Abstract
Background:
Leishmaniasis is important vector-borne parasitic disease worldwide, caused by the genus Leishmania. The objective of the current study was to identify genetic polymorphism in L. major, one of the species causing cutaneous leishmaniasis (CL), isolated from southeastern Iran, using Permissively Primed Intergenic Polymorphic-Polymerase Chain Reaction (PPIP-PCR) method.
Methods:
Overall, 340 patients with suspected CL were examined. They referred to the Central Laboratory in Chabahar, Iran during Apr 2013 to Feb 2014. Microscopic examination of Giemsa-stained slides from lesions as well as aspirates cultured in Novy- Mac Neal-Nicolle (NNN) Media was employed in order to diagnose CL in these patients. Our analyses detected 86 suspected subjects as having CL from which 35 isolates were cultured successfully. PPIP-PCR method was performed on extracted genomic DNA from selected isolates in order to determine the genetic polymorphism among L. major isolates.
Results:
The electrophoresis patterns demonstrated two genetic profiles including A or A1 patterns between all samples tested. Frequency of A and A1 sub-types were 33 (94.3%) and two (5.7%), respectively.
Conclusion:
Both host and parasite factors may contribute to the clinical profile of human leishmaniasis in the endemic foci of the disease. Here we showed that genetic variations pertaining to the Leishmania parasites might determine, in part, the clinical outcomes of human leishmaniasis.
Keywords: PPIP-PCR, Leishmania major, Genetic polymorphism, Iran
Introduction
Three forms of human leishmaniasis, including zoonotic cutaneous leishmaniasis (ZCL), anthroponotic cutaneous leishmaniasis (ACL), and visceral leishmaniasis (VL) are of public health importance in Middle East, especially in Iran, Afghanistan, Pakistan and Saudi Arabia. However, it seems that ZCL is endemic to the Eastern and Southeastern foci of Iran, and is the most frequently reported from these regions. “There are 350 million people in 98 countries at risk of contracting the disease, between 200,000 and 400,000 cases of VL, and between 700,000 and 1.2 million cases of CL occur annually throughout the world” (1–3).
PPIP-PCR is one of the methods that it used for determination of polymorphism in species and subspecies of Leishmania parasites belonging to the old world including L. major, L. tropica, L. donovani and L. aethiopica. The primers designed for this test do not amplify non-kinetoplastid DNA. Besides, this test is quick and reproducible, generating products, which can be easily contrasted to reference strains, PPIP–PCR, can be used for the reliable characterization and identification of cultured parasites (4).
Chabahar is situated in the Sistan and Baluchestan Province of Iran. L. major, as the agent of ZCL, has been reported previously in this endemic focus of the disease (5, 6).
The aim of this study was to characterize genetic variations in L. major isolated from patients with cutaneous leishmaniasis using a PPIP-PCR method.
Materials and Methods
Study area and population studied
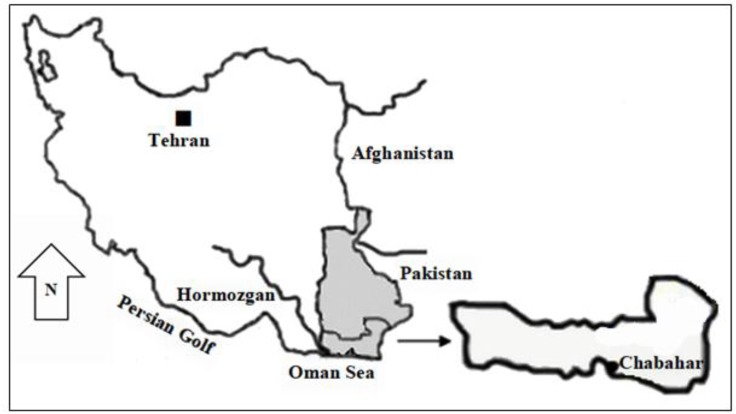
Chabahar (25° 17′ N, 60° 38′ E) has a population of 264051 according to the last census. This area consists of two urban (Chabahar and Negor), and six rural districts with over 508 villages (Fig. 1). The climates considered as very warm and the average annual temperature and humidity are 36.4 °C and 75.9%, respectively.
Fig. 1:
Map of Iran, showing the location of Chabahar City, Sistan and Baluchestan Province
Of 340 patients with suspected CL referred to the Central Laboratory in Chabahar during a period of 11 months from Apr 2013 to Feb 2014 (Fig. 2), eighty-six patients were diagnosed as having cutaneous leishmaniasis by microscopy.
Fig. 2:
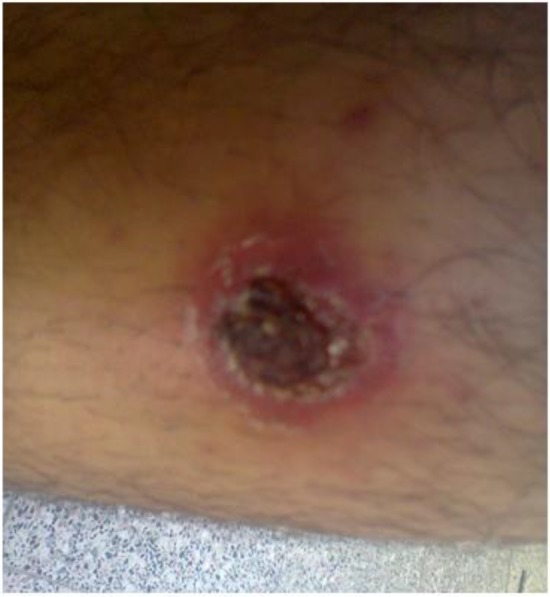
Typical macroscopic lesion of cutaneous leishmaniasis caused by L.major was prepared from the patient (Original)
The study was approved by the Ethical Committee of Kerman University of Medical Sciences and the Iranian Ministry of Health, Treatment and Medical Training Protection Code of Human Subjects in Medical Research. Written informed consents were obtained from subjects that participated in this study.
Aspirates from skin lesions were cultured in the biphasic media, Novy-MacNeal-Nicole (NNN). Primary-isolated parasites were subsequently sub-cultured in RPMI1640 (Gibco, Life technologies GmbH, Frankfurt, Germany) media enriched with L-glutamine (Merck, Germany), inactivated 10% fetal bovine serum (Gibco, Germany), 100μg/ml streptomycin and 100 U/ml penicillin (Gibco, Germany). Thirty-five positive were maintained in culture successfully and were selected for further molecular analyses.
DNA Extraction
Promastigotes were harvested by centrifugation at 600 g for 15 min, and washed twice in sterile PBS, pH 7.2. DNA extraction was conducted using a High Pure PCR Template Preparation Kit (Roche, City, Germany) as recommended by the manufacturer. The extracted DNA was eluted in 70 μl elution buffer and stored at −20 °C until use,
PPIP-PCR
Primers used were 2B (5′-CAG GAG CGC GCA CAC GCA CAC ACG -3′) and 2A (5′-ATT AAC CCT CAC TAA ACC CAA CAA -3′) (4). Amplification reactions were performed in 50 μl containing 25 μl Tag Master Mix RED (Ampliqon, Denmark)), 2 μl of each primer (10 pmole concentration; MWG Germany), 5 μl Template DNA and 18 μl of distillated water. Amplification was started with one cycle at 94 °C for 5 min, followed by 30 cycles including, 75 sec at 94 °C, 1 min at 63 °C, 2 min at 72 °C, and one cycle at 72 °C for 5 min, followed by a final cycle at 15 °C for 3 min using a thermocycler (Bioer thermal cycler TC-XP-G, Germany). The PCR products were visualized using a 1.5% gel agarose electrophoresis, and compared with the patterns of two previously known Iranian Leishmanias species (L. major; MRHO/IR/75/ER) and L. tropica (IR/09 /Khamesipour_ Masha) No DNA template were used as the negative control.
Results
Comparison of banding pattern of each isolate with reference stains revealed that all isolates from the foci were characterized as L. major. From 340-suspected patients, 86 cases confirmed by microscopy (1000x) and 35 cases selected for culture exams and molecular method, PPIP-PCR. Additionally, further analyses demonstrated that the frequency of A and A1 sub-types were 33 (94.3%) and two (5.7%), respectively. Besides comparing the patterns with those of reference DNAs, the PPIP-PCR assay showed genetic variation with special genetic patterns, related to L. major. However, the PCR product profiles in this area showed specific band 500 bp for L. major and majority with subtype A (Fig. 3). These cases were 15 men (42.9%) and 20 women (57.1%) and their ages were between 1 to 60 yr (average age: 13.6 yr).
Fig. 3:
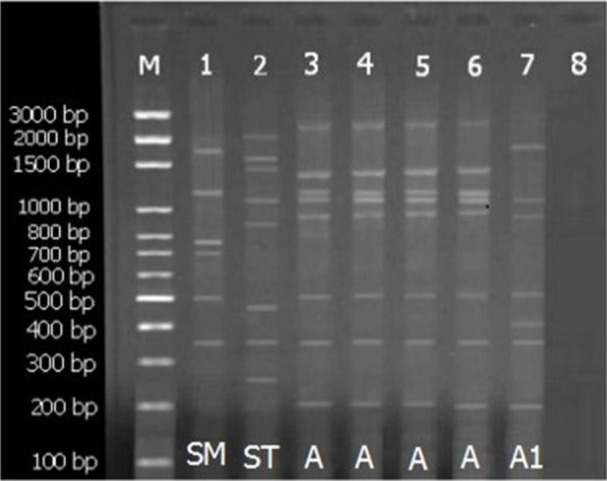
The pattern of agarose gel electrophoresis of Leishmania major isolates. lane M, DNA size marker 100 bp; lane 1, L. major (MRHO/IR/75/ER) as a positive control; lane 2, L. tropica (IR/09/Khamesipour_ Masha) as a positive control; lane 8, negative control; lanes 3, 4, 5, 6 and 7 related to strains of L. major isolated from skin lesions of the patients, Chabahar, southeast of Iran
The frequency of lesions of face, hand, foot, body and coexistence of hand and body were 16.7, 22.8, 2.9 and 5.7%, respectively. Lesions were most frequently diagnosed in patients in the face (48.6 %) with lowest frequency observed in lesions on body trunk (2.9%). Numbers of lesions varied between 1 to 3, and the minimum and maximum lesion sizes were 1.5 and 5 cm in diameter, respectively (Fig. 2).
Discussion
Since zoonotic leishmaniasis is the most important endemic disease in hyper endemic areas of Iran, identification of L. major genetic variations seems unavoidable for designing appropriate control programs. Microscopy is still considered as a gold standard method for the diagnosis of CL. However, the sensitivity of these methods is variable from 27 to 85% depending on parameters such as clinical presentations, numbers of parasites in specimens, technical skills, how skilled the examiner is as well as the parasite species (7–11).
In past decade, PCR-based techniques have been improved for the identification of Leishmania parasites at species or strain levels (7–9). Differential diagnosis of Leishmania parasites is very important for implementing appropriate strategies such as therapies and epidemiologic studies, especially in foci where different genetic types of parasite are present (10, 11).
In the present study, we applied a PPIP-PCR method to identify Leishmania parasites. PPIPPCR analysis showed that among the positive samples, all isolates produced fragments heavy and light. In this size and number of fragments divided to 2 group, named LmA and LmA1 (12). Profile A consisted of 515 bp as a major band and 350 bp and 220 bp as a light molecular weight band and four heavy molecular weight bands the frequency of this profile band was more than others (Fig. 2).
Profile A in this region separated from Isfahan and Ahvaz profile A. As those profiles, banding has not 400 and 180 bp bands but heavy bands of all of them were specific with four molecular bands. Profile A1 was the same as Isfahan and Ahvaz province, central and southwest of Iran, respectively.
In the last decade, there are few documents showing L. major polymorphism in Israel (4) and same studies showed polymorphism in L. major in two hyper endemic region of Iran, Isfahan and Ahvaz (12).
The results of the current study are consistent with recent studies from these countries. Our study could confirm the genetic variation among L. major in Chabahar City, Southeast of Iran
In our study, maximum frequency of lesions was observed in face with 17 cases (48.6 %) and minimum frequency was with one case (2.9%) in body. Previous studies were showed that the most common lesion site was the face (13,14).
Based on our results it is clear that most of the lesions in open areas of the body can be seen because the areas are availability to mosquitoes.
To our knowledge, this work is the first application of PPIP-PCR for genetic variation within L. major in Chabahar City and tertiary application in the worldwide.Various developmental mechanisms such as selection, genetic drift and migration play a basic role in the genetic distribution in populations of natural, however reproduction is the fundamental biological process that influences the genetic structure of populations (15).
Several studies were reported evidence of hybrid formation in Leishmania (16, 17), reinforcing the idea that sexual reproduction may occur in Leishmania, however at a level yet undefined. Certainly, rare or occasional bouts of sexual recombination in usually asexual organisms can have an extreme effect on the genetic diversity extent.
Conclusion
L. major in zoonotic form in Chabahar is genetically a highly polymorphic species. For consolidate these findings, the L. major extra isolates from patients, reservoir hosts, and vectors from various locations require to be collected for further examination.
Acknowledgments
We are grateful to Dr Javad Sharifi-Rad for critically reading the manuscript and thank the Leishmaniasis Research Center, Kerman, Iran for financial support. In addition, Chabahar Laboratory Center for all their cooperation in this study.
This study was financially supported by Kerman University of Medical Sciences grant No. 92/299. This paper was part of the MSc. thesis of Mr. Mehdi Sharifi-Rad supervised by Dr. Zahra Babaei and Dr. Mansour Dabirzadeh. The authors declare that there is no conflict of interests.
References
- 1. WHO Control of the Leishmaniasis . Report of a meeting of the WHO expert committee on the control of Leishmaniasis; Geneva. Important document summarizing the epidemiology and latest diagnostic and treatment recommendations for all forms of leishmaniasis, per region. 2010.
- 2. Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, Jannin J, den Boer M. Leishmaniasis worldwide and global estimates of its incidence. PLoS One. 2012; 7( 5): e35671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sharifi I, Ardehali S, Motazedian H, Aflatoonian MR, Fekri AR. Identification and characterization of Leishmania isolates in school children in Bam, southeastern Iran. Iran J Med Sci. 1997; 22( 3,4): 82– 8. [Google Scholar]
- 4. Eisenberger CL, Jaffe CL. Leishmania: Identification of old world species using a Permissively Primed Intergenic Polymorphic - Polymerase Chain Reaction. Exp Parasitol. 1999; 91: 70– 7. [DOI] [PubMed] [Google Scholar]
- 5. Fazaeli A, Fouladi B, Hashemi-Shahri SM, Sharifi I. Clinical Features of Cutaneous Leishmaniasis and Direct PCR-Based Identification of Parasite Species in A New Focus in Southeast of Iran. Iran J Public Health. 2008; 37( 3): 44– 51. [Google Scholar]
- 6. Kassiri H, Naddaf SR, Javadian EA, Mohebali M. First Report on Isolation and Characterization of Leishmania major from Meriones hurrianae (Rodentia: Gerbillidae) of A Rural Cutaneous leishmaniasis Focus in South-Eastern Iran. Iran Red Crescent Med J. 2013; 15( 9): 789– 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Veland N, Boggild AK, Valencia C, Valencia BM, Llanos-Cuentas A, Van der Auwera G, Dujardin JC, Arevalo J. Leishmania (Viannia) species identification on clinical samples from cutaneous leishmaniasis patients in Peru: Assessment of a molecular stepwise approach. J Clin Microbiol. 2012; 50: 495– 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parhizkari M, Motazedian MH, Asqari Q, Mehrabani D. The PCR-based detection of Leishmania major in Mus musculus and other rodents caught in southern Iran: A guide to sample selection. Ann Trop Med Parasitol. 2011; 105: 319– 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carvalho EM, Donelson JE, Wilson ME. Serial quantitative PCR assay for detection, species discrimination, and quantification of Leishmania spp. in human samples. J Clin Microbiol. 2011; 49: 3892– 904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schönian G, Nasereddin A, Dinse N, Schweynoch C, Schallig HD, Presber W. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn Microbiol Infect Dis. 2003; 47: 349– 58. [DOI] [PubMed] [Google Scholar]
- 11. Jaffe CL, Baneth G, Abdeen ZA, Schlein Y, Warburg A. Leishmaniasis in Israel and the Palestinian Authority. Trends Parasitol. 2004; 20: 328– 32. [DOI] [PubMed] [Google Scholar]
- 12. Dabirzadeh M, Mirmohammad Sadeghi H, Baghaie M, Hejazi H. Genetic Polymorphism of Leishmania major in Two Hyper Endemic Regions of Iran Revealed by PPIP-PCR and ITS-RFLP. Arch Iran Med. 2012; 15( 3): 151– 6. [PubMed] [Google Scholar]
- 13. Layegh P, Moghiman T, Ahmadian Hoseini SA. Children and cutaneous leishmaniasis: a clinical report and review. J Infect Dev Ctries. 2013; 7( 8): 614– 7. [DOI] [PubMed] [Google Scholar]
- 14. Talari SA, Talaei R, Shajari G, Vakili Z, Taghaviardakani A. Childhood cutaneous leishmaniasis: report of 117 cases from Iran. Korean J Parasitol 2006; 44: 355– 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tibayrenc M. Population genetics of parasitic protozoa and other microorganisms. Adv Parasitol. 1995; 36: 47– 115. [DOI] [PubMed] [Google Scholar]
- 16. Evans D. Handbook on Isolation, Characterization and Cry Preservation of Leishmania UNDP/World Bank/World Health Organization. Geneva, Switzerland: 1989. [Google Scholar]
- 17. Kelly JM, Law JM, Chapman CJ, Van Eys GJ, Evans DA. Evidence of genetic recombination in Leishmania. Mol Biochem Parasitol. 1991; 46: 253– 63. [DOI] [PubMed] [Google Scholar]