tor signalling in bugs, brain and brawn. Nat Rev Mol Cell Biol. 2003;4(2):117–126. doi: 10.1038/nrm1018. [DOI] [PubMed] [Google Scholar]
- 75.Shives KD, et al. West nile virus-induced activation of mammalian target of rapamycin complex 1 supports viral growth and viral protein expression. J Virol. 2014;88(16):9458–9471. doi: 10.1128/JVI.01323-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Le Sage V, Cinti A, Amorim R, Mouland AJ. Adapting the stress response: Viral subversion of the mTOR signaling pathway. Viruses. 2016;8(6):E152. doi: 10.3390/v8060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jung CH, Ro SH, Cao J, Otto NM, Kim DH. mTOR regulation of autophagy. FEBS Lett. 2010;584(7):1287–1295. doi: 10.1016/j.febslet.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weisman R, Roitburg I, Schonbrun M, Harari R, Kupiec M. Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics. 2007;175(3):1153–1162. doi: 10.1534/genetics.106.064170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123(1):131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 80.Dick GW, Kitchen SF, Haddow AJ. Zika virus. I. Isolations and serological specificity. Trans R Soc Trop Med Hyg. 1952;46(5):509–520. doi: 10.1016/0035-9203(52)90042-4. [DOI] [PubMed] [Google Scholar]
- 81.Dick GW. Zika virus. II. Pathogenicity and physical properties. Trans R Soc Trop Med Hyg. 1952;46(5):521–534. doi: 10.1016/0035-9203(52)90043-6. [DOI] [PubMed] [Google Scholar]
- 82.Nkeze J, Li L, Benko Z, Li G, Zhao RY. Molecular characterization of HIV-1 genome in fission yeast Schizosaccharomyces pombe. Cell Biosci. 2015;5:47. doi: 10.1186/s13578-015-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 84.Madeo F, Fröhlich E, Fröhlich KU. A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol. 1997;139(3):729–734. doi: 10.1083/jcb.139.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huard S, et al. HIV-1 Vpr-induced cell death in Schizosaccharomyces pombe is reminiscent of apoptosis. Cell Res. 2008;18(9):961–973. doi: 10.1038/cr.2008.272. [DOI] [PubMed] [Google Scholar]