Abstract
Truncating ASXL3 mutations were first identified in 2013 by Bainbridge et al. as a cause of syndromic intellectual disability in four children with similar phenotypes using whole-exome sequencing. The clinical features – postulated by Bainbridge et al. to be overlapping with Bohring–Opitz syndrome – were developmental delay, severe feeding difficulties, failure to thrive and neurological abnormalities. This condition was included in OMIM as ‘Bainbridge–Ropers syndrome' (BRPS, #615485). To date, a total of nine individuals with BRPS have been published in the literature in four reports (Bainbridge et al., Dinwiddie et al, Srivastava et al. and Hori et al.). In this report, we describe six unrelated patients with newly diagnosed heterozygous de novo loss-of-function variants in ASXL3 and concordant clinical features: severe muscular hypotonia with feeding difficulties in infancy, significant motor delay, profound speech impairment, intellectual disability and a characteristic craniofacial phenotype (long face, arched eyebrows with mild synophrys, downslanting palpebral fissures, prominent columella, small alae nasi, high, narrow palate and relatively little facial expression). The majority of key features characteristic for Bohring–Opitz syndrome were absent in our patients (eg, the typical posture of arms, intrauterine growth retardation, microcephaly, trigonocephaly, typical facial gestalt with nevus flammeus of the forehead and exophthalmos). Therefore we emphasize that BRPS syndrome, caused by ASXL3 loss-of-function variants, is a clinically distinct intellectual disability syndrome with a recognizable phenotype distinguishable from that of Bohring–Opitz syndrome.
Introduction
The availability of next-generation sequencing techniques (NGS) has greatly enabled the elucidation of genetic causes underlying clinically well-characterized syndromes as well as those accounting for isolated intellectual disability (ID, IQ<70) or newly described ID syndromes. More than 800 genes causative for ID have been identified up to now (821 confirmed ID genes and 424 candidate genes; SysID-database as of 24 March 20161).
Applying whole-exome and whole-genome sequencing techniques, Bainbridge et al.2 identified in 2013 for the first time truncating variants in the gene ASXL3 (MIM *615115) in four individuals with ID and overlapping clinical phenotypes.2 This condition was then included in OMIM as 'Bainbridge–Ropers syndrome' (BRPS, MIM #615485). Clinical features were mainly feeding difficulties, failure to thrive, global developmental delay (DD) with muscular hypotonia, ID, mild ulnar deviation of the hands and facial dysmorphism. In three out of these four patients with ASXL3 variants, Bohring–Opitz syndrome (BOPS, MIM #605039) was also initially considered as tentative clinical diagnosis. Therefore, the clinical features in patients with ASXL3 variants were postulated by Bainbridge et al.2 to be overlapping with those of patients with Bohring–Opitz syndrome. BOPS is a severe developmental disorder mainly characterized by intrauterine growth retardation, severe DD and ID, feeding difficulties (often requiring tube feeding), failure to thrive, microcephaly, a specific craniofacial phenotype with trigonocephaly, exophthalmus, nevus flammeus of the forehead, upslanting palpebral fissures and a peculiar posture of the arms (known as 'BOPS posture') with flexion of elbow and wrist, ulnar deviation of the wrist and of the metacarpophalangeal joints.3, 4, 5 In 2011, germline mutations in ASXL1 (MIM *612990) were shown to be causative for Bohring–Opitz syndrome.6 ASXL3 and ASXL1 belong to the same gene family. Together with ASXL2 (MIM *612991), they constitute the ASXL family of additional sex combs like genes, human homologs of the Asx gene in Drosophila,7 which serve as epigenetic scaffolding proteins.8, 9 Screening of a small cohort of individuals with clinically suspected BOPS did not reveal ASXL3 variants.2
Only five additional children with a pathogenic ASXL3 variant and BRPS have been published along with detailed clinical descriptions, that is, one by Dinwiddie et al.,10 three by Srivastava et al.11 and one by Hori et al.12 All nine variants were exclusively truncating (five nonsense, one splice site and three frameshift mutations).
Here, we report on six unrelated patients in whom we newly identified heterozygous de novo ASXL3 loss-of-function (LoF) variants as the most likely cause of their syndromic ID and thus diagnosed BRPS. We comprehensively describe their concordant clinical features that we consider as distinguishable from those in Bohring–Opitz syndrome.
Materials and methods
Patients
Written informed consent to the study was obtained from the legal representatives of each participant and written consents for publication of the clinical photographs were given. The investigations were performed in accordance with the Declaration of Helsinki protocols and were approved by the local institutional review boards (ethics vote 08-3663 for MRNET and 5360/13 for the Technische Universität München).
Two of the six individuals described here (patients 3, 6) were detected in a whole-exome sequencing (WES) cohort of 311 individuals with ID that could not be attributed to a clinically recognizable syndrome by experienced clinical geneticists. Inclusion criteria to this cohort were DD/ID with an IQ<70 either as non-syndromic ID (isolated, without additional features, present in 88 out of 311 individuals) or as syndromic ID with additional features (present in 223 out of 311 individuals). Additional features were, for example, craniofacial dysmorphism (present in 70% of individuals), epilepsy (present in 26%), organ malformations (eg, congenital heart defects found in 7% or brain malformations found in 14%) or aberrant body measurements (eg, micro- or macrocephaly, present in 33% manuscript in preparation). In addition, clinically relevant chromosomal aberrations had to be excluded previously by chromosomal microarray analysis, and fragile-X testing had to be normal.
By clinical collaboration we collected four further individuals with newly diagnosed ASXL3 variants identified in different WES or NGS approaches. Patient 1 was investigated as a single trio by WES. Patient 2 was detected in a clinical exome sequencing setting, in which 189 individuals were included. Patient 4 was identified in a cohort of 793 individuals with a suspected mitochondriopathy. WES was performed under the same conditions as in patients 3 and 6. Patient 5 was identified in a WES cohort of 32 patients with marfanoid features.
Exome sequencing, data analysis
The exome of patient 1 was sequenced in a trio design. Briefly, target regions were enriched using SureSelect Target Enrichment v5 (Agilent Technologies, Santa Clara, CA, USA) and sequenced on a HiSeq2500 device (Illumina, San Diego, CA, USA). Sequences were aligned to the human reference genome hg19 (GRCh37) using Burrows Wheeler Aligner (BWA, http://bio-bwa.sourceforge.net/). Variants were filtered for a coverage of ≥15 × , a novel allele count of ≥3, a MAF of <0.1% in data of the Exome Aggregation Consortium and calling by ≥2 calling algorithms. Only non-synonymous variants including splice sites were taken into account, and we filtered for three modes of inheritance: autosomal dominant (de novo), autosomal recessive and X-linked recessive. Variants were filtered for known and candidate ID genes using a published database.1
For patient 2, we used the 'Kingsmore panel'. About 1222 genes were enriched (Illumina TruSeq Custom Enrichment Kit) and sequenced (Illumina MiSeq 2 × 100 bp paired-end), and 526 known ID-associated genes13 of the 1222 genes were specifically analyzed.
For patients 3, 4 and 6, exome sequencing was performed as described in Kuechler et al.14 In brief, exomes were enriched using the SureSelect XT Human All Exon 50 Mb kit, versions 3 or 5 (Agilent Technologies), sequencing was performed on HiSeq2000/2500 systems (Illumina). Image analysis and base calling was performed using Illumina Real-Time Analysis. Reads were aligned against the human assembly hg19 (GRCh37) using BWA v 0.5.9. We performed variant calling using SAMtools (v 0.1.18; http://samtools.sourceforge.net/), PINDEL (v 0.2.4t; http://gmt.genome.wustl.edu/pindel/current/), ExomeDepth (v 1.0.0; http://www.stats.bris.ac.uk/R/web/packages/ExomeDepth/) and custom scripts. Subsequently the variant quality was determined using the SAMtools varFilter script. To discover putative de novo variants, we queried the database to show only those variants of a child that were not found in the corresponding parent.
WES in patient 5 was performed as trio analysis as part of a research project focused on patients with marfanoid habitus and ID. Methods have been described elsewhere.15
Sanger sequencing of ASXL3 was performed on whole-blood genomic DNA in the patients and their parents in order to confirm the pathogenic variants and their de novo status. Primer sequences are available upon request. ASXL3 reference sequence was NM_030632.1, ENST00000269197, and all genomic positions are denoted according to GRCh37/hg19. All six variants are publically available at the DECIPHER database (https://decipher.sanger.ac.uk; accession numbers 327923–327928).
Results
Clinical reports and identification of ASXL3 pathogenic variants
The main clinical findings of the six novel patients are summarized in the following case reports; additional detailed information is provided in Table 1.
Table 1. Detailed clinical data of the six novel patients with Bainbridge–Ropers syndrome and ASXL3 variants.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | |
|---|---|---|---|---|---|---|
| Gender | Male | Male | Female | Female | Female | Male |
| Gestation, birth | Normal | Normal | After ICSI, oligohydramnios, C-section | Oligohydramnios | Premature labor from week 22; C-section | |
| Gestational weeks | 36 | 39 | 37 | 39 | 39 | 37 |
| Birth weight (g)/ | 2850 | 3420/ | 2750 | 2950 | 3600 | 2660 |
| (SD) | 0.0 | −0.2 | −0.5 | −0.9 | 0.6 | −1.0 |
| Birth length (cm)/ | 51 | 52 | 47 | 50 | 51 | 46 |
| (SD) | 0.7 | 0.0 | −1.0 | −0.5 | −0.6 | −1.7 |
| Birth OFC (cm)/ | 33 | 35 | 32 | 34 | 37.5 | 35 |
| (SD) | −0.5 | −0.2 | −1.4 | −0.5 | 2.2 | 0.4 |
| Age (y) at last exam | 1 8/12 | 15 3/12 | 4 | 3 3/12 | 22 | 10 |
| Height (cm)/ | 82 | ~130 (a) | 96 | 91.5 | 156 | 136 |
| (SD) | −0.3 | (−3.4) | −1.9 | −1.3 | −1.5 | −0.7 |
| Weight (kg)/ | 9.1 | 27 | 12.3 | 12 | 35 | 22 |
| BMI (kg/m2) | 13.5 | 16.0 | 13.3 | 14.3 | 14.4 | 11.9 |
| (Percentiles) | <3rd | 3rd – 10th | 3rd – 10th | 10th – 25th | <3rd | <3rd |
| OFC (cm)/ | 44.5 | 51.5 | 49 | 48 | 54 | 52.7 |
| (SD) | −3.0 | −1.7 | −0.9 | −1.3 | −0.3 | −0.3 |
| Gastrointestinal | ||||||
| Feeding difficulties | Poor sucking, extreme feeding difficulties, gastric tube | Poor sucking, extreme feeding difficulties, gastric tube | Poor sucking (no breast feeding possible), failure to thrive | Poor sucking, feeding difficulties, failure to thrive | Extreme feeding difficulties | Insufficient sucking reflex, feeding difficulties |
| Other | Vomiting, hypersalivation | Vomiting, prone to infections, tachy-/apneas | Vomiting | Hypersalivation | ||
| Craniofacial | ||||||
| Long face/temporal narrowing | +/+ | +/+ | +/+ | +/+ | +/+ | +/+ |
| Prominent forehead | + | + | + | + | − | + |
| Arched eyebrows | + | + | + (mild synophrys) | (+) | + | + |
| Downslanting palpebral fissures | (+) | (+) | + | + | (+) | + |
| Strabism | − | + | + | + | + | + |
| High, narrow palate | ? | + Submucous cleft palate, bifid uvula | + | + | + | + |
| Prominent columella and small alae nasi | + | + | + | + | + | + |
| Anteverted nares | + | − | + | + | + | + |
| Downturned corners of the mouth | + | + | + | + | + | + |
| Little facial expression/open mouth appearance | (+)/++ | +/+ | +/+ | +/+ | +/+ | +/+ |
| Microcephaly | + | − | − | − | − | − |
| Muscular | ||||||
| Hypotonia or hypertonia | Pronounced hypotonia of trunk>extremities | Hypotonia of the trunk, hypertonia of the extremities | Hypotonia | Hypotonia of the trunk, hypertonia of the extremities | Hypotonia | Hypotonia |
| Psychomotor development | ||||||
| Motor delay | + | + | + | + | + | + |
| Free walking | Not yet | No | Not yet | Not yet | At 6 years | At 4 years |
| ID | +? (too young) | + | + | + | + Severe | + |
| Speech impairment | No words yet | Severe, no words | Severe, no words yet | Severe, no words yet | Few words | Severe, few single words, uses talker/ gestures |
| Basic speech comprehension | +? (too young) | Poor | Little | Poor | Little | Good |
| Other | Short eye contact and short moment of smiling | Poor eye contact | Poor eye contact; end-position nystagmus | Self-injuries Behavior troubles Obsessive compulsive disorder | Stereotypic movements and poor eye contact in infancy | |
| EEG | n.p. No seizures | Pathologic No seizures | Normal No seizures | Normal No seizures | Pathologic Febrile seizures in childhood, seizures at 18 years | Disposition to seizures at 7 years; first seizures (complex focal) at 8 years |
| Cranial MRI | Normal | Hypoplasia of cerebellar vermis | Normal | Mild hypoplasia of corpus callosum, slightly dilated lateral ventricles | Thin corpus callosum, Mild white matter volume loss | Mildly enlarged ventricles |
| Skeletal | ||||||
| Abnormalities of the hands | Bent fingers and thumbs, stiff finger joints | Frequently clenched fists, ulnar deviation | Long, slender fingers, mild ulnar deviation | Ulnar deviation, frequently clenched hands in infancy, long, slender fingers | Ulnar deviation Arachnodactyly | Frequently clenched fists in infancy, broad terminal phalanges, broad thumbs, slender fingers |
| Craniosynostosis/ trigonocephaly | – | – | – | – | – | – |
| Other | Mild pectus carinatum, small sacral dimple and Mongolian spot | – | Mild pectus excavatum | – | Kyphoscoliosis Pectus carinatum Limited extension of elbows, flat feet | – |
| Skin | ||||||
| Hirsutism | – | + | Mild synophrys | – | – | 'hairy elbows' |
| Pathogenic ASXL3 variant (b) | chr18:g.31,318,587delA c.1219delA. p.(Ser407AlafsTer2) | chr18:g.31,318,737G>T c.1369G>T p.(Glu457Ter) | chr18:g.31,322,918C>T c.3106C>T p.(Arg1036Ter) | chr18:g.31,323,306_31,323,307delGT c.3494_3495delGT p.(Cys1165Ter) | chr18:g.31,323,425G>Tc. 3613G>T p.(Glu1205Ter) | chr18:g.31,323,884_31323885delGT c.4072_4073delGT p.(Val1358LeufsTer8) |
Abbreviations: BMI, body mass index; C-section, caesarian section; ID, intellectual disability; MRI, magnetic resonance imaging; n.p., not performed; OFC, occipito-frontal circumference.
Mutations newly described in this study (patients 3–6) are located in the last exon 12.
Difficult to measure due to spasticity, last measurements at age 12 10/12 years.
ASXL3 reference sequence was NM_030632.1, ENST00000269197.
Patient 1 was the first child of a mother of Philippine and a father of German origin. The pregnancy was uneventful; the mother reported fetal movements. Birth was spontaneously in the 37th week of gestation. Due to an inability to suck, breast feeding was impossible. Controls of weight gain were performed starting at the third day of life. Physiotherapy was started for muscular hypotonia. The boy had severe feeding difficulties, vomited regularly and showed hypersalivation. Retrospectively, the parents noticed that he never complained of hunger nor did he cry within the first months of life. At age 14 months, he obtained a gastric tube. At age 2½ years, he was fed 6 × per day by gastric tube, had been started on small amounts of food by spoon and was able to drink up to 150 ml of milk. Cranial MRI scan, extensive metabolic testing, echocardiography, analyses of creatine kinase and ENGs were normal. An EMG at an upper and at a lower extremity was normal without movements; under movement, it revealed partly shortened units at both extremities in terms of myopathic patterns. A mitochondrial disorder was suspected, as the activity of complex I was slightly decreased in a muscle biopsy (11.8 U/g NCL (non-collagenous proteins), reference range: 15.8–42.8 U/g NCL). Further enzymatic tests of mitochondrial function and molecular genetic analyses of mitochondrial sequence were normal as was (molecular) karyotyping and preceding diagnostic tests for Prader–Willi syndrome, multi-mini-core disease (SEPN1 gene) and myotonic dystrophy type I. When he was presented at age 18/12 years, he was unable to sit or crawl, but changed toys between his hands. He was tired due to an infection of the upper airway, but had short moments of eye contact and smiling. His face was long with temporal narrowing, arched eyebrows, an open mouth with hypersalivation and large ears. His fingers and thumbs were in bent position, and the phalangeal joints were stiff. He had cryptorchidism.
A heterozygous de novo frameshift variant in exon 11 of ASXL3 [chr18:g.31,318,587delA, c.1219delA., p.(Ser407AlafsTer2)] was identified by WES trio analysis.
Patient 2 (Figures 1a and f) is a 15-year-old Turkish boy, born to healthy non-consanguineous parents. He had no pre- or perinatal abnormalities (Table 1). He had severe feeding problems requiring a gastric tube. He developed a postnatal short stature, severe ID, hypertrichosis, hypotonia of the trunk and alternating periods of apnea and hyperventilation. He did not speak and was unable to walk. He had frequently clenched fists and ulnar deviation of hands but defining features for Bohring–Opitz syndrome like trigonocephaly, nevus flammeus of the forehead, seizures and intrauterine growth restriction were not present. His facial features were characterized by a long face with arched eyebrows, synophrys, prominent columella and downturned corners of the mouth. He had a high narrow palate with bifid uvula and a submucous cleft and crowded teeth. Laboratory work-up for Smith-Lemli-Opitz, ATRX and CDG syndromes and metabolic disorders gave normal results.
Figure 1.
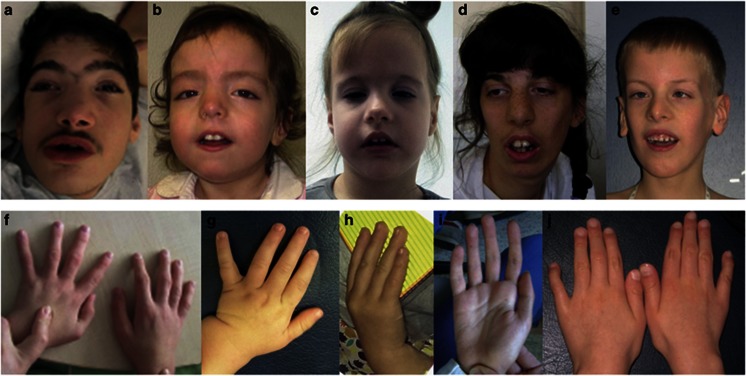
Facial phenotypes and hands of five individuals with ASXL3 mutation. All six individuals show overlapping craniofacial features characterized by a longish face with a prominent forehead and temporal narrowing, arched eyebrows, downslanting palpebral fissures, a prominent columella and small alae nasi, downturned corners of the mouth and little facial expression with open mouth appearance. A slender habitus with long and slender fingers was a frequent sign. Photographs of the face and the hands are shown for five of the six individuals: patient 2 at age 15 years (a and f), patient 3 at age 2½ years (b and g), patient 4 at age 3¼ years (c and h), patient 5 at age 20 years (d and i) and patient 6 at age 7½ years (e and j).
Finally, a de novo heterozygous nonsense variant in exon 11 (chr18:g.31,318,737G>T, c.1369G>T; p.(Glu457Ter) of the ASXL3 gene was detected.
Patient 3 (Figures 1b and g) is the first child born to non-consanguineous parents of Turkish and Italian background. The pregnancy was conceived after ICSI and complicated by suspected intrauterine growth retardation and oligohydramnios. The girl was born with normal measurements by secondary Cesarean section after induction of labor (Table 1). After birth, inflected index fingers, abnormal reflexes and a high palate were remarkable. The girl could not be breast fed but did not need tube feeding. She showed a muscular hypotonia and a failure to thrive. Divergent strabism of the left eye was treated with occlusion therapy. Cranial MRI and myosonography to investigate causes for her muscular hypotonia showed normal results. At age 13 months, the patient could roll from a ventral to a dorsal position. She could not sit or crawl and had poor head control, but demonstrated canonical babbling. Physical examination at age 7 months showed dystrophy with all measurements below the third percentile. At re-evaluations at ages 13 months, 27 months and 4 years, measurements normalized approaching the low normal range (Table 1). Her stature was thin, her fingers long and slender. She had a prominent forehead, arched eyebrows, hypertelorism, downslanting palpebral fissures, prominent columella, small alae nasi, small lips and downturned corners of the mouth. Her palate was high and narrow, and she had crowded teeth. She had no active speech and an impaired speech comprehension.
WES revealed a de novo truncating variant in exon 12 of the ASXL3 gene [chr18:g.31,322,918C>T, c.3106C>T, p.(Arg1036Ter)].
Patient 4 (Figures 1c and h) is the only child of a healthy, non-consanguineous Russian couple. Two paternal half-siblings are healthy. The pregnancy was complicated by oligohydramnios and suspected intrauterine growth retardation, but birth measurements were normal (Table 1). The girl showed abnormally clenched hands with ulnar deviation and a high, narrow palate. Due to feeding difficulties, breast feeding was impossible, but a gastric tube was not required. Physical examination at age 3 4/12 years showed low normal body measurements (Table 1). The girl was not able to walk yet, had no expressive speech and little speech comprehension. Her face was long with a prominent forehead, temporal narrowing, downslanting palpebral fissures, a prominent columella and small alae nasi and downturned corners of the mouth. She had long and slender hands/fingers, still with some intermittent ulnar deviation, muscular hypotonia of the trunk and rather hypertonic extremities.
A de novo truncating variant in exon 12 of the ASXL3 gene was identified by WES [chr18:g.31,323,306_31,323,307delGT, c.3494_3495delGT, p.(Cys1165Ter)].
Patient 5 (Figures 1d and i) is a 20-year-old lady identified in a series of patients recruited because of marfanoid habitus and ID. She had suffered from extreme feeding difficulties and vomiting in infancy. Her measurements at birth and later remained basically normal (Table 1). Her development was significantly delayed; she started unsupported walking at age 6 years, had severe ID and was able to speak a few single words. Her EEG was pathologic. She had febrile seizures in childhood and suffers from seizures since the age of 18 years (treated by valproate). She had behavioral abnormalities with sleeping disturbance, self-injuries and obsessive compulsive disorder. Puberty was normal. Although she was not tall, she displayed some marfanoid features – arachnodactyly, kyphoscoliosis, pectus carinatum, limited extension of elbows, ulnar deviation of hands and flat feet. Facial features presented by the patient were downslanting palpebral fissures, a prominent nasal bridge and columella with hypoplastic alae nasi, downturned corners of the mouth, crowded teeth and micrognathia.
WES disclosed a de novo truncating variant in exon 12 of the ASXL3 gene [chr18:g.31,323,425G>T, c.3613G>T, p.(Glu1205Ter).
Patient 6 (Figures 1e and j) is the second son of non-consanguineous German parents, born from a twin pregnancy with a healthy sister. The pregnancy was complicated by severe nausea and premature labor starting at week 22. At birth by Cesarean section, the patient's measurements were normal (Table 1) but a bit lower than those of his twin sister. In contrast to his sister, he had poor sucking and feeding difficulties, an uneasy sleep, poor eye contact and frequently laid in an asymmetrical (fencing) posture with closed fists. His psychomotor development was significantly delayed (head control at 11/2 years, crawling at 2 years, walking without support at 4 years). At age 3 years, he started to speak a few single words but mainly relied on an electronic talker and gestures to communicate. He had a good speech comprehension. At age 7 years, a disposition to seizures was suspected in the EEG, 1 year later first seizures (complex focal) occurred, which were initially difficult to control. Upon physical examinations at 7½ and 10 years of age (Table 1), he showed measurements appropriate to his age. He had a slender build, 'hairy elbows', his hands showed short distal phalanges, fifth finger clinodactyly, mildly broad thumbs, big toes were also mildly broad. His face was long with temporal narrowing, open mouth appearance, little facial expression, downslanting palpebral fissures, strabismus, prominent columella with small alae nasi and a high narrow palate.
WES revealed a de novo frameshift variant in exon 12 of ASXL3 [chr18:g.31,323,884_31323885delGT, c.4072_4073delGT, p.(Val1358LeufsTer8)].
Discussion
The human ASXL genes (ASXL1 at 20q11.21, ASXL2 at 2p23.3 and ASXL3 at 18q12.1) are orthologs of the Drosophila additional sex combs (Asx) gene that encodes a regulator of the Polycomb-group repressor complex (PRC).7 Somatic mutations in all three human genes are associated with various types of malignancies.9 Furthermore, germline LoF variants in ASXL1 are causative of the Bohring–Opitz syndrome (BOPS, MIM #605039), and germline LoF variants in ASXL3 cause the Bainbridge–Ropers syndrome (BRPS, MIM #615485), whereas ASXL2 has not yet been associated with a disorder of development.9
Until recently, only nine different de novo truncating ASXL3 germline variants have been identified in individuals with the clinical diagnosis of BRPS.2, 10, 11, 12 Thus, available information about BRPS is still limited. In addition, three different LoF variants have been identified in a large autism cohort.16 Unfortunately, these authors did not provide any additional clinical data on their patients.
In this study, we add six additional individuals (three females, three males) with overlapping clinical phenotypes caused by de novo ASXL3 LoF variants.
Clinical characterization of patients with ASXL3 LoF variants
Comparing the clinical features of all 15 individuals with BRPS (six males, eight females; for one (subject 1 in Bainbridge et al.2) information of the sex is not available) known so far (Table 2), the most significant finding was a profound DD with ID present in all individuals. About 13/15 individuals had a muscular hypotonia with frequent drooling and 14/15 had serious feeding difficulties in infancy. However, only two of our six patients required tube feeding, in contrast to seven out of nine published individuals. In all 15 individuals, motor milestones were achieved with significant delay; one of our patients never gained the ability to walk, two started walking at ages 4 and 6 years, respectively, and three have not started walking until now (aged 2, 3 and 4 years, respectively). The patient by Dinwiddie et al. started walking at age 3 years,10 one of the four patients by Bainbridge et al.2 was not yet able to sit at age 3.5 years, one died in the first year of life, and for the two other individuals no specific data were mentioned. Patient 1 by Srivastava et al.11 patient 1 was not able to sit, crawl or stand at age 6 years, patient 2 was not able to walk at age 4 years and patient 3 was unable to roll over or sit upright at age 13 months.11 The patient published by Hori et al.12 was able to sit without support at age 2 years and to stand holding on to things at age 3 years. Speech development was also impaired in all 12 patients in whom this information was available. Twelve out of twelve patients had an absent speech or spoke only single words (Table 1).
Table 2. Summary of the main clinical features of all known patients (15) with Bainbridge–Ropers syndrome and ASXL3 LoF variant identified so far.
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5 | Patient 6 | Indiv. 1–4 Bainbridge etal2 | Dinwiddie et al10 | Indiv. 1–3 Srivastava et al11 | Hori et al12 | Total n=15 (a) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Characteristic facial dysmorphism (b) | + | + | + | + | + | + | + (4/4) | n.a. | + (2/3) | n.a. | 12/13 |
| Intellectual disability | (too young) | ++ | ++ | ++ | ++ | ++ | ++ (4/4) | ++ | + (2/2) | + | 14/14 |
| Feeding difficulties | + | + | + | + | + | + | + (3/4) | + | + (3/3) | + | 14/15 |
| Muscular hypotonia | + | + | ++ | + | + | ++ | + (3/4) | + | + (3/3) | − | 13/15 |
| Motor delay | + | + + | ++ | ++ | + + | ++ | + (4/4) | ++ | + (3/3) | + | 15/15 |
| Speech impairment | + | ++ | ++ | ++ | ++ | ++ | ?? (1/1) | ++ | + (3/3) | + | 12/12 |
| Seizures | − | − | − | − | + | + | −(0/4) | − | + (1/3) | − | 3/15 |
| Birth measurements | Normal | Normal | Normal | Normal | Normal | Normal | SGA (2/4) | Normal | SGA (1/3) | SGA | Normal in 11/15, SGA in 4/15 |
| Postnatal growth | Normal | Short | Normal | Normal | Normal | Normal | Short (2/4) | Normal | Normal (1/1) | Short | Normal in 9/13, short in 4/13 |
| Postnatal OFC | ↓ | Normal | Normal | Normal | Normal | Normal | ↓ (2/3) | ↓ | Normal (3/3) | ↓ | Normal in 9/14, ↓ in 5/14 |
Abbreviations: n.a., not applicable; OFC, occipito-frontal circumference; SGA, small for gestational age.
Not every feature was documented for all patients; therefore the total number of patients can differ.
Characteristic craniofacial features comprise a longish face with prominent forehead, temporal narrowing, arched eyebrows, (synophrys), downslanting palpebral fissures, prominent columella and small alae nasi, downturned corners of the mouth and little facial expression/open mouth appearance.
Another consistent finding was the craniofacial phenotype (Figure 1), characterized by a long face with a prominent forehead and temporal narrowing, arched eyebrows, downslanting palpebral fissures, a prominent columella and small alae nasi, downturned corners of the mouth and little facial expression with open mouth appearance.
Due to phenotypic overlap, patient 4 was initially given the tentative diagnosis of a mitochondriopathy and enrolled in a respective exome sequencing project. Interestingly, in most of our six patients a mitochondriopathy was clinically considered at some stage. Patient 5 was identified in a cohort of individuals with marfanoid features. Evaluation of our patients with regard to this aspect revealed a slender habitus with long and slender fingers as a frequent sign (Figure 2). Moreover, in most individuals, an ulnar deviation of the hands was observed at younger age which was, however, mild and not comparable to the typical BOPS posture.3, 4, 5
Figure 2.
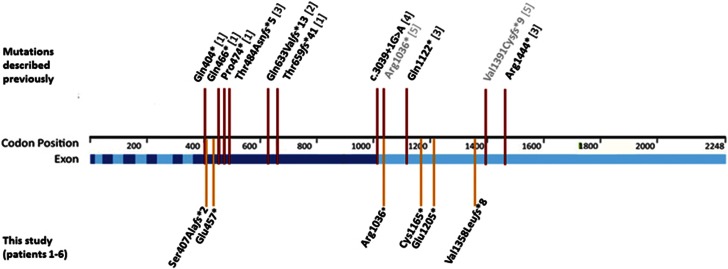
Localization of ASXL3 variants of all known patients with Bainbridge–Ropers syndrome15 (adapted from Bainbridge et al.2). The nine previously published variants are marked with red bars and references are given in square brackets ([1] subjects 1–4 by Bainbridge et al, [2] the patient by Dinwiddie et al, [3] subjects 1–3 by Srivastava et al. and [4] the patient by Hori et al). Two ASXL3 variants detected in individuals with autism are depicted in gray ([5] De Rubeis et al). The six variants identified in this study (patients 1–6) are depicted by yellow bars. Positions of the 12 exons are marked in light blue/blue. Notably, six of the nine previously published variants as well as two of the newly identified variants (patients 1 and 2) are located in exon 11; 2 mutations described by Srivastava et al,11 and 4 of the 6 mutations newly described in this study (patients 3-6) are located in the last exon 12.
Bainbridge et al.2 as well as Dinwiddie et al.2, 10 postulated that their patients carrying an ASXL3 mutation resembled those with BOPS (also reviewed by Russell and Graham),17 as they showed clinical overlap with severe feeding difficulties (4/5), neurological abnormalities with significant DD (5/5), small birth measurements (2/4) and microcephaly (3/4). The same findings were observed in the girl reported by Hori et al.,12 but especially because of the typical posture being absent, her phenotype was evaluated as distinct from those in BOPS. One patient10 had a metopic craniosynostosis. For patient 3 by Srivastava et al.,11 a trigonocephaly was mentioned. Thus, there are only two of 15 patients with BRPS that share the feature of trigonocephaly reported as characteristic for BOPS.
As the majority of key features characteristic for BOPS were absent in our patients (eg, typical BOPS posture of the arms (ie, flexion of elbow and wrist, ulnar deviation of wrist and metacarpophalangeal joints), intrauterine growth retardation, microcephaly (absent in 5/6), trigonocephaly, typical facial gestalt with nevus flammeus and prominent eyes/proptosis), we did not find much clinical overlap between these two entities. In fact, in none of our six patients the tentative diagnosis of Bohring–Opitz syndrome was clinically considered at any time.
In two patients,10, 12 autism was a reported finding. Some features overlapping with the autistic spectrum such as poor eye contact or stereotypies were also observed in our patients, but no formal diagnosis of autism was made. This assessment might be influenced by their severe developmental impairment. It remains unclear whether there is more clinical overlap (apart from autism) with the three autistic individuals published by De Rubeis et al.16 carrying ASXL3 LoF variants.
Mutational spectrum
ASXL3 shares a common domain architecture with its family members ASXL1 and ASXL2, and although sequence variants in both, the ASXL3 and ASXL1 genes cause disorders with DD and ID, ASXL3 gene and protein exhibit unique features. In contrast to ASXL1 (1541 amino acids, aa) and ASXL2 (1435 aa), the ASXL3 protein is 2248 aa long. The difference in coding potential is almost exclusively due to the different lengths of the protein-coding parts of the two last exons of the genes, that is, exons 11 (1957 bp) and 12 (3708 bp) in ASXL3 versus exons 12 (634 and 718 bp) and 13 (2907 and 2448 bp) of ASXL1 and ASXL2, respectively. The reference sequences used for this comparison are for ASXL1 ENST00000375687, ASXL2 ENST00000435504 and ASXTL3 ENST00000269197 (http://www.ensembl.org/index.html).
Despite its largest open reading frame (6747 bp) within this gene family, the occurrence of apparently benign LoF variants in ASXL3 in apparently healthy controls is strikingly lower than in ASXL1 (4626 bp) and ASXL2 (4308 bp). As of April 2016, the database of the Exome Aggregation Consortium (ExAC, http://exac.broadinstitute.org/, assessed 28 April 2016) lists only three LoF variants (Arg322Ter, Lys926SerfsTer9 and Glu1933Ter) and one potential splice variant (c.715+1G>A) for ASXL3 (each variant with a single occurrence), but 28 LoF and four potential splice variants for ASXL1 and four LoF and two potential splice variants for ASXL2. This difference is also reflected in the database of the 1000 Genomes Project (The 1000 Genomes Project Consortium, 2015, http://www.1000genomes.org/home). As of 25 August 2016, this database lists only 34 stop gain, 9 frameshift, 1 stop lost and no splice variants for ASXL3, but 101 stop gain, 159 frameshift and 16 splice region variants for ASXL1, and 38 stop gain, 14 frameshift, 23 splice region and 2 start loss variants for ASXL2. Almost all LoF described in these genes listed in the 1000 Genomes Project are somatic and not germline mutations.
This low frequency of apparently benign LoF variants in ASXL3 in apparently healthy individuals is an indication of a LoF intolerance making haploinsufficiency with high penetrance the most likely mechanism of inheritance for BRPS, in contrast to a proposed much lower penetrance of ASXL1 LoF variants in BOPS.18, 19 This is in line with a haploinsufficiency score of ASXL3 of 13,78, that is, a very high probability for haploinsufficiency compared with a haploinsufficiency score of ASXL1 of 20,76, that is, a lower probability for haploinsufficiency than ASXL3 (https://decipher.sanger.ac.uk).20
Together with the six cases presented in this study, 15 different truncating variants in 15 unrelated individuals with clinically approved BRPS are known by now. De Rubeis et al.16 published three LoF variants, two truncating mutations and one whole-gene deletion in individuals with autism. One of these sequence variants (c.3106C>T; p.(Arg1036Ter)) is identical to the one of our patient 3. This means that up to now in total 17 different LoF variants of the ASXL3 gene have been identified as putatively disease causing. The 16 different point mutations are scattered over the two largest exons that represent 84% of the entire ASXL3 protein-coding region (Figure 2). Thus, there is no evidence for a mutational hotspot in ASXL3.
A high number of different missense variants, spread almost evenly over the entire coding sequences of the respective genes, has been found in the germline of healthy individuals in all three genes (eg, ExAC database: ASXL1: 522, ASXL2: 411 and ASXL3: 671). Thus, variations of the amino acids sequence of these proteins seem to exert much lesser pathogenic effects than a reduction of the total protein amount as expected for heterozygous truncating mutations does.
Frequency of pathogenic ASXL3 variants
With only nine published individuals diagnosed with BRPS until recently, this seemed to represent a rare cause of syndromic ID. In our primary cohort of 311 individuals with ID, we identified two patients (0.6%) carrying an ASXL3 LOF variant suggesting that this might be a more frequent cause of ID than previously assumed. By clinical collaboration we were able to collect four additional patients. This again emphasizes the importance of clinical collaboration to characterize new entities.21 The frequencies of ASXL3 mutations in different cohorts varies (about 0.5% in the cohort of patient 2, about 0.6% in the cohort of patients 3 and 6; 0.13% in the cohort of patient 4 and about 3% in the cohort of patient 5), and seems to depend on the inclusion criteria.
Conclusion
By adding the largest cohort so far, we have substantially increased the number of patients with BRPS due to LoF variants in ASXL3, and further delineated the clinical phenotype. We regard BRPS as distinct entity with a recognizable clinical phenotype, which is distinguishable from BOPS. BRPS is characterized by distinct craniofacial features, muscular hypotonia, feeding difficulties in infancy, significant motor delay, profound speech impairment and ID. Analysis of further cohorts of patients with precisely characterized clinical features are necessary to further systematically delineate the clinical phenotype, the underlying mutational spectrum and the frequency of ASXL3 variants among individuals with syndromic ID or related entities.
Acknowledgments
We are grateful to the patients and their families for participating in this study and for giving consent to publish data and photographs. We thank Sabine Kaya and Daniela Falkenstein for excellent technical assistance. Parts of this study were supported by a grant from the Interdisciplinary Center for Clinical Research (laboratory rotation) of the Clinical Center Erlangen of the Friedrich-Alexander-Universität Erlangen-Nürnberg (to UH), the German Ministry of Research and Education (grant numbers 01GS08164, 01GS08167, 01GS08163, German Mental Retardation Network) as part of the National Genome Research Network (to HE), the German Network for Mitochondrial Disorders (mitoNET; 01GM1113 to HP), the E-Rare project GENOMIT (01GM1207 to HP) and the Juniorverbund in der Systemmedizin 'mitOmics' (FKZ 01ZX1405C to TBH). We thank the PARI 2012 Marfanoid habitus and intellectual deficiency (Regional Council of Burgundy and Dijon University Hospital, France) for its financial support.
Footnotes
The authors declare no conflict of interest.
References
- Kochinke K, Zweier C, Nijhof B et al: Systematic phenomics analysis deconvolutes genes mutated in intellectual disability into biologically coherent modules. Am J Hum Genet 2016; 98: 149–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Hu H, Muzny DM et al: De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med 2013; 5: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberklaid F, Danks DM: The Opitz trigonocephaly syndrome. A case report. Am J Dis Child 1975; 129: 1348–1349. [DOI] [PubMed] [Google Scholar]
- Bohring A, Silengo M, Lerone M et al: Severe end of Opitz trigonocephaly (C) syndrome or new syndrome? Am J Med Genet 1999; 85: 438–446. [DOI] [PubMed] [Google Scholar]
- Hastings R, Cobben JM, Gillessen-Kaesbach G et al: Bohring-Opitz (Oberklaid-Danks) syndrome: clinical study, review of the literature, and discussion of possible pathogenesis. Eur J Hum Genet 2011; 19: 513–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoischen A, van Bon BWM, Rodríguez-Santiago B et al: De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat Genet 2011; 43: 729–731. [DOI] [PubMed] [Google Scholar]
- Katoh M, Katoh M: Identification and characterization of ASXL3 gene in silico. Int J Oncol 2004; 24: 1617–1622. [PubMed] [Google Scholar]
- Katoh M: Functional and cancer genomics of ASXL family members. Br J Cancer 2013; 109: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M: Functional proteomics of the epigenetic regulators ASXL1, ASXL2 and ASXL3: a convergence of proteomics and epigenetics for translational medicine. Expert Rev Proteomics 2015; 12: 317–328. [DOI] [PubMed] [Google Scholar]
- Dinwiddie DL, Soden SE, Saunders CJ et al: De novo frameshift mutation in ASXL3 in a patient with global developmental delay, microcephaly, and craniofacial anomalies. BMC Med Genomics 2013; 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava A, Ritesh KC, Tsan YC et al: De novo dominant ASXL3 mutations alter H2A deubiquitination and transcription in Bainbridge-Ropers syndrome. Hum Mol Genet 2016; 25: 597–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori I, Miya F, Ohashi K et al: Novel splicing mutation in the ASXL3 gene causing Bainbridge-Ropers syndrome. Am J Med Genet 2016; 170: 1863–1867. [DOI] [PubMed] [Google Scholar]
- Gillissen C, Hehir-Kwa JY, Thung DT et al: Genome sequencing identifies major causes of severe intellectual disability. Nature 2014; 511: 344–347. [DOI] [PubMed] [Google Scholar]
- Kuechler A, Zink AM, Wieland T et al: Loss-of-function variants of SETD5 cause intellectual disability and the core phenotype of microdeletion 3p25.3 syndrome. Eur J Hum Genet 2015; 23: 753–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenon J, Duffourd Y, Masurel-Paulet A et al: Diagnostic odyssey in severe neurodevelopmental disorders: towards clinical whole-exome sequencing as a first-line diagnostic test. Clin Genet 2016; 89: 700–707. [DOI] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP et al: Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014; 515: 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell B, Graham JMJr: Expanding our knowledge of conditions associated with the ASXL gene family. Genome Med 2013; 5: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ropers HH, Wienker T: Penetrance of pathogenic mutations in haploinsufficient genes for intellectual disability and related disorders. Eur J Med Genet 2015; 58: 715–718. [DOI] [PubMed] [Google Scholar]
- Arunachal G, Danda S, Omprakash S, Kumar S: A novel de-novo frameshift mutation of the ASXL1 gene in a classic case of Bohring-Opitz syndrome. Clin Dysmorphol 2016; 25: 101–105. [DOI] [PubMed] [Google Scholar]
- Huang N, Lee I, Marcotte EM, Hurles ME: Characterising and predicting haploinsufficiency in the human genome. PLoS Genet 2010; 6: e1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden EC: Data barriers limit genetic diagnosis. Nature 2013; 494: 156–157. [DOI] [PubMed] [Google Scholar]