Abstract
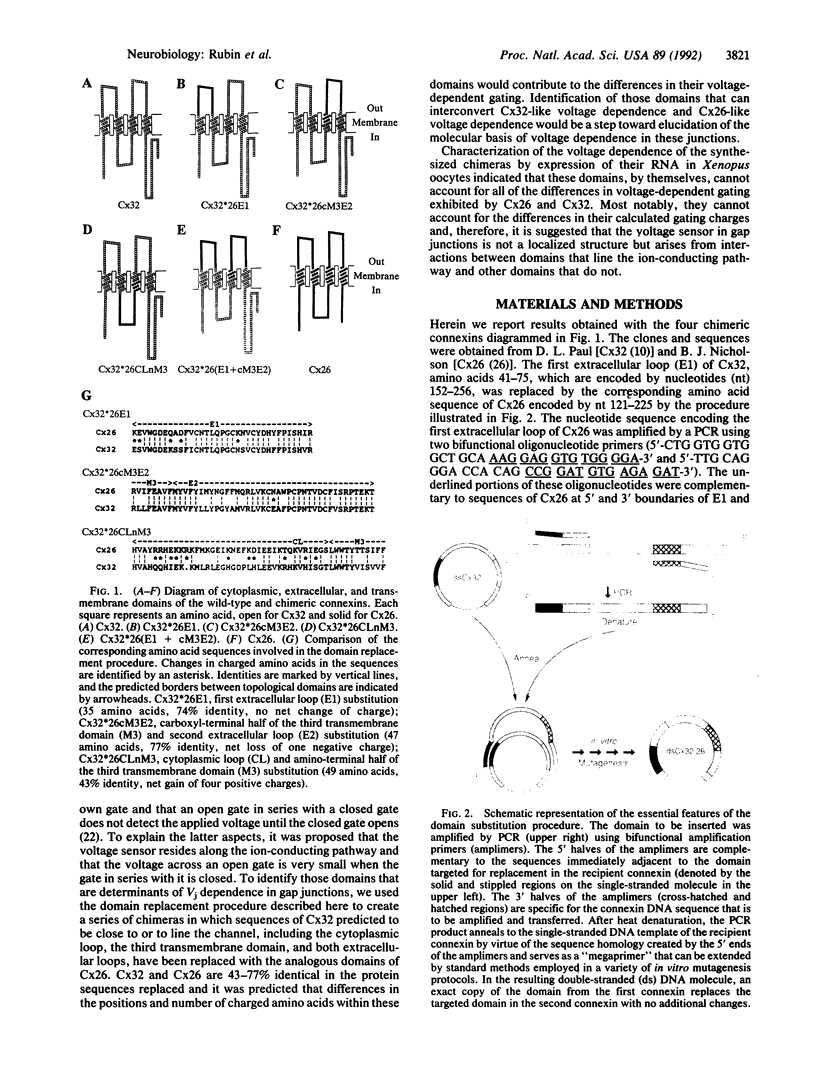
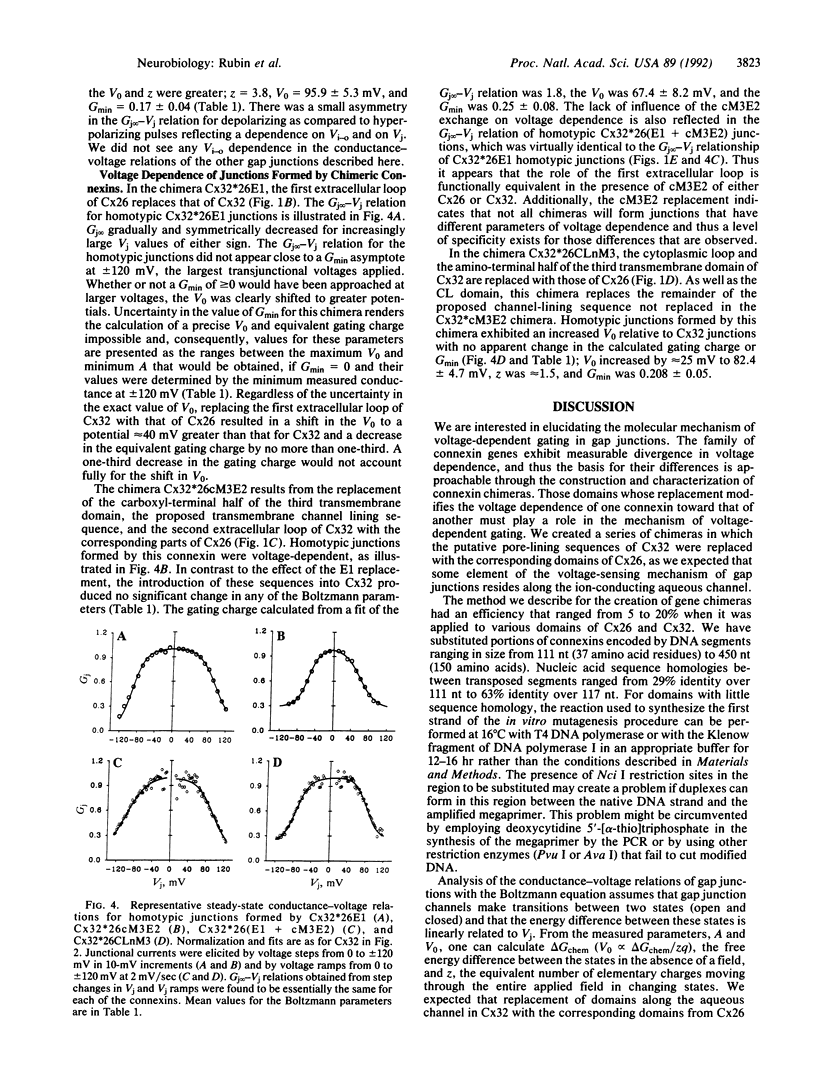
We have developed a procedure for the replacement of defined domains with specified domains from other proteins that we used to examine the molecular basis for the differences in voltage-dependent gating between connexins 26 (Cx26) and 32 (Cx32). This technique does not depend on sequence homology between the domains to be exchanged or the presence of restriction endonuclease sites. Rather, it makes use of a PCR strategy to create an adhesive "band-aid" that directs the annealing of the amplified sequence to the correct location in the recipient clone. With this technique we created a series of chimeras involving the replacement of topologically defined protein domains of Cx32 with the corresponding sequences of Cx26. We focused on domains that are predicted to line the gap junction channel as we expect that a component of the voltage-sensing mechanism resides there. Differences between Cx26 and Cx32 in the sequences of their first and second extracellular loops, the cytoplasmic loop, and the third transmembrane domain did not account for the difference in their calculated gating charges. Shifts along the voltage axis in the steady-state conductance-voltage relations of the chimeric connexins were produced by replacement of the first extracellular loop or the cytoplasmic loop and the amino-terminal half of the third transmembrane domain. These data suggest that the voltage-sensing mechanism arises from the interaction of domains lining the aqueous channel and domains deeper in the channel wall.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barrio L. C., Suchyna T., Bargiello T., Xu L. X., Roginski R. S., Bennett M. V., Nicholson B. J. Gap junctions formed by connexins 26 and 32 alone and in combination are differently affected by applied voltage. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8410–8414. doi: 10.1073/pnas.88.19.8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett M. V., Barrio L. C., Bargiello T. A., Spray D. C., Hertzberg E., Sáez J. C. Gap junctions: new tools, new answers, new questions. Neuron. 1991 Mar;6(3):305–320. doi: 10.1016/0896-6273(91)90241-q. [DOI] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin family of gap junction proteins. J Membr Biol. 1990 Jul;116(3):187–194. doi: 10.1007/BF01868459. [DOI] [PubMed] [Google Scholar]
- Dermietzel R., Traub O., Hwang T. K., Beyer E., Bennett M. V., Spray D. C., Willecke K. Differential expression of three gap junction proteins in developing and mature brain tissues. Proc Natl Acad Sci U S A. 1989 Dec;86(24):10148–10152. doi: 10.1073/pnas.86.24.10148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebihara L., Beyer E. C., Swenson K. I., Paul D. L., Goodenough D. A. Cloning and expression of a Xenopus embryonic gap junction protein. Science. 1989 Mar 3;243(4895):1194–1195. doi: 10.1126/science.2466337. [DOI] [PubMed] [Google Scholar]
- Fishman G. I., Hertzberg E. L., Spray D. C., Leinwand L. A. Expression of connexin43 in the developing rat heart. Circ Res. 1991 Mar;68(3):782–787. doi: 10.1161/01.res.68.3.782. [DOI] [PubMed] [Google Scholar]
- Fishman G. I., Moreno A. P., Spray D. C., Leinwand L. A. Functional analysis of human cardiac gap junction channel mutants. Proc Natl Acad Sci U S A. 1991 May 1;88(9):3525–3529. doi: 10.1073/pnas.88.9.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimlich R. L., Kumar N. M., Gilula N. B. Differential regulation of the levels of three gap junction mRNAs in Xenopus embryos. J Cell Biol. 1990 Mar;110(3):597–605. doi: 10.1083/jcb.110.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimlich R. L., Kumar N. M., Gilula N. B. Sequence and developmental expression of mRNA coding for a gap junction protein in Xenopus. J Cell Biol. 1988 Sep;107(3):1065–1073. doi: 10.1083/jcb.107.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. L., Spray D. C., Bennett M. V. Kinetic properties of a voltage-dependent junctional conductance. J Gen Physiol. 1981 Jan;77(1):95–117. doi: 10.1085/jgp.77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann H. A., Kirsch G. E., Drewe J. A., Taglialatela M., Joho R. H., Brown A. M. Exchange of conduction pathways between two related K+ channels. Science. 1991 Feb 22;251(4996):942–944. doi: 10.1126/science.2000495. [DOI] [PubMed] [Google Scholar]
- Hertzberg E. L., Disher R. M., Tiller A. A., Zhou Y., Cook R. G. Topology of the Mr 27,000 liver gap junction protein. Cytoplasmic localization of amino- and carboxyl termini and a hydrophilic domain which is protease-hypersensitive. J Biol Chem. 1988 Dec 15;263(35):19105–19111. [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Goodenough D. A. Gap junction structures. II. Analysis of the x-ray diffraction data. J Cell Biol. 1977 Aug;74(2):629–645. doi: 10.1083/jcb.74.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks L. C., Kumar N. M., Houghten R., Unwin N., Gilula N. B. Topology of the 32-kd liver gap junction protein determined by site-directed antibody localizations. EMBO J. 1988 Oct;7(10):2967–2975. doi: 10.1002/j.1460-2075.1988.tb03159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D. L. Molecular cloning of cDNA for rat liver gap junction protein. J Cell Biol. 1986 Jul;103(1):123–134. doi: 10.1083/jcb.103.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson K. I., Jordan J. R., Beyer E. C., Paul D. L. Formation of gap junctions by expression of connexins in Xenopus oocyte pairs. Cell. 1989 Apr 7;57(1):145–155. doi: 10.1016/0092-8674(89)90180-3. [DOI] [PubMed] [Google Scholar]
- Unwin N. The structure of ion channels in membranes of excitable cells. Neuron. 1989 Dec;3(6):665–676. doi: 10.1016/0896-6273(89)90235-3. [DOI] [PubMed] [Google Scholar]
- Verselis V. K., Bennett M. V., Bargiello T. A. A voltage-dependent gap junction in Drosophila melanogaster. Biophys J. 1991 Jan;59(1):114–126. doi: 10.1016/S0006-3495(91)82204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willecke K., Heynkes R., Dahl E., Stutenkemper R., Hennemann H., Jungbluth S., Suchyna T., Nicholson B. J. Mouse connexin37: cloning and functional expression of a gap junction gene highly expressed in lung. J Cell Biol. 1991 Sep;114(5):1049–1057. doi: 10.1083/jcb.114.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey S. B., John S. A., Lal R., Austin B. J., Revel J. P. The 43-kD polypeptide of heart gap junctions: immunolocalization, topology, and functional domains. J Cell Biol. 1989 Jun;108(6):2241–2254. doi: 10.1083/jcb.108.6.2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yellen G., Jurman M. E., Abramson T., MacKinnon R. Mutations affecting internal TEA blockade identify the probable pore-forming region of a K+ channel. Science. 1991 Feb 22;251(4996):939–942. doi: 10.1126/science.2000494. [DOI] [PubMed] [Google Scholar]
- Zhang J. T., Nicholson B. J. Sequence and tissue distribution of a second protein of hepatic gap junctions, Cx26, as deduced from its cDNA. J Cell Biol. 1989 Dec;109(6 Pt 2):3391–3401. doi: 10.1083/jcb.109.6.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer D. B., Green C. R., Evans W. H., Gilula N. B. Topological analysis of the major protein in isolated intact rat liver gap junctions and gap junction-derived single membrane structures. J Biol Chem. 1987 Jun 5;262(16):7751–7763. [PubMed] [Google Scholar]