Abstract
Introduction:
Extended-spectrum beta-lactamases (ESBLs) are the major cause of resistance to beta-lactam antibiotics such as penicillins, cephalosporins, and monobactams. They are derived from the narrow-spectrum beta-lactamases (TEM-1, TEM-2, or SHV-1) by mutations that alter the amino acid configuration around the enzyme active site.
Aim:
To determine the prevalence of ESBL (blaTEM, blaCTX-M, and blaSHV) genes among the members of Enterobacteriaceae.
Methodology:
The present prospective study was carried out from January 2015 to June 2015 in the Department of Microbiology and Molecular Medicine of a Teaching Tertiary Care Hospital. A total of 526 urine samples were studied. Seventy-eight isolates were subjected to polymerase chain reaction for detection of ESBL genes.
Results:
In our study, ESBL genes were detected among 18 (45%) phenotypically confirmed ESBL producers and 20 (52.5%) phenotypically confirmed non-ESBL producers. The gene that predominated was blaTEM (48.7%), followed by blaCTX-M (7.6%) and blaSHV (5.1%).
Conclusion:
Definitive identification of ESBL genes is only possible by molecular detection methods. Phenotypic tests need to be evaluated periodically as their performance may change with the introduction of new enzymes.
Key words: Beta-lactam antibiotics, extended-spectrum beta-lactamases, polymerase chain reaction
INTRODUCTION
Beta-lactamases are enzymes that are major cause of bacterial resistance to the beta-lactam family of antibiotics such as penicillins, cephalosporins, cephamycins, and carbapenems. These enzymes catalyze the hydrolysis of the amide bond of four-membered beta-lactam ring and render the antibiotic inactive against its original cellular target, the cell wall transpeptidase. On the basis of their primary structure, beta-lactamases are grouped into four classes A, B, C, and D enzymes. Enzymes of classes A, C, and D have serine at the active site, whereas the class B enzymes are zinc-metalloenzymes.
Extended-spectrum beta-lactam antibiotics have widely been used for treatment of serious Gram-negative infections. However, bacterial resistance has emerged due to production of extended-spectrum beta-lactamases (ESBLs). These enzymes are capable of hydrolyzing extended-spectrum beta-lactam antibiotics such penicillins, cephalosporins along with a monobactam (aztreonam) but are inhibited by suicide inhibitors such as clavulanic acid, sulbactam, and tazobactam. ESBLs are derived from genes for the narrow-spectrum beta-lactamases (TEM-1, TEM-2, or SHV-1) by mutations that alter the amino acid configuration around the enzyme active site. They are typically encoded by plasmids that can be exchanged readily between bacterial species. These enzymes are most commonly produced by the members of the Enterobacteriaceae, especially Escherichia coli and Klebsiella. To date, more than 350 different natural ESBL variants are known that have been classified into nine distinct structural and evolutionary families based upon their amino acid sequence comparisons such as TEM, SHV, CTX-M, PER, VEB, GES, BES, TLA, and OXA.[1,2,3,4] Since no data have been documented regarding the prevalence of genes responsible for beta-lactam resistance in our region, the present study was initiated to determine the prevalence of ESBL genes in our geographical region.
METHODOLOGY
The present prospective study was carried out in the Department of Microbiology and Molecular Medicine of a Teaching Tertiary Hospital located in Central India. It was approved by the Institutional Ethical Committee.
A total of 526 urine samples collected from patients with suspected urinary tract infection during the 6-month study period (January 2015–June 2015) were considered for the study. A total of 160 samples were found to be culture positive. Out of the 80 members of the Enterobacteriaceae isolated from the culture positive urine samples, a total of 78 E. coli and Klebsiella isolates were subjected to phenotypic and genotypic studies.
Identification of bacterial isolates was carried out using conventional biochemical methods,[5,6] an automated system (Vitek-2 compact, BioMerieux, France) and by 16s rRNA sequencing (Yaazh Xenomics, Madurai, India) under special conditions. Following identification, the isolates were stored at 4°C on nutrient agar. All the E. coli and Klebsiella isolates were phenotypically tested for ESBL production by double disk synergy test.[7] Phenotypic detection of ESBL was included in the routine susceptibility test.[8] While performing antibiotic sensitivity testing, ceftazidime plus clavulanic acid (30/10 mcg) and cefotaxime plus clavulanic acid (30/10 mcg) discs were also included along with ceftazidime (30 mcg) and cefotaxime (30 mcg) discs on Muller-Hinton agar. An organism was considered as ESBL producer if there was a ≥5 mm increase in the zone diameter of ceftazidime/clavulanic acid disc and that of ceftazidime disc alone and/or ≥5 mm increase in the zone diameter of cefotaxime/clavulanic acid disc and that of cefotaxime disc alone. E. coli 25922 and a known in-house ESBL producer were used as negative and positive controls, respectively. The test was done in accordance with the CLSI 2013 and 2014 guidelines.[9] The phenotypically confirmed ESBL and non-ESBL isolates were tested genotypically by performing polymerase chain reaction (PCR), using primers specific for the detection of blaSHV, blaTEM, and blaCTX-M genes.
DNA isolation was done; briefly, 1 ml of 24-h old bacterial broth culture was transferred into 1.5 ml sterile Eppendorf microcentrifuge tubes and centrifuged at 5000 rpm for 5 min at 4°C. The pallets were dissolved in 300 μl of tris-ethylenediaminetetraacetic acid (EDTA) buffer (tris-HCL 1.0 M, pH 8.0; 3 μl of 0.5 M EDTA, pH 8.0; and 40 μl of 10% sodium dodecyl sulfate) and incubated at 65°C for 5 min. Following incubation, 750 μl of isopropanol was added, mixed, and centrifuged at 14,000 rpm for 10 min at 15°C.
The resulting pallets were resuspended in 500 μl of TE buffer and 2 μl of mRNAase A. This was incubated at 65°C for 30 min, followed by addition of 2 μl of prokinase K. It was further incubated at 37°C for 15 min. This was followed by addition of phenol-chloroform (1:1). The upper phase was transferred to another clean tube and equal volume of chloroform was added. After shaking, the tube was centrifuged at 14,000 rpm for 5 min at 15°C. The supernatant was then treated with 40 μl of 5 M sodium acetate (pH 5.2) and 1 ml of ethanol. It was left at room temperature for 1 h after centrifugation at 7000 rpm for 5 min at 4°C. The DNA pallet was washed with 70% ethanol and suspended in 50 μl of TE buffer. DNA purity was confirmed using a spectrophotometer (260/280).
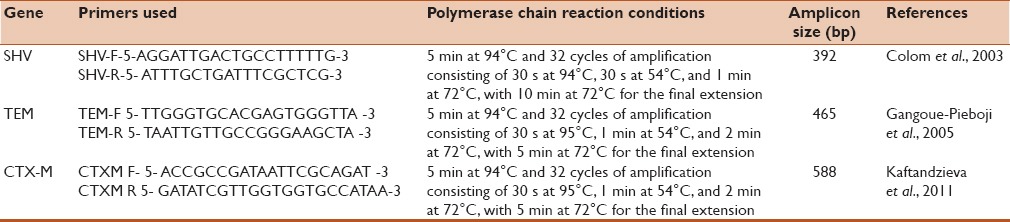
PCR was carried out in 50 μl PCR volumes containing 20 ng of template DNA, 0.5 mM of dNTPs, 1.25 μM of each primer (for TEM, SHV, and CTX-M gene detection), and 3 μl of Taq polymerase (Bangalore Genei, Bangalore, India) in 1× PCR buffer. Amplification of DNA was performed in master cycler, a personal thermocycler (Eppendorf, Germany)[1,3,4,10,11,12] with cycling parameters and primers used as described in Table 1.[13,14,15]
Table 1.
Cycling parameters and Primers used in a master cycler
PCR products were analyzed in 1% agarose gel containing 25 μg of ethidium bromide in tris-EDTA buffer, and the gel was photographed under ultraviolet illuminator using gel documentation system (Bio-Rad, USA). Further, 100 bp DNA ladder was included in each run [Figure 1].
Figure 1.
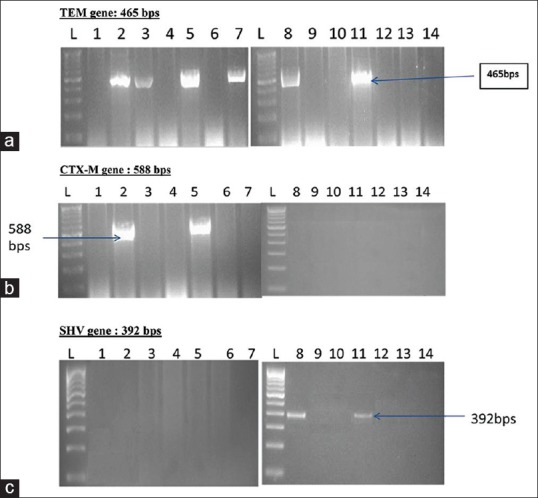
Gel pictures of amplified products (a) TEM gene: 465 bps. (b) CTX-M gene: 588 bps. (c) SHV gene: 392 bps. *L: DNA ladder of 100 bps
RESULTS
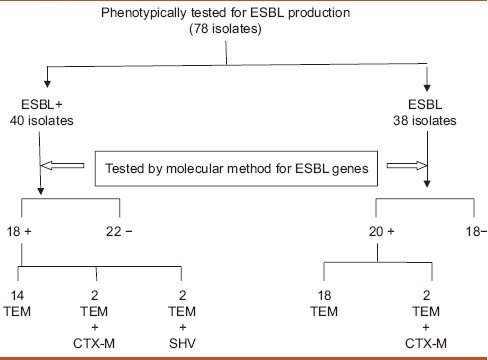
In the present prospective study, 78 bacterial isolates including 7 Klebsiella pneumoniae and 71 E. coli were studied genotypically. Among them, 40 isolates were phenotypically confirmed ESBL producers while 38 were phenotypically confirmed non-ESBL producers. Out of the three beta-lactamase (bla) genes studied, blaTEM was detected among 38 (48.7%), followed by blaCTX-M in six (7.6%) and blaSHV in four (5.1%) phenotypically confirmed ESBL and non-ESBL isolates out of the total 78 isolates studied. Out of the 40 phenotypically confirmed ESBL producers, 18 (45%) isolates were found to be positive for the presence of any of the three ESBL genes (blaSHV, blaTEM, and blaCTX-M). All these 18 (45%) isolates had blaTEM as one of the ESBL genes out of which 14 (77.7%) isolates had blaTEM alone as the ESBL gene. None of the isolates carried blaSHV or blaCTX-M genes alone. Among these 18 genotypically positive isolates, 2 (11.1%) isolates carried blaSHV gene and 4 (22.2%) of them carried blaCTX-M gene. Both these genes were present in association with the blaTEM gene. None of the 18 isolates carried all the three genes together. In addition, none of the isolates carried blaCTX-M and blaSHV genes together. Out of the 40 phenotypically confirmed ESBL producers, 22 (55%) isolates were not detected for the presence of any of the three ESBL genes (blaSHV, blaTEM, and blaCTX-M). Out of the 38 phenotypically non-ESBL isolates, 20 (52.6%) isolates were positive for the presence of ESBL genes and 18 (47%) were negative for any of the three ESBL genes [Table 2].
Table 2.
The phenotypic and genotypic extended spectrum beta-lactamases detection results along with the presence and absence of blaTEM, blaSHV, and blaCTX-M genes
DISCUSSION
In our tertiary care hospital settings, TEM (48.7%) gene predominated the SHV (7.6%) and CTX-M (5.1%) genes responsible for ESBL production. The results are in accordance with a study by Yazdi et al., 2012 (87.1% TEM, followed by 70.6% SHV)[12] but disagreed with the results of studies by Eftekhar et al., 2012, in which SHV (43.1%) exceeded TEM (35.2%),[4] by Shahid et al., 2011, in which CTX-M (28.8%) exceeded SHV (13.7%),[3] and by Ahmed et al., 2013, in which CTX-M (71.4% in E. coli and 68.4% in Klebsiella) exceeded TEM (55.1% E. coli and 58% Klebsiella).[11] Several other studies performed throughout the world showed variable results. In a Chinese study, the TEM gene predominated followed by SHV. A report from Canada showed SHV as the main group of ESBLs. However, reports from South America, Israel, Spain, New York, the United Kingdom, and several parts of Indian subcontinent revealed CTX-M as the predominant gene. Until the year 2000, TEM was the most prevalent ESBL gene in the Indian bacterial population but was replaced by CTX-M in the following decade. In urine isolates in our setting, TEM is again predominant. The differences between our study results and those of other authors indicated that the prevalence and type of ESBL genes may vary from one geographical region to another. The present study clearly demonstrates the dramatic change in the gene pool in Indian Enterobacteriaceae. In our study, 22 (55%) of phenotypically positive ESBL strains lacked TEM, SHV, and/or CTX-M genes, which can be explained by the possible presence of other ESBL-encoding genes in the studied Indian bacterial population.[3,4,11]
Occurrence of more than one beta-lactamase within the same isolate has been detected in our study. In our isolates of Enterobacteriaceae from urine isolates, not the TEM gene was observed both singly and in two other combinations. their wider dissemination may be due to the involvement of genetic elements in mobilization of these genes as was suggested by Shahid et al.[3]
Further, in the present study, all 20 (52.6%) (genotypically positive) out of 38 phenotypically non-ESBL isolates carried TEM gene. Of these, 18 (90%) isolates carried TEM gene alone while 2 (10%) of them carried TEM in association with CTX-M. The difference observed in the detection of ESBL-positive isolates by two different methods (phenotypic and genotypic) may reflect the lower sensitivity of phenotypic method and the influence of environmental factors on the incidence of resistance. It may also depend on the phenotypic methods chosen for detection of ESBL among bacterial isolates. Some ESBLs may fail to reach a level to be detectable by disk diffusion tests, thus result in treatment failure in the infected patients. The lack of correlation between ESBL production and disk diffusion susceptibility results was evident in the present study and reinforced the need for an improved method of ESBL detection to be incorporated in routine susceptibility procedures.[1] In contrast, genotypic method using specific PCR amplification of resistance genes seems to have 100% specificity and sensitivity. Our study showed the presence of blaTEM in all 38 genotypically positive isolates. None of the isolates was detected to carry SHV or CTX-M gene alone. In addition, none of the isolates had SHV and CTX-M together or TEM, SHV, and CTX-M genes in combination. Our study showed the presence of TEM gene (52.6%) in all genotypically positive isolates. The presence of TEM gene in all genotypically positive isolates indicates that this gene can be an appropriate candidate for molecular screening of ESBL-positive samples in our settings.[12]
The CTX-M enzymes are known as an increasingly serious public health concern worldwide and have been noted to be the cause of outbreaks throughout the world including India. They constitute a distinct phylogenetic lineage of molecular class A beta-lactamases that exhibit a higher preference for cefotaxime and ceftriaxone, than ceftazidime, and a higher susceptibility to tazobactam than to clavulanate. They still exist among the Indian bacterial population especially in our settings and hence are a matter of concern for clinicians.[3]
Out of the 38 phenotypically non-ESBL isolates, irrespective of whether they carried ESBL gene (TEM, SHV, or CTX-M), 20 (52.6%) isolates were totally sensitive while 18 (47.3%) isolates exhibited multidrug resistance. They were chosen as phenotypically non-ESBL because they did not follow the CLSI rules for confirmation of ESBL. It was observed that 14 (77.7%) out of 18 isolates exhibited another drug resistance mechanisms phenotypically. Four (22.2%) carried AMP C while ten (55.5%) were carbapenemase producers. Among the organisms that were phenotypically ESBL-negative isolates, eight (57.1%) carried one or more of the three ESBL genes. This may mean that the presence of ESBLs can be masked by the expression of chromosomal or plasmid-mediated AMP C beta-lactamases. Therefore, ESBL-producing strains with AMP C beta-lactamases can cause a false negative ESBL production. The same can be true with other simultaneous mechanisms of drug resistance such as carbapenemase production. Therefore, expression of beta-lactamase genes depends upon the environmental conditions such as the presence of antibiotics, and gene presence detected by PCR does not necessarily indicate its expression. Studies have revealed that some of these “hidden ESBL” genes can be detected through modifications in the phenotypic ESBL confirmatory tests.[4,16]
Out of the 38 phenotypically ESBL-negative isolates, ESBL genes were detected in twenty, indicating higher sensitivity of genotypic methods. Such presentations during phenotypic tests can be masked due to varying reasons but can often be revealed genotypically. However, those that lacked SHV, TEM, or CTX-M may have actually been negative or might have carried some other “hidden” gene for ESBL production.
CONCLUSION
Incorrect identification of antibiotic resistance may lead to inappropriate antibiotic prescription, which may in turn select for new resistance genes. Phenotypic tests for ESBL detection only confirm whether an ESBL is produced but cannot detect the ESBL subtype and cannot detect those genes whose expression is hidden or masked. Therefore, the genotypic method is suggested as the method of choice for detection of ESBL-producing strains of Enterobacteriaceae. Molecular methods are sensitive, but they are expensive and require specialized equipment and expertise. Furthermore, genotypic methods can only detect those genes with known sequences. Phenotypic tests need to be evaluated periodically: Their performance may change with the introduction of a new enzyme, and they may detect new enzymes not included within the laboratory's test algorithm. For best results, phenotypic methods of ESBL detection should be improved.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
We wish to thank the Chairperson and Dean of the Institute for providing laboratory facilities and healthy working atmosphere during the study period. We are also thankful to the technical staff of the Institute for providing necessary helping hand during the endeavor.
REFERENCES
- 1.Sharma J, Sharma M, Ray P. Detection of TEM and SHV genes in Escherichia coli and Klebsiella pneumoniae isolates in a tertiary care hospital from India. Indian J Med Res. 2010;132:332–6. [PubMed] [Google Scholar]
- 2.Naseer U, Sundsfjord A. The CTX-M conundrum: Dissemination of plasmids and Escherichia coli clones. Microb Drug Resist. 2011;17:83–97. doi: 10.1089/mdr.2010.0132. [DOI] [PubMed] [Google Scholar]
- 3.Shahid M, Singh A, Sobia F, Rashid M, Malik A, Shukla I, et al. Bla (CTX-M), bla (TEM), and bla (SHV) in Enterobacteriaceae from North-Indian tertiary hospital: High occurrence of combination genes. Asian Pac J Trop Med. 2011;4:101–5. doi: 10.1016/S1995-7645(11)60046-1. [DOI] [PubMed] [Google Scholar]
- 4.Eftekhar F, Rastegar M, Golalipoor M, Mansour Samaei N. Detection of extended spectrum beta-lactamases in urinary isolates of Klebsiella pneumonia in relation to Bla SHV, Bla TEM, Bla CTX-M gene carriage. Iran J Public Health. 2012;41:127–32. [PMC free article] [PubMed] [Google Scholar]
- 5.Collee JG, Fraser AG, Marmian BP, Simmons A, editors. Mackie and McCartney Practical Medical Microbiology. 14th ed. New York: Churchill Livingstone; 1996. [Reprinted 1999] [Google Scholar]
- 6.Cheesbrough M. Medical Laboratory Manual for Tropical Countries. Vol. 2. Cambridgeshire, England: Tropical Health Technology, Norfolk; 1984. Microbiology; p. 985. [Google Scholar]
- 7.Thomson KS. Extended-spectrum-β-lactamase, AmpC, and carbapenemase issues. J Clin Microbiol. 2010;48:1019–25. doi: 10.1128/JCM.00219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 9.24th Information Supplement. NCCLS Document M100-S23-24. Wayne, Pennsylvania, USA: Clinical Laboratory Standards Institute; 2013-2014. Clinical Laboratory Standards Institute. Performance Standard for Antimicrobial Susceptibility Testing. [Google Scholar]
- 10.Moosavian M, Deiham B. Distribution of TEM, SHV and CTX-M genes among ESBL-poducing Enterobacteriaceae isolates in Iran. Afr J Microbiol Res. 2012;6:5433–9. [Google Scholar]
- 11.Ahmed AB, Omar AO, Asghar AH, Elhassan MM. Prevalence of TEM, SHV and CTX-M genes in Escherichia coli and Klebsiella spp. urinary isolates from Sudan with confirmed ESBL phenotype. Life Sci J. 2013;10:191–5. [Google Scholar]
- 12.Yazdi M, Nazemi A, Mirinargasi M, Jafarpour M, Sharifi SH. Genotypic versus phenotypic methods to detect extended-spectrum beta lactamases (ESBL's) in uropathogenic Escherichia coli. Ann Biol Res. 2012;3:2454–8. [Google Scholar]
- 13.Colom K, Pérez J, Alonso R, Fernández-Aranguiz A, Lariño E, Cisterna R. Simple and reliable multiplex PCR assay for detection of blaTEM, bla (SHV) and blaOXA-1 genes in Enterobacteriaceae. FEMS Microbiol Lett. 2003;223:147–51. doi: 10.1016/S0378-1097(03)00306-9. [DOI] [PubMed] [Google Scholar]
- 14.Gangoué-Piéboji J, Bedenic B, Koulla-Shiro S, Randegger C, Adiogo D, Ngassam P, et al. Extended-spectrum-beta-lactamase-producing Enterobacteriaceae in Yaounde, Cameroon. J Clin Microbiol. 2005;43:3273–7. doi: 10.1128/JCM.43.7.3273-3277.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaftandzieva A, Trajkovska-Dokic E, Panovski N. Prevalence and molecular characterization of ESBLs producind Escherichia coli and Klebsiella pneumonia. Biol Med Sci. 2011;32:129–41. [PubMed] [Google Scholar]
- 16.Poulou A, Grivakou E, Vrioni G, Koumaki V, Pittaras T, Pournaras S, et al. Modified CLSI extended-spectrum ß-lactamase (ESBL) confirmatory test for phenotypic detection of ESBLs among Enterobacteriaceae producing various ß-lactamases. J Clin Microbiol. 2014;52:1483–9. doi: 10.1128/JCM.03361-13. [DOI] [PMC free article] [PubMed] [Google Scholar]


