Abstract
Autosomal dominant (AD) familial Meniere's disease (FMD) is a rare disorder involving the inner ear defined by sensorineural hearing loss, tinnitus and episodic vertigo. Here, we have identified two novel and rare heterozygous variants in the SEMA3D and DPT genes segregating with the complete phenotype that have variable expressivity in two pedigrees with AD-FMD. A detailed characterization of the phenotype within each family illustrates the clinical heterogeneity in the onset and progression of the disease. We also showed the expression of both genes in the human cochlea and performed in silico analyses of these variants. Three-dimensional protein modelling showed changes in the structure of the protein indicating potential physical interactions. These results confirm a genetic heterogeneity in FMD with incomplete penetrance and variable expressivity.
Introduction
Meniere's disease (MD (MIM 156000)) is a chronic multifactorial disorder affecting the inner ear that is characterized by episodes of vertigo, associated with low- to middle-frequency sensorineural hearing loss (SNHL), tinnitus and aural fullness.1 The disorder may affect both ears in 25–40% of patients,2 and bilateral MD is associated with an increase of the duration of vestibular symptoms and a negative impact on health-related quality of life.3 According to the clinical presentation and progression of the symptoms, there is great variability in the phenotype of MD that may overlap with other vestibular disorders such as vestibular migraine4, 5 or autoimmune inner ear disease.6 Epidemiological studies support an association with autoimmune disorders7, 8 and allergic background,9, 10 and genetic susceptibility is widely recognized.11, 12 MD has a female preponderance and is more commonly found in Caucasian and Asian than in African descendant populations.13 Most of the cases of MD are considered sporadic, but familial clustering has been reported.12, 14, 15, 16 Familial MD (FMD) is observed in 8–10% of sporadic cases,17 and FAM136A and DTNA genes have been found in a single family with incomplete penetrance.18 Although most cases present an autosomal dominant (AD) pattern of inheritance, different patterns have been observed,14, 15 suggesting a genetic heterogeneity in FMD.
Whole-exome sequencing (WES) has been widely used for the identification of novel disease genes in rare Mendelian disorders. However, if the phenotype is heterogeneous with a variable expressivity, as occurs in FMD, multiple genes or an interaction between genetic and environmental factors may complicate the interpretation of WES data. Recently, complex disorders have been deciphered by using WES.19 In the present study, we describe the clinical phenotype of two families with AD-FMD showing incomplete penetrance and variable expressivity. We also report two missense variants in the DPT and SEMA3D genes in FMD, suggesting genetic heterogeneity.
Materials and methods
Patients
Two multicase families with AD-FMD originally from the southeast of Spain were selected for WES (Figure 1). The Institutional Review Board for Clinical Research approved this study, and an informed consent for donor biological samples was obtained from all subjects. The diagnosis of FMD was established according to the diagnostic criteria defined by the Bárány Society.1 A complete neurootology assessment was carried out in all cases, and a brain magnetic resonance imaging was performed to exclude other possible causes of neurological symptoms. Patients were followed with serial pure tone audiograms at each visit to monitor hearing loss from the initial diagnosis.
Figure 1.
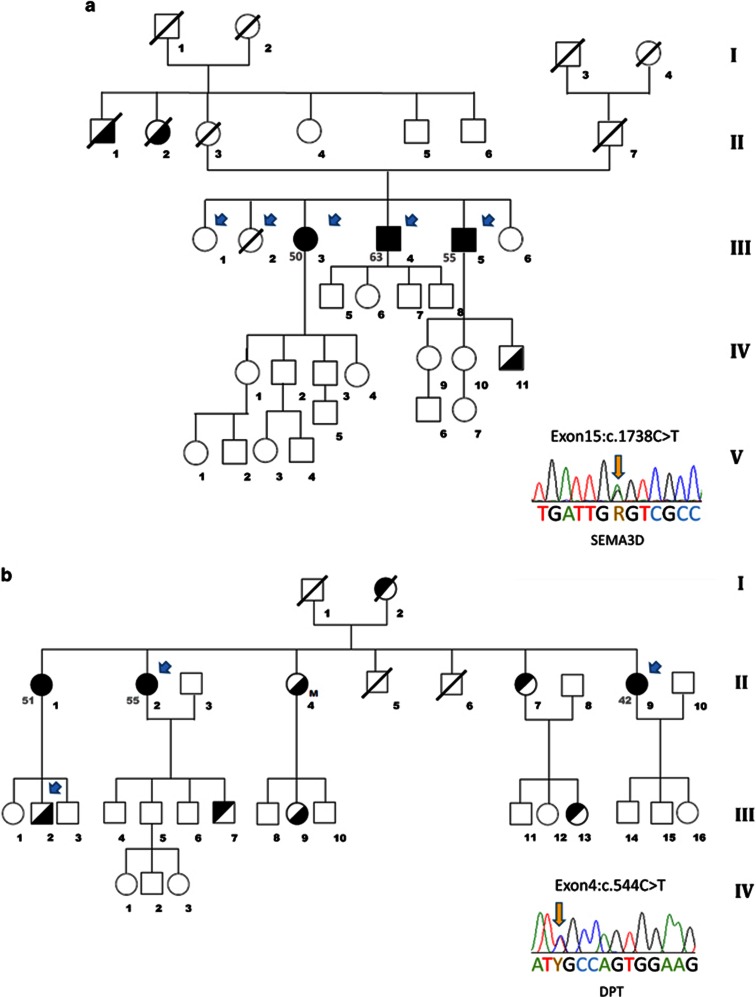
Spanish families with Meniere's disease and SEMA3D and DPT genes. (a) Pedigree of a family presenting AD-FMD in three affected individuals with complete phenotype in the same generation and Sanger sequence of the variant affecting SEMA3D gene. (b) Pedigree of a family with three women affected with MD in the same generation with an autoimmune background and sequence of the variant in the DPT gene. Symbols in the pedigree represent: full black square or circle, complete MD phenotype (numbers at the left of the symbol indicate age of onset); partial black square or circle, incomplete phenotype (upper-left black: vestibular phenotype; lower-right black: cochlear phenotype). M, migraine.
Whole-exome sequencing
DNA samples from patients and controls selected from each family were sequenced with the SOLiD (Life Technologies, Carlsbad, CA, USA) 5500xl platform. Exons and flanking intronic regions were selected and captured by Agilent (Santa Clara, CA, USA) All Exon 50 MB capture kit. The conditions and primer sets are available on commercial websites. The sequences were analysed with Lifescope software 2.5 (https://www.thermofisher.com/es/es/home/technical-resources/software-downloads/lifescope-genomic-analysis-software.html). The results obtained were mapped with the GRCh37/hg19 human assembly. Files with single-nucleotide variants (SNVs), called Variant Call Format files (VCF), containing ~50 000 SNVs per exome sequenced, were generated for each sample sequenced.
Bioinformatics analyses
To identify variants associated with the MD phenotype, we first filtered them using exome data from our in-house control database (N=35) and 578 controls from the CIBERER Spanish Variant server (http://csvs.babelomics.org/). We used a minor allelic frequency (MAF) >0.0001, according to the criteria defined for AD hearing loss genes,20 and the SNVs previously annotated in dbSNP 146 and the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/). The analysis strategy to prioritize candidate variants scored them according to their effect on protein structure and phylogenetic conservation by using a seven-point scoring system (Pathogenic Variant or PAVAR score) with the annotation of the following tools: SIFT (Sort Intolerant from Tolerant), PolyPhen2 (Polymorphism Phenotyping v.2), Grantham's Matrix, GERP+ (Genomic Evolutionary Rate Profiling), Mutation Taster, PhastCons and PhyloP. After annotation with wANNOVAR (http://wannovar.usc.edu/), SeattleSeq (http://snp.gs.washington.edu/SeattleSeqAnnotation137/), PROVEAN (http://provean.jcvi.org/genome_submit_2.php) and Mutation Taster (http://www.mutationtaster.org/StartQueryEngine.html) web tools, we filtered SNVs with a PAVAR score >5.18 To improve the accuracy of the variant prioritization, we combined the previous results with other bioinformatics tools that include phenotype information such as Exomiser v.2,21 and Variant Annotation Analysis and Search Tool (VAAST)+Phevor that prioritize the variants and the genes affected using a ranking system.22, 23 We used linkage information derived from the WES-common SNVs within each pedigree to reduce the list of candidate variants, according to the method described by Gazal et al.24
Validation by Sanger sequencing
The candidate variants were validated by Sanger sequencing in a 3130 Genetic Analyzer (Applied Biosystems, Waltham, MA, USA) and visualized with Sequence Scanner Software v1.0 (Applied Biosystems) to confirm WES results. Primer pairs flanking the SNV, whose sequences are shown in Supplementary Table S1, were designed using Primer3 software (http://primer3.sourceforge.net/) and checked with Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast/).
RNA extraction from cochleae and semicircular canals
Cochlear and semicircular canal human tissues were collected from schwannoma surgery patients (N=2), and were preserved immediately in RNAlater (Ambion, Austin, TX, USA). The tissue was homogenized mechanically by adding 700 μl of Qiazol (QIAGEN, Hilden, Germany) and a steel bead in a TyssueLyser LT (QIAGEN) for 5 min at 50 Hz. RNA was isolated using the QIAamp RNA miRNeasy Mini Kit (QIAGEN) and a Nanodrop (Thermo Fisher Scientific, Wilmington, DE, USA) spectrophotometer, and an Agilent RNA 6000 Nano Bioanalyzer chip was used to determine the integrity, quality and quantity of RNA. cDNA was synthesized with the QuantiTect Reverse Transcription Kit (QIAGEN) according to the manufacturer's instructions.
Expression analysis
Quantitative real-time PCR was performed in a 7900HT PCR System using SYBR Green RT-PCR techniques (Life Technologies) to confirm the expression of candidate genes in human cochlea and vestibular samples. The data were analysed with ABI RQ Manager Software (Applied Biosystems). HPRT1 was used as housekeeping gene to determine relative expression levels. Each sample was run in triplicate to calculate the ΔCt values for each sample with their SD. Statistical analysis was performed with IBM SPSS software (SPSS Inc., Armonk, NY, USA). The primers used to validate the expression of candidate genes are given in Supplementary Table S1.
Protein modelling
The elaboration of 3D models for Dermatopontin (Q07507) and Semaphorin-3D (O95025) proteins was performed by comparing the known protein sequence and its homologues in the RCSB Protein Data Bank (http://www.rcsb.org/pdb/), using them as templates for the predicted models in SwissModel Deep View Software (http://swissmodel.expasy.org) and for generating.pdb files. Prediction software calculates the P-value for the global quality of the model assembly, GDT/uGDT (global distance test and unnormalized global distance test), and RMSD for the absolute local quality of each residue in the model. Visualization and comparison between models were made on PyMOL software (http://pymol.org). PyMOL software was used to analyse RMS deviation between native model and mutant model by superimposing both.pdb files with Theseus software (http://www.theseus3d.org/). In addition, possible interactions between amino-acid residues in the position involved and other residues were determined by PyMOL software tool. The quality of the model was checked by Ramachandran plots using RAMPAGE webtool software (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php). Ramachandran plots analyse residue-by-residue geometry and overall structure geometry, detailing when a residue is out of the predicted zone by using its own stereochemical properties.
Variant data submission
All candidate variants have been submitted to ClinVar database (http://www.ncbi.nlm.nih.gov/clinvar/). The accession numbers for the variants in SEMA3D, GRHL2 and DPT genes are SCV000266468, SCV000266470 and SCV000266469. Rest of variants not annotated previously and not covered in this study were submitted to ClinVar and their accession numbers are SCV000323086, SCV000323087, SCV000323083, SCV000323085, SCV000323084 and SCV000323088. Exons were numbered according to the GenBank reference sequence files of each gene shown in Table 1.
Table 1. Candidate SNVs obtained with the different bioinformatics tools after filtering and prioritization process.
| family | CHR | POS | GENE | Ref Sequence | cDNA | Protein | PAVAR score | VAAST | PHEVOR | EXOMISER | MAF |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 7 | 84642128 | SEMA3D | NM_152754.2 NG_051329.1 | c.1738C>T | p.(P580S) | 6 | 10 | 5 | 0.62 | NR |
| 8 | 102555482 | GRHL2 | NM_024915.3 NG_011971.1 | c.34G>T | p.(V12L) | 5 | 26 | 1 | 1 | NR | |
| 3 | 52455673 | PHF7 | NM_016483.6 | c.583C>T | p.(H195Y) | 5 | 22 | 11 | 0.84 | 2.48E−05 | |
| 4 | 170634382 | CLCN3 | NM_001243372.1 NG_029731.1 | c.2221A>G c.2302A>G | p.(M741V) p.(M768V) | 5 | 38 | 23 | 0.86 | 8.24E-05 | |
| 3 | 42264873 | TRAK1 | NM_001042646.2 | c.2506G>A | p.(V836M) | 6 | 6 | 5 | 0.58 | 1.63E−05 | |
| 6 | 89974214 | GABRR2 | NM_002043.4 NG_033977.2 | c.1003G>A | p.(V335M) | 6 | 42 | 26 | 0.86 | 8.24E−06 | |
| 2 | 1 | 168665849 | DPT | NM_001937.4 | c.544C>T | p.(R182C) | 6 | 12 | 2 | 0.86 | 2.50E−05 |
| 2 | 109086855 | GCC2 | NM_181453.3 | c.1070A>G | p.(Y357C) | 6 | 96 | NS | 0 | NR | |
| X | 49034780 | PRICKLE3 | NM_006150.4 NG_017135.1 | c.609C>A | p.(S203R) | 5 | 33 | 11 | 1 | NR |
Abbreviations: NR, not reported; NS, not scored.
Positions are related to the ‘Genome Reference Consortium GRCh37'. Reference Sequences for each variant according to RefSeq file and GenBank file (if available). PAVAR score is a composite score including the following tools: SIFT, PolyPhen2, Grantham's Matrix, GERP+, Mutation Taster, PhastCons and PhyloP. Genes we cover in this study are in bold.
Results
Familial phenotype
Family 1
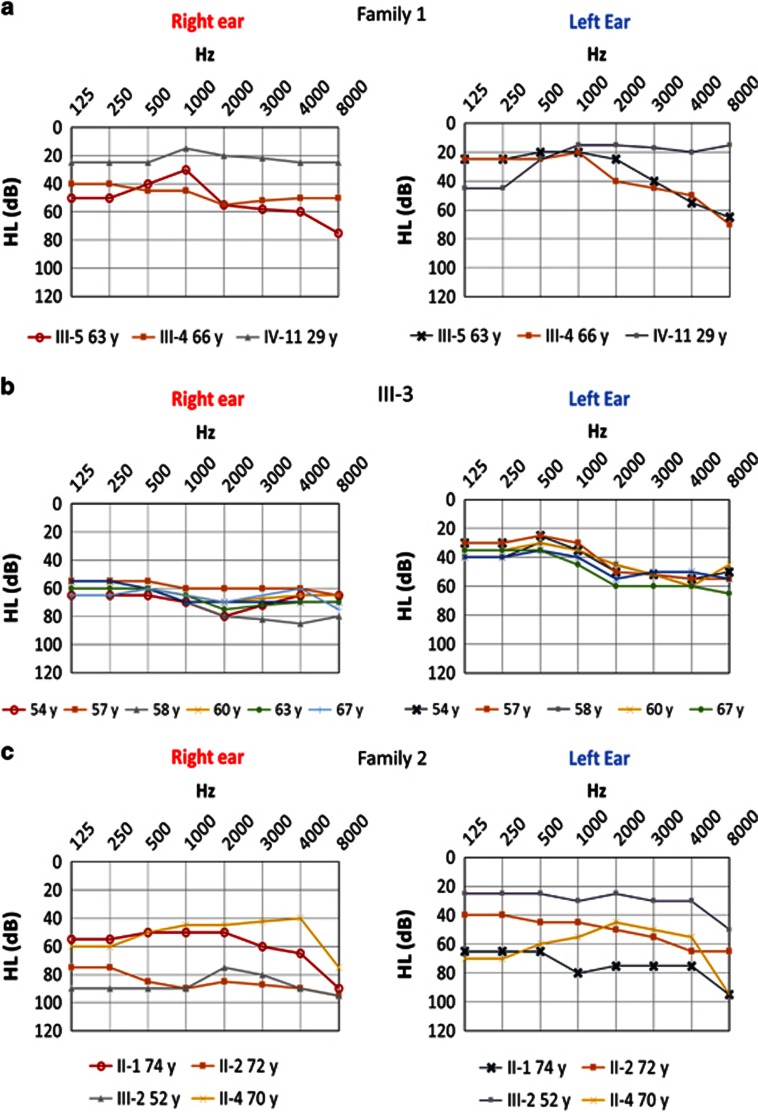
A family consisting of three siblings (two men and one woman) in the same generation with the complete phenotype was selected. None of the patients in this family has a history of migraine. The initial case (III: 3) was a 68-year-old woman with bilateral MD with an onset at 50 years of age. She has an asymmetrical bilateral pantonal SNHL since the onset of the symptoms, with a mild SNHL at low frequencies and moderate hearing loss from 2000 to 8000 Hz in the left ear. In the right ear, her hearing loss was severe with a pure tone average (PTA) of 58–70 dB. During the first 4 years of the disease, the patient showed a hearing loss fluctuation in both ears at all frequencies (Figure 2). She developed a progressive bilateral vestibular hypofunction and, after 15 years, she had reached a caloric areflexia in the right ear and severe hyporeflexia in the left ear. Her follow-up was over 19 years and she was controlled by low-sodium diet, high water intake and betahistine. She also experienced occasional episodes of sudden falls that were considered Tumarkin otolithic crises. Although her father did not have a confirmed history of vertigo or early-onset hearing loss, she had two maternal uncles with a history of SNHL (II: 1, II: 2). Her older brother (III: 4) developed a right ear MD at 63 years of age and he has been followed for 3 years. This patient has a history of high blood pressure and severe obesity (body mass index (BMI)=40 kg/m2). He initially presented a unilateral SNHL in the right ear and, a few months later, he started with episodes of vertigo and ipsilateral fluctuating auditory symptoms. Magnetic resonance imaging ruled out a brainstem or labyrinthine infarction, and he was diagnosed with delayed MD in the right ear, a clinical variant of the MD phenotype. Cervical vestibular evoked myogenic potentials (cVEMPs) showed an absence of vestibulocollic reflexes in both ears, confirming the impairment of saccular function, and caloric testing confirmed a bilateral vestibular hypofunction. The younger brother (III: 5) presented a left ear MD that started at 55 years of age and he has a follow-up of 9 years. His hearing profile was also a pantonal SNHL in the right ear and a mid- to high-frequency SNHL in the left ear. cVEMPs confirmed a bilateral loss of saccular function and caloric tests demonstrated a right vestibular hypofunction in the horizontal canal. He also has high blood pressure and is severely overweight (BMI=35 kg/m2) and was treated with betahistine. This patient experienced a sudden drop of hearing that was treated with oral steroids, showing a sustained recovery of SNHL over several years. During the past 2 years, he has presented several episodes of sudden falls, highly suggestive of Tumarkin crisis. His 29-year-old son has a low-tone SNHL without vestibular symptoms.
Figure 2.
Air-conduction audiograms. (a) Family 1 show a severe sensorineural hearing loss (SNHL). (b) Case III-3 has several audiograms at different ages showing hearing loss fluctuation (age symbols shown below the chart). (c) Family 2 show severe to profound SNHL. Upper x axis represents pure tone frequencies in Hz, and y axis represents hearing level (HL) in dB.
Family 2
The second family consisted of three women with MD in the same generation with an autoimmune background. None of the patients in this family has a history of migraine. The initial proband was the younger sister (II: 9), who started with a right ear MD at 42 years of age, with a low-frequency fluctuating SNHL and tinnitus. Her follow-up was 19 years. She experienced a large number of attacks with no response to betahistine. At 8 years after onset, she was treated with intratympanic gentamicin in the right ear with an immediate relief of vertigo attacks. cVEMPs and bithermal caloric testing confirmed an ipsilateral vestibular areflexia with normal response in the left ear. Hearing levels were maintained in the right ear with a PTA of 25 dB. Autoimmune screening found anti-ribonucleoprotein antibodies, suggesting a concomitant immune disorder without criteria for systemic lupus erythematosus. The elder sister (II: 1) developed a bilateral MD at 51 years of age. She had a history of atrial fibrillation, high blood pressure and type II diabetes. She described episodes of vertigo, tinnitus and fluctuating hearing loss. Her bilateral SNHL showed a rapid progression in the left ear and later in the right ear involving all frequencies reaching a PTA of 70 and 55 dB, respectively, after 5 years of follow-up. She has been maintained on a low-sodium diet and high water intake for the past 2 years without new episodes of vertigo. The third sibling (II: 2) was a woman who started with MD at 55 years of age. She also had Sjögren syndrome, high blood pressure, type II diabetes and ischaemic cardiac syndrome. She presented initially with a bilateral diachronic SNHL affecting all frequencies that started in the right ear and advanced a few years later to include the left ear. She was treated with oral steroids producing a partial recovery of 25 dB HL in both ears. After 19 years of follow-up, hearing loss was permanent with a PTA of 85 and 48 dB in the right and left ears, respectively. Two additional members of this pedigree presented SNHL without any vestibular symptoms. The first was a sister of the three affected patients (II: 4) who developed a progressive bilateral, synchronic, low- to middle-frequency SNHL in her late fifties. She also had high blood pressure, type II diabetes and cardiac failure secondary to an aortic valve double lesion. After 11 years of follow-up, the hearing loss was permanent with a PTA of 45 and 60 dB in the right and left ears, respectively. The last patient was the second son of the elderly patient with MD (III: 2), who presented with sudden tinnitus with SNHL in the right ear and a PTA of 80 dB at 51 years of age. He was treated with oral steroids with no recovery. Currently, he has a permanent hearing loss, persistent tinnitus in the right ear and hyperacusis.
Candidate variants
A list of candidate SNVs for each family is shown in Table 1. After the filtering and prioritizing process, the three individuals with MD in family 1 segregated one novel missense variant in the exon 15 of SEMA3D gene (NM_152754.2). In addition, missense variants in the GRHL2 (NM_024915.3) and TRAK1 (NM_001042646.2) genes were identified in the three cases with definite MD and in one familial control (III: 2). However, linkage analysis using WES data on this pedigree excluded the variants in the GRHL2 and TRAK1 genes (Supplementary Table S2). In the second family, the three sisters with MD shared a rare missense variant in exon 4 of the DPT gene (NM_001937.4). This variant (chr1:g.168665849 G>A, c.544C>T) segregates the hearing loss phenotype, as it was also found in two individuals with incomplete phenotype (II: 4 and III: 2) presenting progressive bilateral SNHL and sudden SNHL, respectively (Supplementary Table S3).
Protein modelling
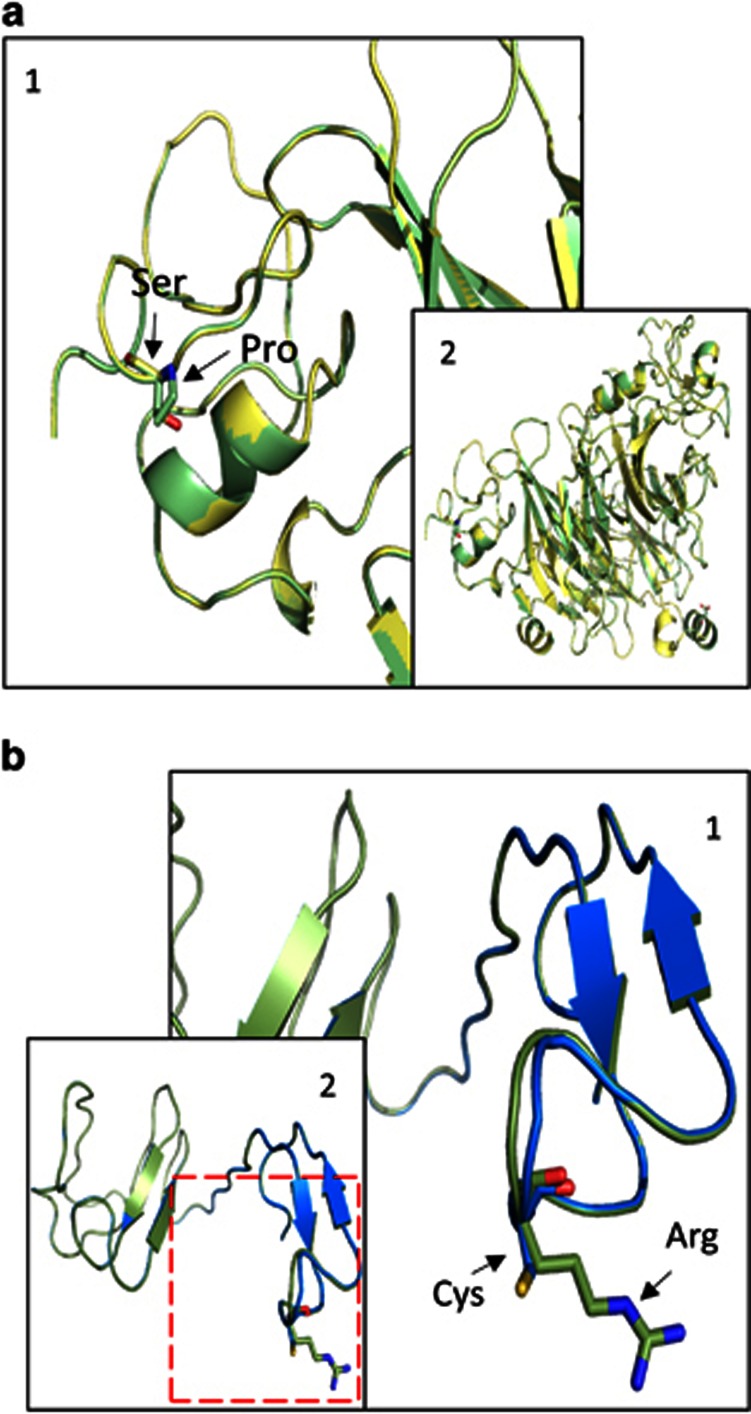
The protein sequences of Semaphorin-3D and Dermatopontin were aligned along with their orthologue sequences from different species to compare the degree of conservation of the involved domains for both proteins (Supplementary Figure S1). We observed that our amino-acid substitutions occur on highly conserved regions for each reference alignment. Three-dimensional protein models were constructed following previously crystallized related proteins from human in Protein Data Bank. In the case of Dermatopontin, the closest model described including our involved region was 1m8uA (Bovine Gamma E model) that shares a 31% sequence identity with our query sequence, including a partial high confidence identity with our region of interest. To model Semaphorin-3D we used a crystallized model for Semaphorin-3A, 1q47, with a sequence identity of 58% that includes our amino-acid substitution. A Ramachandran plot for Dermatopontin model shows 11% of the residues (9 from the total query) found in a region disallowed for being considered correct by stereochemical quality, but all of them were found far from our region of interest (Supplementary Figure S2). Similar considerations were made on a Semaphorin-3D protein model after finding 1.5% residues (8 from the total query) in the disallowed region (Supplementary Figure S3). Our protein models show how variants can affect tertiary structure of proteins by changing a single amino-acid residue. In the Semaphorin-3D model, the substitution c.1738C>T (p.(Pro580Ser)) does not induce a major structural change because of the similar biochemical properties of both amino-acid residues (Figure 3a). However, the substitution c. 544C>T (p.(Arg182Cys)) in the Dermatopontin predicted model represents a relevant structural change in the properties of the protein (Figure 3b). Superposition for both the Dermatopontin and Semaphorin-3D models shows structural differences between the mutant and wild-type proteins. These tools show a higher variability in Dermatopontin superposition models than in SEMA3D models. Hence, the distribution of the residues close to the variant (p.(Arg182Cys)) is highly affected by the change in the Dermatopontin model (positions 180 to 186 are mostly affected, especially p.Gln183 with a variance of 0.1180). In contrast, (p.(Pro580Ser)) seems to mildly affect the structural variance between the Semaphorin-3D models (higher variance values reach 0.0001 on p.(Asp579)). Although the change in the sequence does not have any phosphate-binding sites close to the involved zone, the (p.(Arg182Cys)) change may induce the disappearance of some steric bonds, according to the Ramachandran Plot Explorer. However, arginine maintains more steric bonds with its proxy environment than cysteine in our Dermatopontin protein model.
Figure 3.
Three-dimensional protein models. (a) Structural differences between predicted mutated Semaphorin-3D protein model. Superimposition of normal (green) and mutant (yellow) Semaphorin-3D protein model. (b) Structural differences between predicted mutant Dermatopontin protein models. Superimposition of normal Dermatopontin protein model (Green) and mutant Dermatopontin protein model (Blue). Amino-acid changes are marked with arrows.
Validation of expression levels of these genes in the cochlea
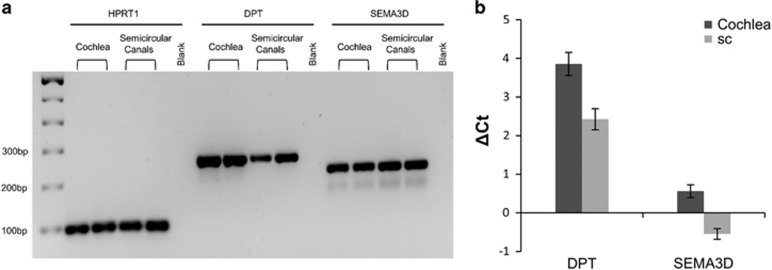
We observed significant expression levels of SEMA3D and DPT genes in human semicircular canals and cochlear tissues (Figure 4).
Figure 4.
Expression of DPT and SEMA3D genes in the human cochlea and semicircular canals (sc) by qPCR. (a) Validation of the expression using primers for DPT (250 bp), SEMA3D (213 bp) and a housekeeping gene HPRT1 (92 bp) in agarose gel. Each sample has a technical replication. (b) ΔCt values of DPT (ΔCt cochlea=3.85±0.3; ΔCt sc=2.42±0.27) and ΔCt values of SEMA3D (ΔCt cochlea=0.56±0.16; ΔCt sc=−0.55±0.13).
Discussion
Familial MD is a rare condition and the estimated prevalence is 6–10% among sporadic cases.15, 17 Some of the described families presented co-segregation with migraine and anticipation,12, 25, 26 but the two Spanish families that we report here do not show these features. Our study shows that AD-FMD has an incomplete penetrance with variable expressivity and it confirms a clinical heterogeneity in FMD. First, the onset of MD in these families may vary from the most common syndrome (episodic vertigo, unilateral low- to mid-frequency SNHL and tinnitus; case II: 9 in family 2) to bilateral diachronic SNHL (the second ear develops the syndrome after several years, eg, II: 1 and II: 2 in family 2), to bilateral pantonal asymmetrical SNHL (III: 3 and III: 5 in family 1) or delayed MD (hearing loss precedes the vestibular symptoms in months or years, eg, III: 4 in family 1). Second, the progression of the hearing loss is also variable within each family, ranging from a rapid progression observed in case II: 1 to a slower one found in II: 9 in family 2 that also had an autoimmune background. These differences may rely on the cumulative effect of several regulatory variants in modifier genes. Therefore, allelic variations in the MICA, NFKB1 or TLR10 genes have been reported to influence the hearing loss outcome in patients with sporadic MD.27, 28, 29 Third, several relatives in these families presented a partial syndrome with different types of SNHL, including sudden hearing loss (III: 2 in family 2) or pantonal SNHL (II: 2 in family 2) and no vestibular symptoms. These findings observed across different families with MD12, 14, 15 suggests that (1) different genes can be involved in the development of the partial or complete phenotype or (2) the interaction of environmental or epigenetic factors can also determine the differences in expressivity within the phenotype.
Our findings have started to define two candidate genes associated with FMD and support the hypothesis of genetic heterogeneity in FMD. Requena et al.18 have described novel variants in two other candidate genes for autosomal dominant FMD, FAM136A and DTNA, in a single family consisting of three women with a MD phenotype showing anticipation in consecutive generations.
In this study, we have identified two new candidate genes for FMD, SEMA3D and DPT, both being extracellular signals30, 31 that may be relevant to the formation or maintaining of inner ear structures.
SEMA3D gene encodes an axonal guiding protein, Semaphorin-3D. The protein is secreted and it inhibits the neural growth cones and the endothelial cell motility and migration, regulating cytoskeletal dynamics and cell adhesion.32 Semaphorins comprise seven different secreted proteins (designated by the letters A–G). These proteins contain a Sema domain involved in the formation of a complex with neuropilin (Nrp) and plexin transmembrane receptors. The Sema3–Nrp–plexin complexes activate the R-ras signalling pathway, and thus decreases the attachment of integrins to the extracellular matrix.33
Family 1 presented a novel missense variant at chr7:g.84642128 G>A (c.1738C>T) that generates a (p.(Pro580Ser)) change in Semaphorin-3D. This variant segregated the complete MD phenotype in three patients within the same generation, suggesting an autosomal dominant inheritance. These patients were bilaterally affected and two of them responded to steroid therapy, but the functional effect of this novel variant is unknown. The amino-acid substitution occurs at the beginning of the a5 helix on the PSI domain. This PSI domain is an important repeated domain rich in disulphide motifs, found in different extracellular receptors like plexins, integrins and semaphorins.34 Our substitution seems to affect a non-highly conserved amino acid on the most conserved part of the sequence (K-R-R-X-R-R-Q-D-V/I-R/K-X-G-D/N-P/A). Although potential interactions cannot be discarded, the sequence seems to be a zone of low probability for protein interactions. We also found in this family a rare missense variant in exon 2 of the GRHL2 gene (chr8:g.102555482 G>T, c.34G>T), a gene associated with AD non-syndromic SNHL (DFNA28), causing low-frequency SNHL without vestibular symptoms.35 However, this variant was also found in an elderly relative without any clinical symptom (III: 2), and it had a lower prioritizing score, according to our pipeline.
The DPT gene, located at 1q12-q23, consists of 4 exons, with a coding region of 1786 bp. DPT encodes Dermatopontin, an extracellular matrix protein of 201 AAs that interacts with integrins of the cell surface and proteoglycans such as dermatan-sulphate. Dermatopontin is expressed in fibroblasts and it binds to TGFβ, regulating its activity by the formation of a complex with decorin, a leucine-rich proteoglycan, that also interacts with type I and II collagens.36 Dermatopontin inhibits the formation of decorin-TGFβ1 complexes and probably also binds TGFβ1 on the surface of endothelial cells, thereby maintaining vascular homoeostasis.36, 37 We have identified a rare missense variant at chr1:g.168665849 G>A, c.544C>T (p.(Arg182Cys)), located at exon 4, in the three women with MD in the same generation, and in two individuals with SNHL in family 2. This variant has a MAF=2.5 × 10–5 in the ExAC and it probably produces a functional change in the protein sequence. It has been found in three individuals of Latino, South Asian and European (Non-Finnish) populations. We cannot determine whether this rare variant changes the interaction with decorin or TGFβ1. However, it is found inside a highly conserved repeating sequence (D-R-E/Q-W-X-F/Y) of the known domain, specifically at the end of the fourth loop structure. Although no biological characteristics have been demonstrated before,36 this could represent important evidence of relevance to the maintenance of the protein functionality. Interestingly, both genes, SEMA3D and DPT, do not show a differential expression along the axis of the cochlea in the mouse transcriptome.38 The absence of a tonotopic gene expression gradient for these genes is consistent with the finding of a pantonal SNHL observed in most of patients and it points to a mechanism involving the entire cochlea subsequent to onset of the disease. However, this is not the case with all MD families. A recently reported family with low-frequency SNHL segregated a novel missense variant in the PRKCB gene, encoding PKCB II protein (Martin-Sierra et al.39). As PKCB II has an expression gradient in the tectal cells along the organ of Corti that is highest at the apical turn of the cochlea, a loss of function in PKCB II may explain the onset of low-frequency SNHL. The observed phenotype in these families confirms a variable expressivity in FMD and the variants identified in the SEMA3D and DPT genes increase the number of candidate genes and suggest a genetic heterogeneity.
Acknowledgments
We thank patients with FMD and their relatives for their participation in this study. This work was supported by grants from Fundación Progreso y Salud (PI0496–14 to TR) and Meniere's Society, UK (to JAL-E). We also acknowledge the COST Action BM1306 TINNET that supports part of our networking activities (http://tinnet.tinnitusresearch.net/). We also thank Dr Anna Lysakowski for reviewing a previous version of the manuscript.
Footnotes
Supplementary Information accompanies this paper on European Journal of Human Genetics website (http://www.nature.com/ejhg)
The authors declare no conflict of interest.
Supplementary Material
References
- Lopez-Escamez JA, Carey J, Chung WH et al: Diagnostic criteria for Meniere's disease. J Vest Res 2015; 25: 1–7. [DOI] [PubMed] [Google Scholar]
- House JW, Doherty JK, Fisher LM, Derebery MJ, Berliner KI: Meniere's disease: prevalence of contralateral ear involvement. Otol Neurotol 2006; 27: 355–361. [DOI] [PubMed] [Google Scholar]
- Lopez-Escamez JA, Viciana D, Garrido-Fernandez P: Impact of bilaterality and headache on health-related quality of life in Meniere's disease. Ann Otol Rhinol Laryngol 2009; 118: 409–416. [DOI] [PubMed] [Google Scholar]
- Lopez-Escamez JA, Dlugaiczyk J, Jacobs J et al: Accompanying symptoms overlap during attacks in Meniere's Disease and vestibular migraine. Front Neurol 2014; 5: 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lempert T, Olesen J, Furman J et al: Vestibular migraine: diagnostic criteria. J Vest Res 2012; 22: 167–172. [DOI] [PubMed] [Google Scholar]
- Greco A, Gallo A, Fusconi M, Marinelli C, Macri GF, de Vincentiis M: Meniere's disease might be an autoimmune condition? Autoimmun Rev 2012; 11: 731–738. [DOI] [PubMed] [Google Scholar]
- Gazquez I, Soto-Varela A, Aran I et al: High prevalence of systemic autoimmune diseases in patients with Meniere's disease. PLoS One 2011; 6: e26759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyrrell JS, Whinney DJ, Ukoumunne OC, Fleming LE, Osborne NJ: Prevalence, associated factors, and comorbid conditions for Meniere's disease. Ear Hear 2014; 35: e162–e169. [DOI] [PubMed] [Google Scholar]
- Derebery MJ: Allergic and immunologic features of Meniere's disease. Otolaryngol Clin North Am 2011; 44: 655–666, ix. [DOI] [PubMed] [Google Scholar]
- Di Berardino F, Cesarani A: Gluten sensitivity in Meniere's disease. Laryngoscope 2012; 122: 700–702. [DOI] [PubMed] [Google Scholar]
- Gabrikova D, Frykholm C, Friberg U et al: Familiar Meniere's disease restricted to 1.48 Mb on chromosome 12p12.3 by allelic and haplotype association. J Hum Gen 2010; 55: 834–837. [DOI] [PubMed] [Google Scholar]
- Morrison AW, Bailey ME, Morrison GA: Familial Meniere's disease: clinical and genetic aspects. J Laryngol Otol 2009; 123: 29–37. [DOI] [PubMed] [Google Scholar]
- Ohmen JD, White CH, Li X et al: Genetic evidence for an ethnic diversity in the susceptibility to Meniere's disease. Otol Neurotol 2013; 34: 1336–1341. [DOI] [PubMed] [Google Scholar]
- Hietikko E, Kotimaki J, Sorri M, Mannikko M: High incidence of Meniere-like symptoms in relatives of Meniere patients in the areas of Oulu University Hospital and Kainuu Central Hospital in Finland. Eur J Med Genet 2013; 56: 279–285. [DOI] [PubMed] [Google Scholar]
- Requena T, Espinosa-Sanchez JM, Cabrera S et al: Familial clustering and genetic heterogeneity in Meniere's disease. Clin Genet 2014; 85: 245–252. [DOI] [PubMed] [Google Scholar]
- Vrabec JT: Genetic investigations of Meniere's disease. Otolaryngol Clin North Am 2010; 43: 1121–1132. [DOI] [PubMed] [Google Scholar]
- Lee JM, Kim MJ, Jung J, Kim HJ, Seo YJ, Kim SH: Genetic aspects and clinical characteristics of familial Meniere's disease in a South Korean population. Laryngoscope 2015; 125: 2175–2180. [DOI] [PubMed] [Google Scholar]
- Requena T, Cabrera S, Martin-Sierra C, Price SD, Lysakowski A, Lopez-Escamez JA: Identification of two novel mutations in FAM136A and DTNA genes in autosomal-dominant familial Meniere's disease. Hum Mol Genet 2015; 24: 1119–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams HJ, Hurst JR, Ocaka L et al: The use of whole-exome sequencing to disentangle complex phenotypes. Eur J Hum Genet 2016; 24: 298–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Eppsteiner RW, Booth KT et al: Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet 2014; 95: 445–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PN, Kohler S, Oellrich A et al: Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res 2014; 24: 340–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandell M, Huff C, Hu H et al: A probabilistic disease-gene finder for personal genomes. Genome Res 2011; 21: 1529–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton MV, Guthery SL, Voelkerding KV et al: Phevor combines multiple biomedical ontologies for accurate identification of disease-causing alleles in single individuals and small nuclear families. Am J Hum Genet 2014; 94: 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazal S, Gosset S, Verdura E et al: Can whole-exome sequencing data be used for linkage analysis? Eur J Hum Genet 2015; 24: 581–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arweiler-Harbeck D, Horsthemke B, Jahnke K, Hennies HC: Genetic aspects of familial Meniere's disease. Otol Neurotol 2011; 32: 695–700. [DOI] [PubMed] [Google Scholar]
- Frykholm C, Larsen HC, Dahl N, Klar J, Rask-Andersen H, Friberg U: Familial Meniere's disease in five generations. Otol Neurotol 2006; 27: 681–686. [DOI] [PubMed] [Google Scholar]
- Requena T, Gazquez I, Moreno A et al: Allelic variants in TLR10 gene may influence bilateral affectation and clinical course of Meniere's disease. Immunogenetics 2013; 65: 345–355. [DOI] [PubMed] [Google Scholar]
- Gazquez I, Moreno A, Aran I et al: MICA-STR A.4 is associated with slower hearing loss progression in patients with Meniere's disease. Otol Neurotol 2012; 33: 223–229. [DOI] [PubMed] [Google Scholar]
- Cabrera S, Sanchez E, Requena T et al: Intronic variants in the NFKB1 gene may influence hearing forecast in patients with unilateral sensorineural hearing loss in Meniere's disease. PLoS One 2014; 9: e112171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn TB, Meberg PJ, Brown MD et al: Regulating actin dynamics in neuronal growth cones by ADF/cofilin and rho family GTPases. J Neurobiol 2000; 44: 126–144. [PubMed] [Google Scholar]
- Kato A, Okamoto O, Ishikawa K et al: Dermatopontin interacts with fibronectin, promotes fibronectin fibril formation, and enhances cell adhesion. J Biol Chem 2011; 286: 14861–14869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghajanian H, Choi C, Ho VC, Gupta M, Singh MK, Epstein JA: Semaphorin 3d and semaphorin 3e direct endothelial motility through distinct molecular signaling pathways. J Biol Chem 2014; 289: 17971–17979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber A, Raz Y: Axon guidance cues in auditory development. Anat Rec A Discov Mol Cell Evol Biol 2006; 288: 390–396. [DOI] [PubMed] [Google Scholar]
- Love CA, Harlos K, Mavaddat N et al: The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D. Nat Struct Biol 2003; 10: 843–848. [DOI] [PubMed] [Google Scholar]
- Vona B, Nanda I, Neuner C, Muller T, Haaf T: Confirmation of GRHL2 as the gene for the DFNA28 locus. Am J Med Genet A 2013; 161A: 2060–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto O, Fujiwara S, Abe M, Sato Y: Dermatopontin interacts with transforming growth factor beta and enhances its biological activity. Biochem J 1999; 337 (Pt 3): 537–541. [PMC free article] [PubMed] [Google Scholar]
- Okamoto O, Fujiwara S: Dermatopontin, a novel player in the biology of the extracellular matrix. Connect Tis Res 2006; 47: 177–189. [DOI] [PubMed] [Google Scholar]
- Yoshimura H, Takumi Y, Nishio SY, Suzuki N, Iwasa Y, Usami S: Deafness gene expression patterns in the mouse cochlea found by microarray analysis. PLoS One 2014; 9: e92547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Sierra C, Requena T, Frejo L, Price SD, Gallego-Martinez A, Batuecas-Caletrio A et al: A novel missense variant in PRKCB segregates low-frequency hearing loss in an autosomal dominant family with Meniere's disease. Hum Mol Genet 2016, e-pub ahead of print 21 June 2016.. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.