Abstract
Myotonic dystrophy type 1 is a multisystemic autosomal dominant disorder caused by the expansion of (CTG) n triplets in the 3'UTR of the DMPK gene, on chromosome 19q13.3. In the last years, few DM1 patients with different patterns of CCG/CTC interruptions at the 3′ end of the DMPK expanded tract have been described. However, the role of these interruptions in DM1 pathogenesis is still unclear. To study the frequency, stability and the structure of DMPK variant expanded alleles in the Italian population, we have re-evaluated 254 Italian DM1 patients using triplet-primed PCR (TP-PCR), at both the 3′ and 5′ ends of the CTG expansion. In addition, three DM1 families were also investigated in order to analyze the intergenerational stability of the interrupted DMPK alleles. Fourteen DM1 patients showed a TP-PCR electrophoretic profile indicating CCG/CTC interruptions within the CTG expansion. Interestingly, interruptions have been detected and, for the first time, sequenced at the 5′ end of the CTG array. Analysis of five intergenerational transmissions revealed a substantial intrafamilial stability of the DM1 mutation among relatives. Our results support the hypothesis that CCG/CTC interruptions within the DMPK expanded alleles have a stabilizing effect on the mutational dynamics and can modulate the severity of symptoms in DM1 patients.
Introduction
Myotonic dystrophy type 1 (DM1, OMIM #160900) is the most common adult onset muscular dystrophy affecting mainly skeletal muscle, heart and the central nervous system.1 The molecular defect at the basis of DM1 is the expansion of an unstable CTG trinucleotide repeat in the 3′ untranslated region of the DMPK gene on chromosome 19q13.3.2, 3 DM1 is a progressive multisystemic disorder characterized by muscle weakness, myotonia, cataracts, cardiac conduction defects and endocrinological disturbances. Similarly to other triplet repeat expansion diseases, DM1 expanded alleles are genetically unstable and tend to expand in both the germline and soma, thus explaining the anticipation phenomenon and the progressive nature of clinical symptoms. Although the CTG repeat tract is usually uninterrupted in non-expanded and expanded alleles, in the last few years, pathological variant expansions containing unstable CCG, CTC and GGC sequence interruptions have been reported in rare instances.4, 5, 6, 7 Complex variant repeats were identified at both ends of the CTG array of DM1 patients of different origin, with an estimated prevalence of 3–5% of cases.4, 5, 6, 7, 8 However, the phenotypical consequences of the interrupted alleles on DM1 patients is still controversial leading either to a complex neurological phenotype or to classical/mild DM1 forms.4, 5, 7 In the present work, we describe the detailed molecular analysis of multiple patients carrying various patterns of interruptions either at the 3′ or 5′ end of the DMPK repeat identified in our cohort of 254 Italian DM1 patients, including three multi-generation families. DM1 patients have been re-evaluated using bidirectional triplet-repeat primed PCR (TP-PCR) method, following by direct sequencing of the PCR product in atypical cases. The stability of interrupted alleles has been determined studying five intergenerational transmissions and the prevalence of variant expansions estimated in our Italian DM1 population was compared with data. Finally genotype–phenotype correlations have been carried out to determine the causal or modifying effects of the interrupted DMPK alleles on the phenotype of their carriers.
Materials and Methods
DM1 patients
Two hundred fifty-four DNA samples belonging to clinically referred Italian DM1 patients with (CTG) repetition number determined by long range-PCR (LR-PCR) and Southern blot analysis of LR-PCR products,9 have been retrospectively analyzed. Major clinical data available from patients with variant DMPK alleles are summarized in Table 1. In three cases in which we identified variant repeats we recruited additional family members to study intergenerational transmissions.
Table 1. Clinical and molecular findings in DM1 patients with CCG/CTC interruptions identified in this study.
| Code | Sex | Age (years) | Family Relationship | Expanded allele length | Interrupted end | Sequence | Age at onset | Muscle weakness | Myotonia | Cataract |
|---|---|---|---|---|---|---|---|---|---|---|
| A1 | M | 66 | Father of A2 | 1000–1400 | 3′ | c.*224_226CTG[(880_1280)]CTG[2]CCG[1]CTG[112]CCG[1]CTG[4] | 58 | + | − | − |
| A2 | F | 39 | Daughter of A1 | 475–640 | 3′ | c.*224_226CTG[(437_602)]CTG[14]CCG[1]CTG[18]CCG[1]CTG[4] | 31 | − | + | − |
| A3 | ? | 0 | Fetus of A2 | 500 | 3′ | c.*224_226CTG[(380)]CTG[28]CCG[1]CTG[40]CTC[1]CTG[36]CCG[1]CTG[8]CCG[1]CTG[4] | – | n.a. | ||
| B1 | F | 55 | Mother of B2 | 740–930 | 3′ | c.*224_226CTG[(699_889)]CCGCTG[2]CCG[2]CTG[3]CCGCTG[3]CTG[26] | 51 | + | + | + |
| B2 | F | 28 | Daughter of B1 | 450–550 | 3′ | c.*224_226CTG[(372_472)]CTG[16]CCG[1]CTG[2]CCGCTG[4]CTG[1]CCGCTG[4]CCG[1]CCGCTG[4]CCG[1]CCGCTG[5]CTG[22] | – | − | − | − |
| C1 | F | 58 | Mother of C2 | 140 | 5′ | c.*224_226CTG[30]CCG[2]CTG[2]CCG[1]CTG[105]a | 58 | − | + | − |
| C2 | F | 40 | Daughter of C1 | 121 | 5′ | c.*224_226CTG[28]CCG[2]CTG[2]CCG[1]CTG[88]a | 37 | − | + | − |
| C3 | ? | 0 | Fetus of C2 | 113 | 5′ | c.*224_226CTG[31]CCG[2]CTG[2]CCG[1]CTG[13]CCG[1]CTG[63]a | – | n.a. | ||
| D | F | 38 | – | 600–700 | 3′ | c.*224_226CTG[(514_614)]CTG[68]CCG[9]CTG[9] | 35 | − | + | − |
| E | F | 56 | – | 500–660 | 3′ | c.*224_226CTG[(404_564)]CTG[33]CCGCTG[28]CTG[7] | 49 | − | + | − |
| F | M | 70 | – | 250 | 3′ | c.*224_226CTG[(208)]CTG[5]CCGCTG[16]CTG[5] | 66 | − | ±b | − |
| G | M | 71 | – | 400–580 | 3′ | c.*224_226CTG[(330_510)]CTG[8]CCGCTG[17]CTG[2]CCG[1]CTG[25] | 61 | + | + | − |
| H | M | 49 | – | 175 | 5′ | c.*224_226CTG[(133)]CTG[8]CCG[1]CTG[5]CCG[2]CTG[1]CCG[4]CTG[2]CCG[4]CTG[1]CCG[2]CTG[2]CCG[1]CTG[9] | 46 | + | + | − |
| I | M | 20 | – | 260–722 | 3′ | c.*224_226CTG[(188_650)]CTG[1]CCG[1]CTG[2]CCG[1]CTG[7]CCG[1]CTG[28]CCG[1]CTG[7]CCG[1]CTG[22] | 15 | − | + | + |
Abbreviation: n.a., not applicable.
For each patient, repeat expansion sizes were determined by a combination of Long range-PCR (LR-PCR) and Southern blot analysis of LR-PCR products. The expansions of the patients C1, C2 and C3 were obtained by LR-PCR and we were able to sequence the whole DMPK expanded allele.
sequences of the entire DMPK expanded allele.
±mild clinical myotnia.
Detection and molecular characterization of the DMPK interrupted alleles
Genomic DNA was extracted from peripheral blood using a salting-out procedure.10 (CTG)n repeat expansion sizes were determined by a combination of long range-PCR (LR-PCR) and Southern blot analysis of LR-PCR products.9 CTG expansions within the DMPK gene were detected through bidirectional TP-PCR as previously described.11 To investigate the presence of variant repeats, variants of the P4 primer have been used accordingly to published protocols.4 The characterization of the interruption motifs has been obtained using different approaches, including direct sequencing of: TP-PCR products with primer P1 for 3′ or P2 for 5′ interruptions (patients A2, B1, D, E, F, G, H, I); gel-purified (Gel Extraction Kit, Qiagen, Germany) TP-PCR, using primers P4hex1 or P4-CC, cloned in pCR2.1 vector (TA Cloning, Invitrogen, Carlsbad, CA, USA) using M13 Reverse vector primer (patients A1, A3 and B2); gel-purified LR-PCR products cloned in pCR2.1 vector (TA Cloning, Invitrogen) (patients C1, C2, C3) using M13 Reverse vector primer. In these latter DM1 patients the entire DMPK expanded allele has been characterized. Primers sequences are reported in Supplementary Table S1.
Results
Detection of CCG and CTC interruptions using TP-PCR analysis at the 3′ and 5′ end of the CTG array and genotype-phenotype correlations
In this study, we have retrospectively analyzed 254 DM1 patients previously characterized by classical LR-PCR analysis.9 Bidirectional TP-PCR with primers P4-CTG at the 3′-end, and P4-CAG at the 5′-end complementary to the CTG repeats revealed an unexpected pattern of the electrophoretic profile in 14 DM1 samples (9 unrelated and 5 familial cases) with remarkable gaps in the normal contiguous peaks pattern detectable by capillary electrophoresis. Interestingly, these interruptions have been mapped at the 3′-end (n=10), and also at the 5′-end (n=4) of the CTG array (Table 1, Figure 1). Bidirectional TP-PCR with primers complementary to the variant repeat motifs reported so far in the literature (CCG, CTC and GGC repeats) indicated the presence of interruptions containing CCG and CTC sequences. A combination of cloning and direct sequencing of the TP-PCR products have been used to determine the structures of the complex variant alleles with CCG and CTC tandem motifs alternating to CTG classical repeats as reported in Table 1. The correlation between the number of CTG repeats and the clinical picture suggests that variant repeat alleles could positively modulate DM1 symptoms and age at onset even in the same family. For example, patient B2 at the time of examination (28 years), did not show any clinical symptoms of DM1 or any EMG abnormalities and her mother (patient B1) developed the first DM1 symptoms at age 51. LR-PCR analysis across the repeat showed a 450–550 CTG-containing expanded allele in the daughter (B2) and a 740–930 CTG repetition in the mother (B1), with TP-PCR profiles indicating interruptions in the 3′-end of the expansions (Table 1). Additionally, the age at onset of patient F is much greater than would be expected for a 250 CTG repeat expanded allele12 with a very mild clinical phenotype characterized only by electrical myotonia (Table 1). The median age at onset of the disease in our cohort of DM1 patients with uninterrupted DMPK alleles classified according to their expansion classes13 is the following: E1 (n=66) (50–199 CTGs) 37.3 years, E2a (n=67) (200–499 CTGs) 27.5 years, E2b (n=60) (500–799 CTGs) 22.7 years, E2c (n=24) (800–999 CTGs) 21.2 years, E3a (n=28) (1.000–1499 CTGs) 17.3 years. If we compare these data with the age of onset of the disease in our patients with interrupted DMPK alleles we found that, in nine out of eleven patients, the DM1 age at onset is higher than expected on the basis of the expansion length.
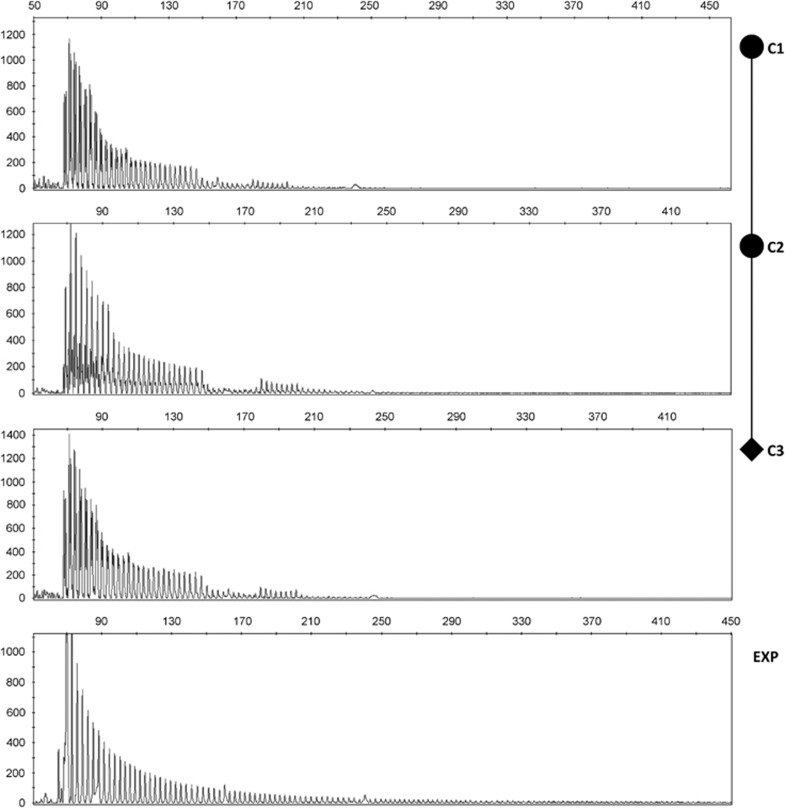
Figure 1.
TP-PCR analysis of 5′ region of the CTG array in members of family C and in a DM1 patient with an uninterrupted DMPK allele as control (bottom panel, EXP). CTG sequence interruptions are visualized as absence of PCR products where the P4-CAG repeated primer cannot bind. Pedigree of family C is added to help to follow the intergenerational dynamics in the pattern of interruptions.
The frequency of expanded interrupted alleles estimated in our Italian study cohort is 3.5% (9/254), in accordance with previous estimates.4, 5
CCG–CTC repeats stabilize the intergenerational variability of the complex DMPK expanded alleles
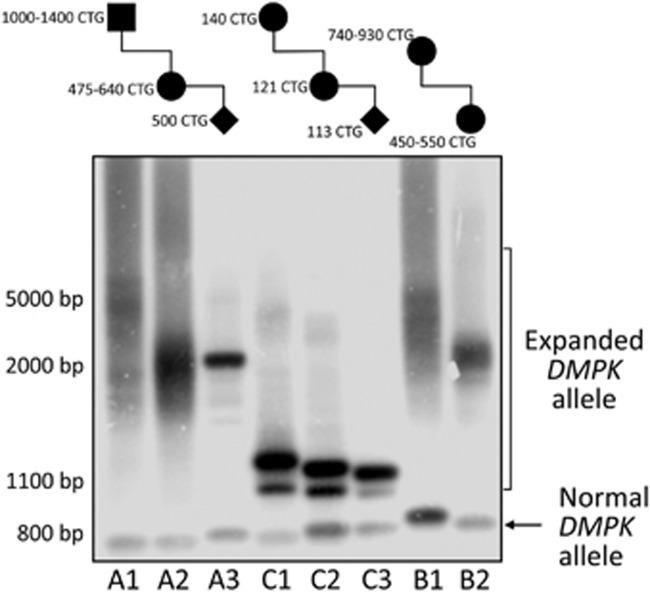
Given the stability of variant repeats in other triplet repeat disorders,14, 15, 16 we decided to study the intergenerational dynamics of the complex DMPK alleles in three DM1 families comprising five intergenerational transmissions (4 maternal and 1 paternal) (Figure 2). Interestingly, in all intergenerational events analyzed in this study, we observed a relative stability of the mutated alleles (family A and C) or a tendency to contraction even when the mutation was transmitted from an adult onset affected DM1 mother (from 740–930 to 450–550 CTGs in family B) (Figure 2). The only paternal transmission available, also lead to a reduction of the CTG repetition length (from 1000–1400 to 475–640 CTGs, family A) (Figure 2).
Figure 2.
Long range-PCR, using primers EX15-Fw and DMK1111-Rv,9 and Southern blot analysis of members belonging to families A, B and C. Pedigrees with estimated sizes of CTG expansions in the analyzed family members are added to the top of the figure. Note that DM1 expansions containing sequence interruptions tend to remain stable or even contract through germline transmissions.
Discussion
The majority of DM1 patients are thought to inherit a pure CTG repeat expansion. In the last few years however, pathological variant expansions containing unstable CCG, CTC and GGC sequence interruptions have been reported in rare cases. Complex variant repeats were identified at the both ends of the CTG array of DM1 patients of different origin, with an estimated prevalence of 3–5%.4, 5, 7, 8 In this study we retrospectively revaluated by bi-directional TP-PCR a cohort of 254 Italian DM1 patients estimating a minimum frequency of interrupted expanded alleles of 3.5%, perfectly in line with previous observations.4, 5 Direct sequencing of the entire or partial interrupted alleles, revealed the presence of complex arrays containing (CTC), (CCG) and/or (CCGCTG) repeats. We were able to identify and, for the first time, characterize four DM1 patients carrying a DMPK expanded allele with a non-pure CTG array at the 5′-end. This is a matter of particular relevance, considering that all the currently published works described DM1 variant repeats strongly clustered at the 3′-end of the array.4, 5, 7 Our study reveal that there is not an absolute polarity in the generation and/or maintenance of non-CTG repeats within the DM1 locus. The identification of sequence interruptions at both ends of the CTG array has also practical consequences for DM1 molecular genetic testing. In case of DMPK interrupted alleles, the detection of the DM1 mutation represents a diagnostic challenge because of the extremely stable secondary structures formed by the repetitive GC-rich sequences within the interruptions,17 which can interfere with LR-PCR amplifications and possibly lead to false negative results.4, 6, 18 Our study further supports the need to perform TP-PCR both at the 5′ and 3′ end of the CTG to detect variant DMPK expanded alleles in putative DM1 patients who have only a single detectable normal allele and when conventional LR-PCR fails.4, 6, 18 The intergenerational transmissions of the variant expanded alleles in our three DM1 families, support the general stabilizing effect reported previously,4, 5 showing even a tendency towards contraction of the DMPK interrupted alleles through intergenerational transmissions. This phenomenon is a matter of particular relevance in DM1 genetic counseling. A highly plausible explanation for the stability of interrupted alleles could be the reduction of slipped strand DNA intermediates that are formed in the CTG repeated region, presumably by the biophysical destabilization of slipped strand structures with misaligned variant repeats.19
Data reported so far suggest that the presence of interruptions in the CTG array does not considerably affect the nuclear accumulation of expanded RNAs since muscle tissues from ‘atypical' (with interrupted alleles DMPK expanded alleles) DM1 patients showed ribonuclear foci and splicing alterations similar to those observed in muscle from ‘classical' (with uninterrupted DMPK expanded alleles) DM1 patients.7 It is therefore possible that variant repeats might mediate an altered pathology through other mechanisms, such as changing the methylation status of the region up and/or down of the repeat, modulating chromatin structure and the nucleosome assembly.20, 21 A recent paper, reported indeed that the presence of CCG/CTC/CGG interruptions at the 3′ end of the CTG array is associated with a highly polarized pattern of CpG methylation at the DM1 locus involving regions downstream of the CTG array.22 Another important issue is the contribution of unusual DMPK alleles to the DM1 clinical picture. Genotype-phenotype correlations studies in variant DM1 patients have highlighted the difficulty to predict disease severity when an atypical interrupted allele is detected. The number of DM1 individuals with interruptions described is however limited and their phenotype range from a complex neurological involvement to the absence of muscular dystrophy associated with a late age of onset.5, 21 Further genetic studies are necessary to understand the possibly modifying effects of these interruptions and to provide patients with improved prognostic information associated with variant DMPK expansions.
Acknowledgments
This work was supported by ‘Uncovering Excellence 2014' grant from Tor Vergata University of Rome assigned to Botta Annalisa (grant number 5-MDESM-PLAT) and by Agenzia Spaziale Italiana (ASI), CoReA project no. 2013-084-R.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Harper PS: Myotonic Dystrophy. Oxford University Press: Oxford, New York, 2009; 106. [Google Scholar]
- Brook JD, McCurrach ME, Harley HG et al: Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 1992; 69: 799–808. [DOI] [PubMed] [Google Scholar]
- Mahadevan M, Tsilfidis C, Sabourin L et al: Myotonic dystrophy mutation: an unstable CTG repeat in the 3′untranslated region of the gene. Science 1992; 255: 1253–1255. [DOI] [PubMed] [Google Scholar]
- Musova Z, Mazanec R, Krepelova A et al: Highly unstable sequence interruptions of the CTG repeat in the myotonic dystrophy gene. Am J Med Genet 2009; 149A: 1365–1374. [DOI] [PubMed] [Google Scholar]
- Braida C, Stefanatos RK, Adam B et al: Variant CCG and GGC repeats within the CTG expansion dramatically modify mutational dynamics and likely contribute toward unusual symptoms in some myotonic dystrophy type 1 patients. Hum Mol. Genet 2010; 19: 1399–1412. [DOI] [PubMed] [Google Scholar]
- Addis M, Serrenti M, Meloni C, Cau M, Melis MA: Triplet-primed PCR is more sensitive than southern blotting-long PCR for the diagnosis of myotonic dystrophy type1. Genet Test Mol Biomarkers 2012; 16: 1428–1431. [DOI] [PubMed] [Google Scholar]
- Santoro M, Masciullo M, Pietrobono R et al: Molecular, clinical, and muscle studies in myotonic dystrophy type 1 (DM1) associated with novel variant CCG expansions. J Neurol 2013; 260: 1245–1257. [DOI] [PubMed] [Google Scholar]
- Dryland PA, Doherty E, Love JM, Love DR: Simple repeat-primed PCR analysis of the myotonic dystrophy type 1 gene in a clinical diagnostic environment. J Neurodegener Dis 2013; 2013: 857564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botta A, Bonifazi E, Vallo L et al: Italian guidelines for molecular analysis in myotonic dystrophies. Acta Myol 2006; 25: 23–33. [PubMed] [Google Scholar]
- Miller SA, Dykes DD, Polesky HF: A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 1998; 16: 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JP, Barron LH, Goudie D et al: A general method for the detection of large CAG repeat expansions by fluorescent PCR. J Med Genet 1996; 33: 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley HG, Rundle SA, MacMillan JC et al: Size of the unstable CTG repeat sequence in relation to phenotype and parental transmission in myotonic dystrophy. Am J Hum Genet 1993; 52: 1164–1174. [PMC free article] [PubMed] [Google Scholar]
- Salehi LB, Bonifazi E, Di Stasio E et al: Risk Prediction for Clinical Phenotype in Myotonic Dystrophy type 1: Data from 2,650 patients. Genet Test 2007; 11: 84–90. [DOI] [PubMed] [Google Scholar]
- Chung MY, Ranum LP, Duvick LA, Servadio A, Zoghbi HY, Orr HT: Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat Genet 1993; 5: 254–258. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Holden JJ, Popovich BW et al: Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat Genet 1994; 8: 88–94. [DOI] [PubMed] [Google Scholar]
- Choudhry S, Mukerji M, Srivastava AK, Jain S, Brahmachari SK: CAG repeat instability at SCA2 locus: Anchoring CAA interruptions and linked single nucleotide polymorphisms. Hum Mol Genet 2001; 10: 2437–2446. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Sinden RR: Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry 1996; 35: 5041–5053. [DOI] [PubMed] [Google Scholar]
- Radvansky J, Ficek A, Minarik G, Palffy R, Kadasi L: Effect of unexpected sequence interruptions to conventional PCR and repeat primed PCR in myotonic dystrophy type 1 testing. Diagnos Mol Pathol 2011; 20: 48–51. [DOI] [PubMed] [Google Scholar]
- Pearson CE, Eichlel EE, Lorenzetti D et al: Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S DNA) formation. Biochemistry 1998; 37: 2701–2708. [DOI] [PubMed] [Google Scholar]
- Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH: Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol 1995; 128: 995–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchin C, Lonigro R, Verriello L, Pellizzari L, Bergonzi P, Damante G: Correlations between individual clinical manifestations and CTG repeat amplification in myotonic dystrophy. Clin Genet 2000; 57: 74–82. [DOI] [PubMed] [Google Scholar]
- Santoro M, Fontana L, Masciullo M et al: Expansion size and presence of CCG/CTC/CGG sequence interruptions in the expanded CTG array are independently associated to hypermethylation at the DMPK locus in myotonic dystrophy type 1 (DM1). Biochim Biophys Acta 2015; 1852: 2645–2652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.