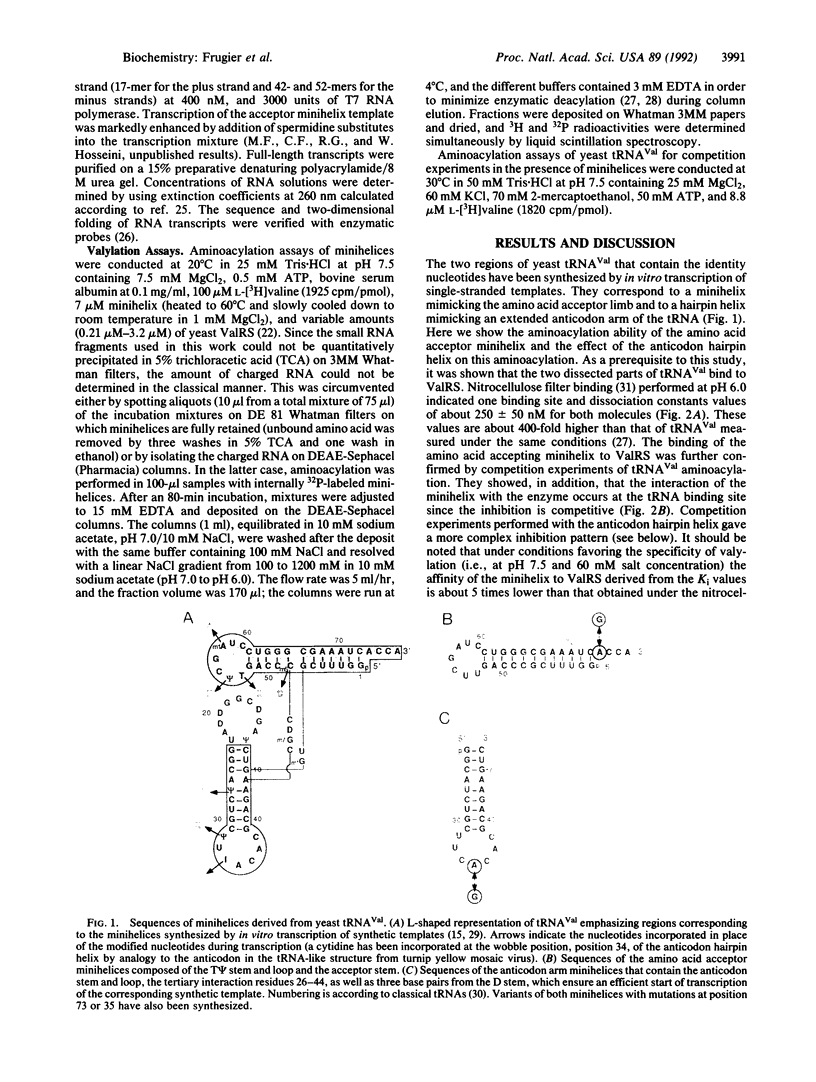
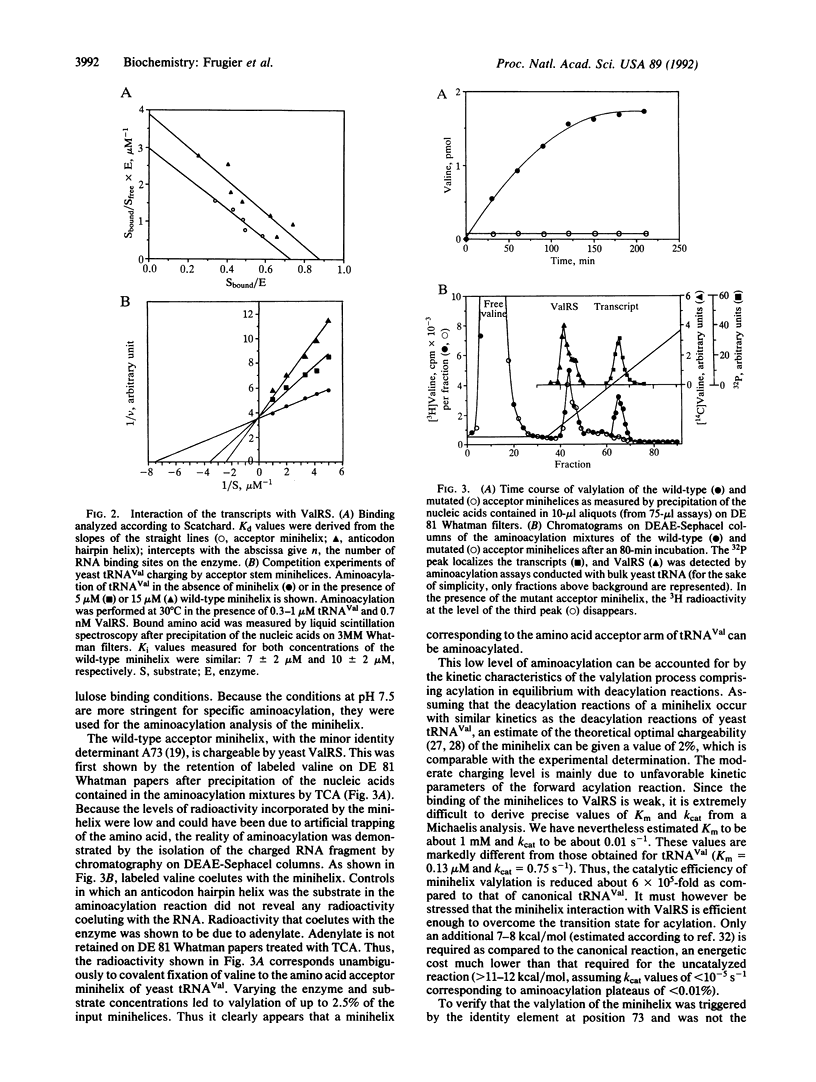
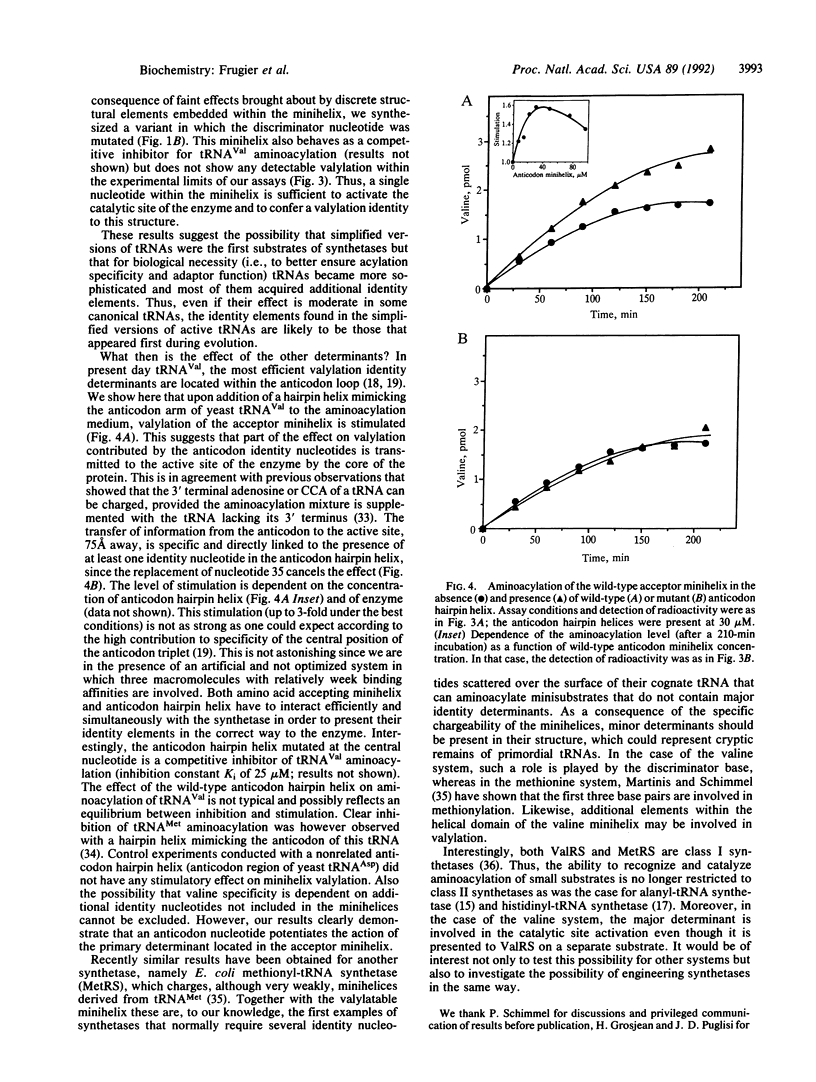
Abstract
Minihelices mimicking the amino acid acceptor and anticodon branches of yeast tRNA(Val) have been synthesized by in vitro transcription of synthetic templates. It is shown that a minihelix corresponding to the amino acid acceptor branch and containing solely a valine-specific identity nucleotide can be aminoacylated by yeast valyl-tRNA synthetase. Its charging ability is lost after mutating this nucleotide. This ability is stimulated somewhat by the addition of a second hairpin helix that mimicks the anticodon arm, which suggests that information originating from the anticodon stem-loop can be transmitted to the active site of the enzyme by the core of the protein.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonnet J., Ebel J. P. Interpretation of incomplete reactions in tRNA aminoacylation. Aminoacylation of yeast tRNA Val II with yeast valyl-tRNA synthetase. Eur J Biochem. 1972 Dec 4;31(2):335–344. doi: 10.1111/j.1432-1033.1972.tb02538.x. [DOI] [PubMed] [Google Scholar]
- Dietrich A., Kern D., Bonnet J., Giegé R., Ebel J. P. Interpretation of tRNA-mischarging kinetics. Eur J Biochem. 1976 Nov 1;70(1):147–158. doi: 10.1111/j.1432-1033.1976.tb10965.x. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Florentz C., Dreher T. W., Rudinger J., Giege R. Specific valylation identity of turnip yellow mosaic virus RNA by yeast valyl-tRNA synthetase is directed by the anticodon in a kinetic rather than affinity-based discrimination. Eur J Biochem. 1991 Jan 1;195(1):229–234. doi: 10.1111/j.1432-1033.1991.tb15698.x. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8655–8659. doi: 10.1073/pnas.87.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giegé R., Briand J. P., Mengual R., Ebel J. P., Hirth L. Valylation of the two RNA components of turnip-yellow mosaic virus and specificity of the tRNA aminoacylation reaction. Eur J Biochem. 1978 Mar;84(1):251–256. doi: 10.1111/j.1432-1033.1978.tb12163.x. [DOI] [PubMed] [Google Scholar]
- Himeno H., Hasegawa T., Ueda T., Watanabe K., Miura K., Shimizu M. Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation. Nucleic Acids Res. 1989 Oct 11;17(19):7855–7863. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y. M., Schimmel P. A simple structural feature is a major determinant of the identity of a transfer RNA. Nature. 1988 May 12;333(6169):140–145. doi: 10.1038/333140a0. [DOI] [PubMed] [Google Scholar]
- Jahn M., Rogers M. J., Söll D. Anticodon and acceptor stem nucleotides in tRNA(Gln) are major recognition elements for E. coli glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):258–260. doi: 10.1038/352258a0. [DOI] [PubMed] [Google Scholar]
- Kern D., Giegé R., Robre-Saul S., Boulanger Y., Ebel J. P. Complete purification and studies on the structural and kinetic properties of two forms of yeast valyl-tRNA synthetase. Biochimie. 1975;57(10):1167–1176. doi: 10.1016/s0300-9084(76)80579-2. [DOI] [PubMed] [Google Scholar]
- Kisselev L. L. The role of the anticodon in recognition of tRNA by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1985;32:237–266. doi: 10.1016/s0079-6603(08)60350-5. [DOI] [PubMed] [Google Scholar]
- Martinis S. A., Schimmel P. Enzymatic aminoacylation of sequence-specific RNA minihelices and hybrid duplexes with methionine. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):65–69. doi: 10.1073/pnas.89.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the acceptor identity of a transfer RNA by altering nucleotides in a "variable pocket". Science. 1988 Sep 30;241(4874):1804–1807. doi: 10.1126/science.2459773. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Changing the identity of a tRNA by introducing a G-U wobble pair near the 3' acceptor end. Science. 1988 May 6;240(4853):793–796. doi: 10.1126/science.2452483. [DOI] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Nucleotides that determine Escherichia coli tRNA(Arg) and tRNA(Lys) acceptor identities revealed by analyses of mutant opal and amber suppressor tRNAs. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9260–9264. doi: 10.1073/pnas.87.23.9260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K., Jenkins R. A., Schneider J. Rapid determination of nucleotides that define tRNA(Gly) acceptor identity. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6147–6151. doi: 10.1073/pnas.88.14.6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain W. H., Foss K. Nucleotides that contribute to the identity of Escherichia coli tRNA(Phe). J Mol Biol. 1988 Aug 20;202(4):697–709. doi: 10.1016/0022-2836(88)90551-7. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Blanquet S., Fayat G. Binding of the anticodon domain of tRNA(fMet) to Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 1991 Jul 20;220(2):205–208. doi: 10.1016/0022-2836(91)90003-o. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Determination of RNA-protein contacts using thiophosphate substitutions. Biochemistry. 1989 Apr 4;28(7):2849–2855. doi: 10.1021/bi00433a016. [DOI] [PubMed] [Google Scholar]
- Normanly J., Ogden R. C., Horvath S. J., Abelson J. Changing the identity of a transfer RNA. Nature. 1986 May 15;321(6067):213–219. doi: 10.1038/321213a0. [DOI] [PubMed] [Google Scholar]
- Perret V., Garcia A., Grosjean H., Ebel J. P., Florentz C., Giegé R. Relaxation of a transfer RNA specificity by removal of modified nucleotides. Nature. 1990 Apr 19;344(6268):787–789. doi: 10.1038/344787a0. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Pütz J., Puglisi J. D., Florentz C., Giegé R. Identity elements for specific aminoacylation of yeast tRNA(Asp) by cognate aspartyl-tRNA synthetase. Science. 1991 Jun 21;252(5013):1696–1699. doi: 10.1126/science.2047878. [DOI] [PubMed] [Google Scholar]
- Renaud M., Bacha H., Remy P., Ebel J. P. Conformational activation of the yeast phenylalanyl-tRNA synthetase catalytic site induced by tRNAPhe interaction: triggering of adenosine or CpCpA trinucleoside diphosphate aminoacylation upon binding of tRNAPhe lacking these residues. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1606–1608. doi: 10.1073/pnas.78.3.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson J. R., DiRenzo A. B., Behlen L. S., Uhlenbeck O. C. Nucleotides in yeast tRNAPhe required for the specific recognition by its cognate synthetase. Science. 1989 Mar 10;243(4896):1363–1366. doi: 10.1126/science.2646717. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Parameters for the molecular recognition of transfer RNAs. Biochemistry. 1989 Apr 4;28(7):2747–2759. doi: 10.1021/bi00433a001. [DOI] [PubMed] [Google Scholar]
- Schulman L. H., Pelka H. Anticodon switching changes the identity of methionine and valine transfer RNAs. Science. 1988 Nov 4;242(4879):765–768. doi: 10.1126/science.3055296. [DOI] [PubMed] [Google Scholar]
- Schulman L. H. Recognition of tRNAs by aminoacyl-tRNA synthetases. Prog Nucleic Acid Res Mol Biol. 1991;41:23–87. [PubMed] [Google Scholar]
- Shi J. P., Schimmel P. Aminoacylation of alanine minihelices. "Discriminator" base modulates transition state of single turnover reaction. J Biol Chem. 1991 Feb 15;266(5):2705–2708. [PubMed] [Google Scholar]
- Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989;17 (Suppl):r1–172. doi: 10.1093/nar/17.suppl.r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt J. R., Chastain M., Puglisi J. D. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991 Dec;11(6):764–769. [PubMed] [Google Scholar]
- Yarus M., Berg P. On the properties and utility of a membrane filter assay in the study of isoleucyl-tRNA synthetase. Anal Biochem. 1970 Jun;35(2):450–465. doi: 10.1016/0003-2697(70)90207-1. [DOI] [PubMed] [Google Scholar]