Abstract
Interaction between short repeats may be a source of genomic rearrangements and deletions. We investigated possible interactions between short (9 base pairs) direct repeats in yeast by using our previously described system for analyzing bacterial transposon Tn5 excision in yeast. Mutations of either POL3 or POL1, the proposed structural genes for polymerases delta and alpha, respectively, yield high levels of excision at semipermissive temperatures. pol2 (corresponding to polymerase epsilon) and pol2 pol3 double mutants do not exhibit enhanced excision. A majority of excision events involve direct repeats and are precise; the remaining imprecise excisions occur within or in the vicinity of the repeats. The three DNA repair pathways identified by rad1, rad6 and rad18, rad50 and rad52 mutations were examined for their possible role in Tn5 excision; no enhancement was observed in mutants. However, the pol3-stimulated Tn5 excision was reduced in rad52 and rad50 mutants. This suggests the potential for interaction between the systems for DNA double-strand break/recombinational repair and DNA synthesis. Based on the suggestion of Morrison et al. [Morrison, A., Araki, H., Clark, A. B., Hamatake, R. H. & Sugino, A. (1990) Cell 62, 1143-1151] that polymerases delta and alpha are responsible for lagging-strand synthesis and that polymerase epsilon is responsible for leading-strand synthesis, we suggest that Tn5 excision is stimulated under conditions of altered lagging-strand synthesis, possibly due to extended opportunities for single-strand interactions between the inverted insertion sequence I550 repeats of Tn5.
Full text
PDF

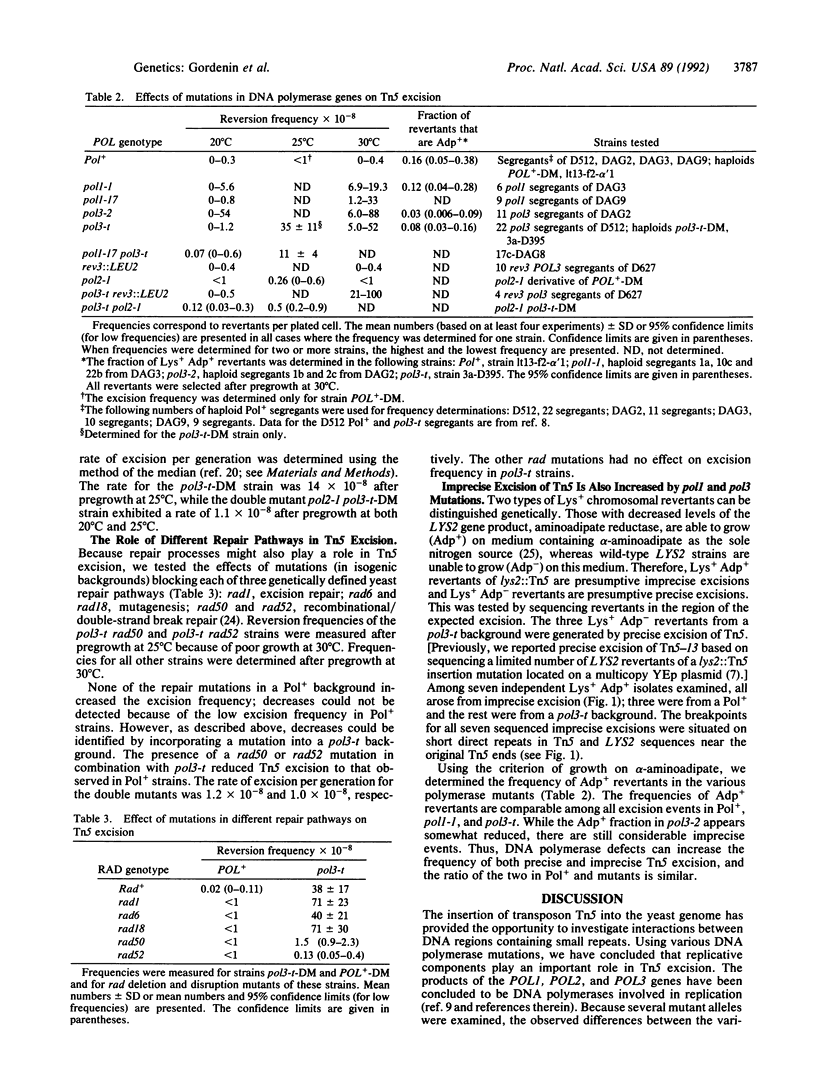
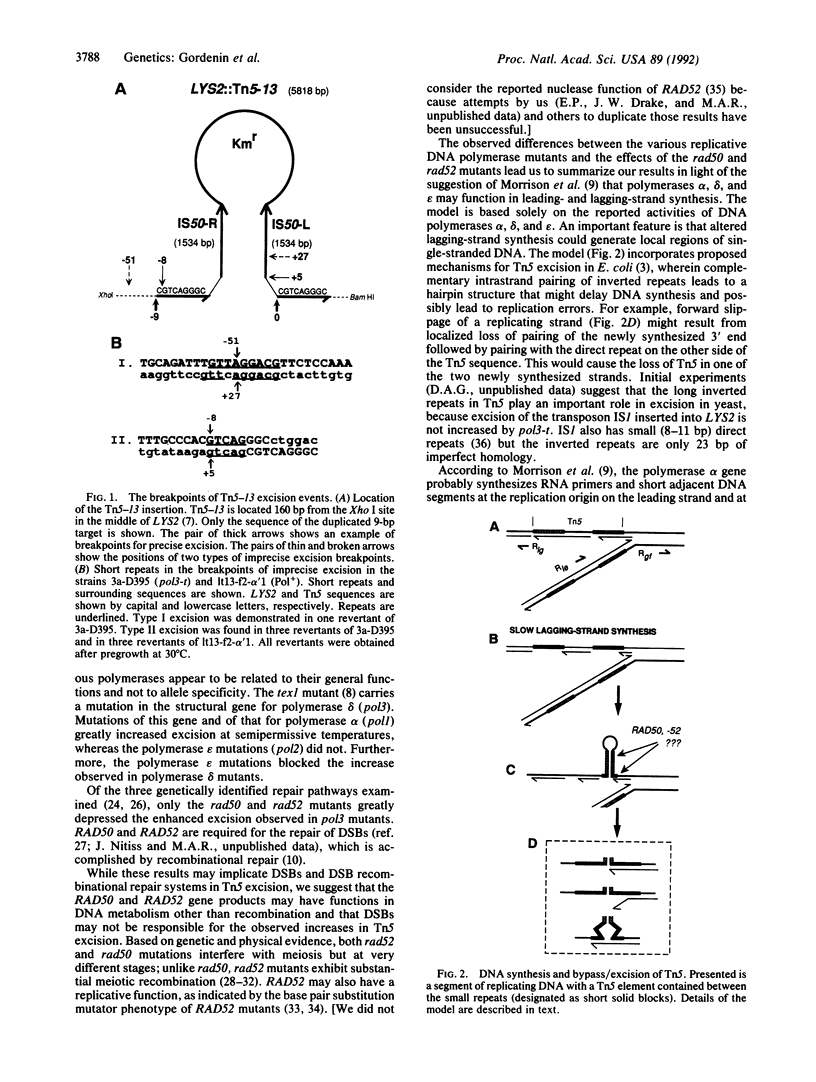

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alani E., Padmore R., Kleckner N. Analysis of wild-type and rad50 mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990 May 4;61(3):419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Alani E., Subbiah S., Kleckner N. The yeast RAD50 gene encodes a predicted 153-kD protein containing a purine nucleotide-binding domain and two large heptad-repeat regions. Genetics. 1989 May;122(1):47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberts B. M. Prokaryotic DNA replication mechanisms. Philos Trans R Soc Lond B Biol Sci. 1987 Dec 15;317(1187):395–420. doi: 10.1098/rstb.1987.0068. [DOI] [PubMed] [Google Scholar]
- Borts R. H., Lichten M., Haber J. E. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics. 1986 Jul;113(3):551–567. doi: 10.1093/genetics/113.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budd M., Campbell J. L. Temperature-sensitive mutations in the yeast DNA polymerase I gene. Proc Natl Acad Sci U S A. 1987 May;84(9):2838–2842. doi: 10.1073/pnas.84.9.2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers P. M., Bambara R. A., Campbell J. L., Chang L. M., Downey K. M., Hübscher U., Lee M. Y., Linn S. M., So A. G., Spadari S. Revised nomenclature for eukaryotic DNA polymerases. Eur J Biochem. 1990 Aug 17;191(3):617–618. doi: 10.1111/j.1432-1033.1990.tb19165.x. [DOI] [PubMed] [Google Scholar]
- Chattoo B. B., Sherman F., Azubalis D. A., Fjellstedt T. A., Mehnert D., Ogur M. Selection of lys2 Mutants of the Yeast SACCHAROMYCES CEREVISIAE by the Utilization of alpha-AMINOADIPATE. Genetics. 1979 Sep;93(1):51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D. S., Bernstein H. Yeast gene RAD52 can substitute for phage T4 gene 46 or 47 in carrying out recombination and DNA repair. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6821–6825. doi: 10.1073/pnas.85.18.6821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad M. N., Newlon C. S. Saccharomyces cerevisiae cdc2 mutants fail to replicate approximately one-third of their nuclear genome. Mol Cell Biol. 1983 Jun;3(6):1000–1012. doi: 10.1128/mcb.3.6.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B., Game J. Repair systems in Saccharomyces. Mutat Res. 1974 Aug;26(4):257–264. doi: 10.1016/s0027-5107(74)80023-0. [DOI] [PubMed] [Google Scholar]
- Efstratiadis A., Posakony J. W., Maniatis T., Lawn R. M., O'Connell C., Spritz R. A., DeRiel J. K., Forget B. G., Weissman S. M., Slightom J. L. The structure and evolution of the human beta-globin gene family. Cell. 1980 Oct;21(3):653–668. doi: 10.1016/0092-8674(80)90429-8. [DOI] [PubMed] [Google Scholar]
- Egner C., Berg D. E. Excision of transposon Tn5 is dependent on the inverted repeats but not on the transposase function of Tn5. Proc Natl Acad Sci U S A. 1981 Jan;78(1):459–463. doi: 10.1073/pnas.78.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T., Alberts B. M. DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell. 1986 Dec 5;47(5):793–806. doi: 10.1016/0092-8674(86)90522-2. [DOI] [PubMed] [Google Scholar]
- Foster T. J., Lundblad V., Hanley-Way S., Halling S. M., Kleckner N. Three Tn10-associated excision events: relationship to transposition and role of direct and inverted repeats. Cell. 1981 Jan;23(1):215–227. doi: 10.1016/0092-8674(81)90286-5. [DOI] [PubMed] [Google Scholar]
- Game J. C., Zamb T. J., Braun R. J., Resnick M., Roth R. M. The Role of Radiation (rad) Genes in Meiotic Recombination in Yeast. Genetics. 1980 Jan;94(1):51–68. doi: 10.1093/genetics/94.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordenin D. A., Proscyavichus Y. Y., Malkova A. L., Trofimova M. V., Peterzen A. Yeast mutants with increased bacterial transposon Tn5 excision. Yeast. 1991 Jan;7(1):37–50. doi: 10.1002/yea.320070105. [DOI] [PubMed] [Google Scholar]
- Gordenin D. A., Trofimova M. V., Shaburova O. N., Pavlov Y. I., Chernoff Y. O., Chekuolene Y. V., Proscyavichus Y. Y., Sasnauskas K. V., Janulaitis A. A. Precise excision of bacterial transposon Tn5 in yeast. Mol Gen Genet. 1988 Aug;213(2-3):388–393. doi: 10.1007/BF00339607. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Kunz B. A., Peters M. G., Kohalmi S. E., Armstrong J. D., Glattke M., Badiani K. Disruption of the RAD52 gene alters the spectrum of spontaneous SUP4-o mutations in Saccharomyces cerevisiae. Genetics. 1989 Jul;122(3):535–542. doi: 10.1093/genetics/122.3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Kleckner N. Mismatch repair mutations of Escherichia coli K12 enhance transposon excision. Genetics. 1985 Jan;109(1):3–19. doi: 10.1093/genetics/109.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundblad V., Taylor A. F., Smith G. R., Kleckner N. Unusual alleles of recB and recC stimulate excision of inverted repeat transposons Tn10 and Tn5. Proc Natl Acad Sci U S A. 1984 Feb;81(3):824–828. doi: 10.1073/pnas.81.3.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990 Sep 21;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Morrison A., Christensen R. B., Alley J., Beck A. K., Bernstine E. G., Lemontt J. F., Lawrence C. W. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989 Oct;171(10):5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A., Chow T., Nitiss J., Game J. Changes in the chromosomal DNA of yeast during meiosis in repair mutants and the possible role of a deoxyribonuclease. Cold Spring Harb Symp Quant Biol. 1984;49:639–649. doi: 10.1101/sqb.1984.049.01.072. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Martin P. The repair of double-strand breaks in the nuclear DNA of Saccharomyces cerevisiae and its genetic control. Mol Gen Genet. 1976 Jan 16;143(2):119–129. doi: 10.1007/BF00266917. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Nitiss J., Edwards C., Malone R. E. Meiosis can induce recombination in rad52 mutants of Saccharomyces cerevisiae. Genetics. 1986 Jul;113(3):531–550. doi: 10.1093/genetics/113.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick M. A. The repair of double-strand breaks in DNA; a model involving recombination. J Theor Biol. 1976 Jun;59(1):97–106. doi: 10.1016/s0022-5193(76)80025-2. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitney K. C., Budd M. E., Campbell J. L. DNA polymerase III, a second essential DNA polymerase, is encoded by the S. cerevisiae CDC2 gene. Cell. 1989 Feb 24;56(4):599–605. doi: 10.1016/0092-8674(89)90582-5. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Borstel R. C., Cain K. T., Steinberg C. M. Inheritance of spontaneous mutability in yeast. Genetics. 1971 Sep;69(1):17–27. doi: 10.1093/genetics/69.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston-Hafer K., Berg D. E. Palindromy and the location of deletion endpoints in Escherichia coli. Genetics. 1989 Apr;121(4):651–658. doi: 10.1093/genetics/121.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]



