Abstract
Background:
Invasion of the bloodstream by microorganisms constitutes one of the most serious situations in infectious disease. Microorganisms present in circulating blood whether continuously, intermittently, or transiently are a threat to every organ in the body. Prevalence and antimicrobial susceptibility of microorganisms vary depending upon the geography and the use of antibiotics.
Methods:
A cross-sectional study to determine the prevalent organisms causing bloodstream infection was conducted. BACTEC BD 9050 system was used to identify the causative organism, and sub-cultures were done on MacConkey Agar and Blood Agar. Antibiotic susceptibility test (AST) was done using Kirby B Disk diffusion method.
Results:
A total of 170 patients were enrolled, and blood samples of 53 patients showed growth of organisms. Staphylococcus aureus was the most commonly isolated organism. Most of the Gram-positive cocci (GPC) were susceptible to vancomycin and linezolid. Most of the Gram-negative bacilli (GNB) showed sensitivity to cefoperazone/sulbactam followed by imipenem.
Keywords: Blood Stream infections, Antibiotic Sensitivity, Antimicrobial resistance BACTEC
Introduction
Invasion of the bloodstream by microorganisms constitutes one of the most serious situations in infectious disease. Microorganisms present in circulating blood whether continuously, intermittently, or transiently are a threat to every organ in the body. Prevalence and antimicrobial susceptibility of microorganism vary depending upon the geography and the use of antibiotics. The excessive and irrational use of antibiotics has led to an increase in the multidrug-resistant bugs and thus worsened the condition. Bloodstream infections have serious consequences like shock, disseminated intravascular coagulation, multiple organ failure, and even death. Increased hospital stay and associated costs are the most troublesome consequences.
Treatment of bloodstream infections is based on the knowledge of prevalent microorganisms and their antimicrobial sensitivity patterns. This information also forms the basis for making recommendations for initial empirical therapy to be started when a bloodstream infection is suspected.
Specific therapy can only be started once the organisms are isolated and their antimicrobial sensitivity patterns are studied. The procedure is time consuming and depends upon the growth of the organisms in culture media. Many faster and automated culture techniques are available. BACTEC 9050 is one of the automated blood culture methods and has a detection rate from 15% to 50%. In this study, we aimed to detect the most prevalent microorganisms in the region and their antimicrobial sensitivity patterns.
Methods
A cross-sectional observational study was carried out from March 2014 to June 2015; blood samples were collected from patients suspected of having bloodstream infection attending and admitted in National Institute of Medical Sciences and Research, NIMS University, Jaipur. Details like hospital identity, number, age, gender of the patients, and type and place of collection of specimen were recorded on a formatted proforma. A total of 170 suspected patients attending the various intensive care units (ICUs), out patient departments (OPDs), and indoor wards of Internal Medicine, Pediatrics, Obstetrics and Gynecology, Orthopedics, and General Surgery were included in the study. Patients of all age groups with fever (both high and low grade) due to infective causes were included. Written and informed consent was taken from all who fulfilled the criteria. Patients having both leukocytosis and leucopenia were included in the study. Those who refused consent were excluded along with patients having autoimmune or chronic diseases like tuberculosis or sarcoidosis. Patients on steroids, having heat stroke, or having a suspected viral or parasitic infection were also excluded.
Blood was collected with all aseptic precautions from the bedside of the patients suspected of having bloodstream infection using a sterile syringe. Approximately 5–10 mL of blood was collected from adult patients, while 1–5 mL blood was collected from pediatric patients, and 1–2 mL from neonates for blood culture. The sample taken was immediately processed for blood culture by BACTEC BD 9050 blood culture system (Figure 1). In the BACTEC BD 9050 Blood Culture System, when growth of bacteria occurred in sufficient amount, the system automatically generated a signal on the front panel. If no growth of bacteria occurred within 5 days of blood culture, then the blood culture was reported as sterile on culture for pyogenic aerobic organisms.
Figure 1.
BACTEC BD 9050 system used for automated cultures.
Identification of the bacterial isolates
Subculture was made on MacConkey Agar and Blood Agar from the BACTEC blood culture bottles, which generated a beep in the Automated BACTEC BD 9050 blood culture system indicating growth of the organism. The organisms were identified as per standard protocol [Ananthanarayan and Paniker, 2013; Collee et al. 1996; Forbes et al. 2014; Winm et al. 2006].
Gram-negative bacilli
The colony character on culture media was observed, and Gram staining, motility, and biochemical tests – indole, methyl red, Voges–Proskauer, citrate utilization, urease test, phenyl pyruvic acid test, triple sugar iron agar, oxidase, amino acids decarboxylase test, and sugar fermentation reaction – were conducted (Figure 2a and b).
Figure 2.
(a) IMVIC tests for Escherichia. coli and (b) fermentation tests for GNB.
GNB, Gram-negative bacilli.
Gram-positive cocci
On the basis of colony character, Gram stain, catalase test, and coagulase test.
Antimicrobial susceptibility testing
This was performed by Kirby Bauer disk diffusion method as per the Clinical and Laboratory Standards Institute (CLSI, [2013]) guidelines. The antibiotics disks (Hi-media, Mumbai) were used as mentioned in Tables 1–3.
Table 1.
Antibiotics for GPC.
| Antibiotic | Potency (µg) | Symbol |
|---|---|---|
| Amikacin | 30 | AK |
| Cefoxitin | 30 | CX |
| Ciprofloxacin | 5 | CIP |
| Clindamycin | 2 | CD |
| Erythromycin | 15 | E |
| Linezolid | 30 | LZ |
| Penicillin G | 10 units | P |
| Vancomycin | 30 | VA |
GPC, Gram-positive cocci.
Table 2.
Antibiotics for GNB.
| Antibiotic | Potency (µg) | Symbol |
|---|---|---|
| Ampicillin | 10 | AMP |
| Ceftriaxone | 30 | CTR |
| Piperacillin/tazobactam | 100/10 | PIT |
| Gentamycin | 10 | GEN |
| Cefoperazone/sulbactam | 75/30 | CFS |
| Chloramphenicol | 30 | C |
| Ofloxacin | 5 | OF |
| Cefepime | 30 | CPM |
| Amikacin | 30 | AK |
| Imipenem | 10 | IPM |
GNB, Gram-negative bacilli.
Table 3.
Antibiotics for Pseudomonas spp.
| Antibiotic | Potency (µg) | Symbol |
|---|---|---|
| Amikacin | 30 | AK |
| Aztreonam | 30 | AT |
| Ceftazidime | 30 | CAZ |
| Ciprofloxacin | 5 | CIP |
| Colistin | 10 | CL |
| Piperacillin/tazobactam | 100/10 | PIT |
| Tobramycin | 10 | TOB |
| Imipenem | 10 | IPM |
The following are quality control strains for antimicrobial sensitivity testing:
(1) Pseudomonas aeruginosa (ATCC27853),
(2) Escherichia coli (ATCC25922),
(3) Staphylococcus aureus (ATCC25923).
Data were entered in excel sheet to prepare a master chart. Quantitative data were shown as mean ± standard deviation (SD), and qualitative data as numbers and percentages.
Results
This study was carried out from March 2014 to June 2015 with 170 non-repetitive blood samples collected from patients suspected of having bloodstream infections attending and admitted in National Institute of Medical Sciences and Research, NIMS University, Jaipur. Details like hospital identity, registration number, laboratory number, age and sex of the patients, and type and place of collection of specimen were recorded in a formatted proforma.
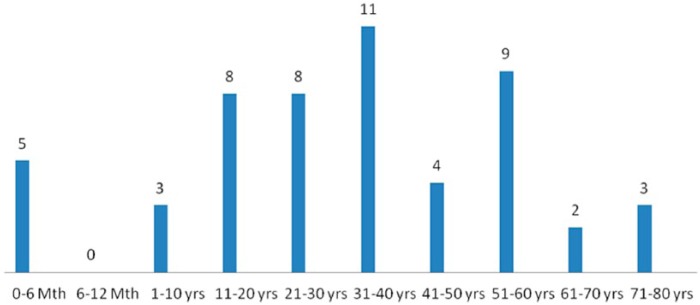
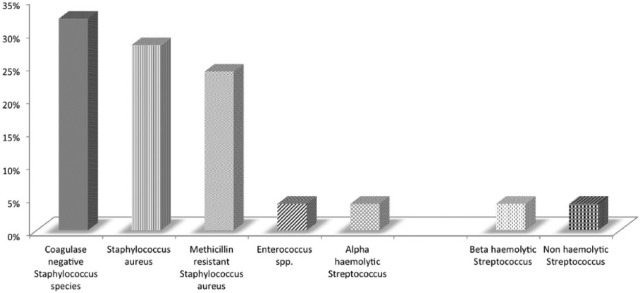
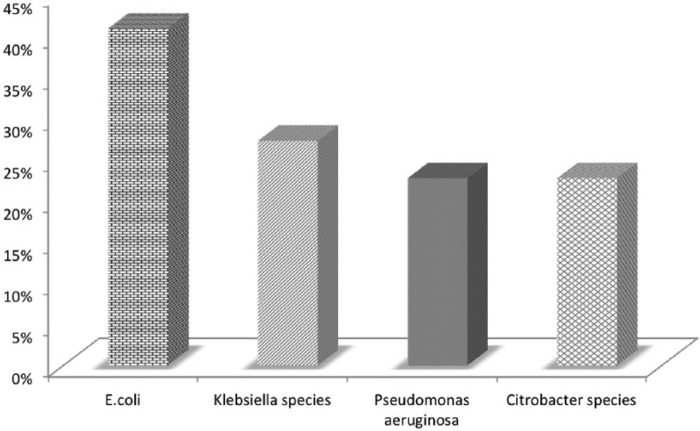
Culture positivity was seen in 53 (31.2%) samples, and 117 (68.8%) samples were sterile as detected with the BACTEC BD 9050 blood culture system. The maximum number of positive blood culture was from age group of 31 to 40 years, and the minimum from age group 11 to 20 years (Figure 3). Of the 53 culture-positive patients, 30 were adults admitted to medical ICUs. The mean time taken to culture positivity was 33.58 ± 21.64 h. Gram-positive cocci (GPC) were the most common organism isolated, with S. aureus being the most common of them (Figure 4), and E. coli was the most commonly isolated Gram-negative bacilli (GNB) (Figure 5).
Figure 3.
Age-wise distribution of culture-positive patients.
Figure 4.
Distribution of GPC isolated from the cultures.
GPC, Gram-positive cocci.
Figure 5.
Distribution of GNB isolated from the cultures.
GNB, Gram-negative bacilli.
There were six Candida fungus isolated, of which four were non albicans species.
Antibiotic sensitivity pattern
The antibiotic sensitivity patterns of GPC are shown in Table 4 and those of GNB are shown in Table 5.
Table 4.
Antibiotic sensitivity pattern of GPC.
| Antibiotics |
Staphylococci |
Enterococcus (n = 2) | Streptococcus. pneumoniae (n = 2) | ||
|---|---|---|---|---|---|
| Coagulase-positive Staphylococcus. aureus (n = 13) |
Coagulase-negative Staphylococcus species (n = 8) | ||||
| MSSA (n = 7) | MRSA (n = 6) | ||||
| Vancomycin | 100% | 100% | 100% | 100% | 100% |
| Linezolid | 100% | 100% | 100% | 100% | 100% |
| Amikacin | 85.71% | 33.33% | 100% | 0% | 100% |
| Penicillin G | 0% | 0% | 0% | 0% | 100% |
| Clindamycin | 85.71% | 50% | 87.5% | 0% | 100% |
| Ciprofloxacin | 85.71% | 16.66% | 87.5% | 0% | 100% |
| Cefoxitin | 100% | 0% | 100% | 100% | 100% |
| Erythromycin | – | – | – | 50% | – |
GPC, Gram-positive cocci; MRSA, methicillin-resistant Staphylococcus aureus; MSSA, methicillin-sensitive Staphylococcus aureus.
Table 5.
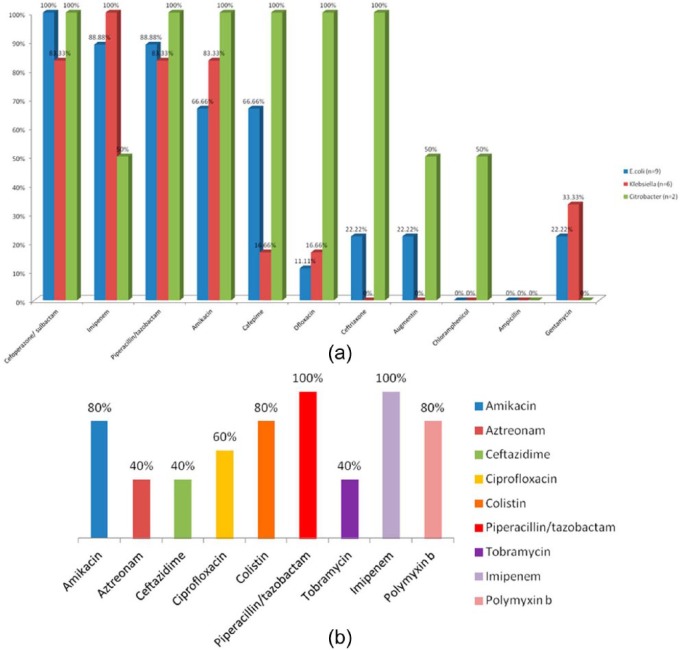
Antibiotic sensitivity pattern of GNB (Figure 6a).
| Antibiotic | Escherichia coli (n = 9) | Klebsiella (n = 6) | Citrobacter (n = 2) |
|---|---|---|---|
| Cefoperazone/sulbactam | 100% | 83.33% | 100% |
| Imipenem | 88.88% | 100% | 50% |
| Piperacillin/tazobactam | 88.88% | 83.33% | 100% |
| Amikacin | 66.66% | 83.33% | 100% |
| Cefepime | 66.66% | 16.66% | 100% |
| Ciprofloxacin | 33.33% | 33.33% | 50% |
| Ceftriaxone | 22.22% | 33.33% | 100% |
| Chloramphenicol | 0% | 0% | 50% |
| Ampicillin | 0% | 0% | 0% |
| Gentamycin | 22.22% | 33.33% | 0% |
GPC, Gram-negative bacilli.
Figure 6.
(a) Antibiotic susceptibility pattern of gram negative bacilli and (b) antibiotic susceptibility pattern of Pseudomonas.
Some other antibiotic sensitivity patterns were also noted for P. aeruginosa, which are listed in Table 6.
Table 6.
Antibiotic sensitivity patterns for Pseudomonas aeruginosa (Figure 6b).
| Antibiotics | P. aeruginosa (n = 5) |
|---|---|
| Amikacin | 80% |
| Aztreonam | 40% |
| Ceftazidime | 60% |
| Ciprofloxacin | 60% |
| Colistin | 80% |
| Piperacillin/tazobactam | 80% |
| Tobramycin | 40% |
| Imipenem | 100% |
Discussion
Bloodstream infection is a challenging problem, and sometimes, it may be life threatening; therefore, timely detection, identification, and antimicrobial susceptibility testing of blood-borne pathogens are one of the most important functions of diagnostic microbiology laboratory. In this study, the culture positivity of the BACTEC BD 9050 blood culture system was 31.2%. This positivity rate was consistent with the study done by Kohli-Kochhar et al. [2011]. This positivity rate was consistent with the Kenyan study by Kohli-Kochhar et al. [2011] in neonates who reported blood culture positivity of 23% but not consistent with Karunakaran et al. [2007] and Viswanathan et al. [2012]. Kolkata whose study yielded positivity rate of 16.7% and 46.3% respectively.
In this study, maximum number of positive cases was found in the middle age group less than 60 years (31–40 years of age). A study done by Prashanth et al. [2011] also reported maximum number of positive cases in age group of 31–45 years.
In this study, men had high culture positivity as compared with women. The result was consistent with the study done by Kaur and Singh [2014] who reported high culture positivity in 65.22% men. The finding was also similar to study by Hussein et al. [2005] that reported 66.66% positivity in men and 33.33% in women, and similar observation of male dominance − 86.92% over 13.08% in female – was reported in study by Salari [2002]. Men are the active and are the main earning members of most families, so they are more privileged to visit physician chamber for treatment. However, Zenebe et al. [2011] reported more high culture positivity in women, 59.2%, than men, 40.8%, in their study.
In the present study, maximum number of blood culture positivity was found in the ICUs, especially medical intensive care unit (MICU) followed by wards and OPD as determined by both blood cultures. This is likely to be due to the admission of the patients more in ICU and wards as bloodstream infections are a serious illnesses and have to be monitored round the clock in the hospital.
In this study, maximum number of GPC which were isolated by the BACTEC BD 9050 blood culture system was of S. aureus 13 (52%) followed by coagulase-negative Staphylococcus spp. 8 (32%), Enterococcus spp. 2 (8%), and Streptococcus pneumoniae 2 (8%), whereas the maximum number of GNB which were isolated were E. coli (40.91%) followed by Klebsiella spp. (27.27%), P. aeruginosa (22.73%), and Citrobacter spp. (9.09%). These above findings were consistent with the studies done by Fayyaz et al. [2013] and Karlowsky et al. [2004] who reported maximum number of E. coli in GNB in their studies. Similar findings were also found in the studies done by Karunakaran et al. [2007] and Aiken et al. [2011] for GNB. However, Karlowsky et al. [2004] and Karunakaran et al. [2007] reported more of coagulase-negative Staphylococci in their study, and Kaur and Singh [2014] reported higher prevalence of Salmonella typhi among GNB in their study.
In this study, Candida spp. 6 (11.32%) were isolated by the BACTEC BD 9050 blood culture system out of which non albicans Candida spp. were 4 (7.55%) and Candida albicans were 2 (3.77%). The results were consistent with study by Karunakaran et al. [2007] who reported positivity rate of 3.8% for Candida spp. with predominance of non albicans Candida spp. and with Kohli-Kochhar et al. [2011] who reported a rate of 3% for yeast isolates.
In this study, it was seen that GPC were 100% sensitive to vancomycin and linezolid. The results were consistent with the studies done by Fayyaz et al. [2013] and Marshall et al. [1998]. However, the results were inconsistent with the study done by Kaur and Singh [2014] who reported sensitivity of 57.14% to vancomycin. The sensitivity rate of amikacin was 85.71% for methicillin-sensitive S. aureus, 33.33% for methicillin-resistant S. aureus, 100% for coagulase-negative Staphylococcus spp. and S. pneumoniae, and 0% for Enterococcus spp. This result was consistent for methicillin-sensitive S. aureus and coagulase-negative Staphylococcus spp. with the study by Fayyaz et al. [2013] who reported sensitivity of amikacin to be 93.33%. The sensitivity rate of clindamycin is 85.71% for methicillin-sensitive S. aureus, 50% for methicillin-resistant S. aureus, and 87.5% for coagulase-negative Staphylococcus spp., which were consistent with the study by Marshall et al. [1998].
In this study, the sensitivity rates of ciprofloxacin was 87.5% for coagulase-negative Staphylococcus spp., 85.71% for methicillin-sensitive S. aureus, 16.66% for methicillin-resistant S. aureus, and 0% for Enterococcus spp., which were consistent with the study by Marshall et al. [1998].
In addition to the above, cefoxitin was 100% sensitive for coagulase-negative Staphylococcus spp., methicillin-sensitive S. aureus, Enterococcus spp., and S. pneumoniae, and 100% resistant for methicillin-resistant S. aureus.
All the GPC showed 100% resistance to penicillin G except S. pneumoniae, which were 100% sensitive. In the present study, among GNB, imipenem showed 88.88% sensitivity to E. coli, 100% to Klebsiella spp., and 50% to Citrobacter spp. The results for Klebsiella spp. were consistent with the study done by Saghir et al. [2009] who reported 96% sensitivity of imipenem for Enterobacteriacae family but was not compatible with E. coli and Citrobacter spp. The results of E. coli and Klebsiella spp. were also consistent with the study done by Jyothi et al. [2013] who reported sensitivity of 93% for E. coli and Klebsiella spp. For Citrobacter spp., the results were not consistent. This might be due to the fact that only two isolates of Citrobacter spp. were isolated. However, the results were not consistent with the findings of Fayyaz et al. [2013] who reported sensitivity of imipenem to be 59.2% for GNB.
In the present study, ceftriaxone showed 22.22% sensitivity to E. coli, 33.33% to Klebsiella spp., and 100% to Citrobacter spp. These findings for E. coli and Klebsiella spp. were consistent with the studies done by Saghir et al. [2009] and Fayyaz et al. [2013] who reported 28% and 22.44%, respectively, sensitivity to Enterobacteriacae family. However, the findings of Citrobacter spp. for ceftriaxone are consistent with the study done by Zenebe et al. [2011] who showed 100% sensitivity to ceftriaxone. The observation of ceftriaxone resistance pattern is suggestive of the fact that 77% E. coli and 66% of Klebsiella spp. isolates were extended spectrum beta-lactamase (ESBL) producers.
In this study, sensitivity to amikacin for E. coli, Klebsiella spp., and Citrobacter spp. was 66.66%, 83.33%, and 100%, respectively. The results for Klebsiella spp. were consistent with the study by Fayyaz et al. [2013] but not consistent for E. coli and Klebsiella spp. showing higher sensitivity of 85.71%, and were consistent with the studies by Kaur and Singh [2014] for Citrobacter spp.
In this study, E. coli, Klebsiella spp., and Citrobacter spp. showed 88.88%, 83.33%, and 100% sensitivity to piperacillin/tazobactam, respectively. This finding was consistent with the study by Karlowsky et al. [2002] who reported a sensitivity of 89.9% for Klebsiella spp., but not compatible for E. coli and Citrobacter spp.
In this study, E. coli and Klebsiella spp. showed 22.22% and 33.33% sensitivity, respectively, to gentamycin, and Citrobacter spp. were 100% resistance to gentamycin; sensitivity to ciprofloxacin was 33.33% for E. coli and Klebsiella spp. and was 50% for Citrobacter spp. The results were consistent for E. coli and Klebsiella spp. with the study done by Fayyaz et al. [2013] for gentamycin and ciprofloxacin, which showed sensitivity of 32.7% and 26.53%, respectively.
In this study, E. coli, Klebsiella spp., and Citrobacter spp. showed 66.66%, 16.66%, and 100% sensitivity, respectively, to cefepime. This was consistent with the study by Saghir et al. [2009] for Klebsiella spp. who reported 20% sensitivity of for Enterobacteriacae family to cefepime. However, the findings were not consistent for E. coli and Citrobacter spp.
In this study, E. coli, Klebsiella spp., and Citrobacter spp showed 100% resistance to ampicillin. The results were consistent with the study done by Jyothi et al. [2013] showing 97% resistance to ampicillin. Also the results were consistent with the study done by Fayyaz et al. [2013] whose study showed that E. coli, Klebsiella spp. and Citrobacter spp. were 93.88% resistant to ampicillin.
In this study, P. aeruginosa showed 40% sensitivity to aztreonam, 60% sensitivity to ciprofloxacin, and 80% sensitivity to amikacin. This finding was consistent with the study by Hafsa et al. [2011] for ciprofloxacin and amikacin, that is, 50% sensitivity to ciprofloxacin and 75% sensitivity to amikacin, and was also consistent with the study by Fayyaz et al. [2013], that is, 37.5% sensitivity to aztreonam, 60% sensitivity to ciprofloxacin, and 72.5% sensitivity to amikacin.
In this study, P. aeruginosa showed 100% sensitivity to imipenem, which was consistent with the study by Hafsa et al. [2011] that reported a sensitivity of 100%.
In this study, P. aeruginosa showed 60% sensitivity to ceftazidime, which was consistent with the study done by Fayyaz et al. [2013], reporting a sensitivity of 60%, and by Rabirad et al. [2014], reporting a sensitivity of 65.8% to ceftazidime. In this study, P. aeruginosa showed 80% sensitivity to piperacillin/tazobactam, which is consistent with the study done by Fayyaz et al. [2013], reporting a sensitivity of 70%.
There was variation in the antibiotic sensitivity rate of various organisms isolated in the present study when compared to different past studies. This may be due to the fact that sensitivity of organisms to antibiotics is variable and depends upon prevalence of strains, antibiotics use, and its resistance patterns in a particular area.
Footnotes
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Conflict of interest statement: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Contributor Information
Nikita Vasudeva, Department of Microbiology, NIMS UNIVERSITY Shobha Nagar, Jaipur-Delhi Highway (NH-11C), Jaipur - 303121, Jaipur, Rajasthan, India.
Prem Singh Nirwan, National Institute of Medical Sciences & Research, NIMS University, Jaipur, India.
Preeti Shrivastava, National Institute of Medical Sciences & Research, NIMS University, Jaipur, India.
References
- Aiken A., Mturi N., Njuguna P., Mohammed S., Berkley J., Mwangi I., et al. (2011) Risk and causes of paediatric hospital-acquired bacteraemia in Kilifi District Hospital, Kenya: a prospective cohort study. Lancet 378: 2021–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayan R., Paniker C. (2013) Ananthanarayan and Paniker’s Textbook of Microbiology, 9th edn. Hyderabad, India: Universities Press, pp. 49–53, pp. 661–663. [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI) (2013) Performance standards for antimicrobial susceptibility testing (Twenty third informational supplement). CLSI document M100-S23, vol. 33 Wayne, PA: CLSI. [Google Scholar]
- Collee J., Fraser A., Marmion B., Simmons A. (1996) Mackie and McCartney Practical Medical Microbiology, 14th edn. New York: Churchill Livingstone, pp. 131–144. [Google Scholar]
- Fayyaz M., Mirza I., Ikram A., Hussain A., Ghafoor T., Shujat U. (2013) Pathogens causing blood stream infections and their drug susceptibility profile in immunocompromised patients. J Coll Physicians Surg Pak 23: 848–851. [PubMed] [Google Scholar]
- Forbes B., Sahm D., Weissfeld A. (2014) Bailey & Scott’s Diagnostic Microbiology, 13th edn. St. Louis, MO: Mosby, Inc, pp. 201–229, pp. 860–876. [Google Scholar]
- Hafsa A., Fakruddin M., Hakim M., Sharma J. (2011) Neonatal bacteremia in a neonatal intensive care unit: analysis of causative organisms and antimicrobial susceptibility. Bangladesh J Med Sci 10: 187–192. [PubMed] [Google Scholar]
- Hussein A., Sayed A., Mohamed A. (2005) Seroepidemiological study on human brucellosis in Assiut Governorate. Egypt J Immunol 12: 49–56. [PubMed] [Google Scholar]
- Jyothi P., Basavaraj M., Basavaraj P. (2013) Bacteriological profile of neonatal septicemia and antibiotic susceptibility pattern of the isolates. J Nat Sci Biol Med 4: 306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlowsky J., Jones M., Draghi D., Thornsberry C., Sahm D., Volturo D. (2004) Prevalence and antimicrobial susceptibilities of bacteria isolated from blood cultures of hospitalized patients in the United States in 2002. Ann Clin Microbiol Antimicrob 3: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunakaran R., Raja N., Ng K., Navaratnam P. (2007) Etiology of blood culture isolates among patients in a multidisciplinary teaching hospital in Kuala Lumpur. J Microbiol Immunol Infect 40: 432–437. [PubMed] [Google Scholar]
- Kaur A., Singh V. (2014) Bacterial isolates and their antibiotic sensitivity pattern in clinically suspected cases of fever of unknown origin. JK Science 16: 105–109. [Google Scholar]
- Kohli-Kochhar R., Omuse G., Revathi G. (2011) A ten-year review of neonatal bloodstream infections in a tertiary private hospital in Kenya. J Infect Dev Ctries 5: 799–803. [DOI] [PubMed] [Google Scholar]
- Marshall S., Wilke W., Pfaller M., Jones R. (1998) Staphylococcus aureus and coagulase-negative staphylococci from blood stream infections: frequency of occurrence, antimicrobial susceptibility, and molecular (mecA) characterization of oxacillin resistance in the SCOPE program. Diagn Microbiol Infect Dis 30: 205–214. [DOI] [PubMed] [Google Scholar]
- Prashanth H., Dominic Saldanha R., Shenoy S., Baliga S. (2011) Predictors of mortality in adult sepsis. Int J Biol Med Res 2: 856–861. [Google Scholar]
- Rabirad N., Mohammadpoor M., Lari A., Shojaie A., Bayat R., Alebouyeh M. (2014) Antimicrobial susceptibility patterns of the gram-negative bacteria isolated from septicemia in Children’s Medical Center, Tehran, Iran. J Prev Med Hyg 55: 23–26. [PMC free article] [PubMed] [Google Scholar]
- Saghir S., Faiz M., Saleem M., Younus A., Aziz H. (2009) Characterization and anti-microbial susceptibility of gram-negative bacteria isolated from bloodstream infections of cancer patients on chemotherapy in Pakistan. Indian J Med Microbiol 27: 341–347. [DOI] [PubMed] [Google Scholar]
- Salari M. (2002) Seroepidemiological survey of brucellosis among animal farmers of Yazd Province. Iranian J Publ Health 31: 29–32. [Google Scholar]
- Viswanathan R., Singh A., Ghosh C., Dasgupta S., Mukherjee S., Basu S. (2012) Profile of neonatal septicaemia at a district-level sick newborn care unit. J Health Popul Nutr 30: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winm W, Jr., Allen S., Janda W., Koneman E., Schreckenberger P., Procop G., et al. (2006) Koneman’s Colour Atlas and Text Book of Diagnostic Microbiology, 6th edn. Philadelphia, PA: Lippincott Williams & Wilkins Company, pp. 97–105. [Google Scholar]
- Zenebe T., Kannan S., Yilma D., Beyene G. (2011) Invasive bacterial pathogens and their antibiotic susceptibility patterns in Jimma University Specialized Hospital, Jimma, Southwest Ethiopia. Ethiop J Health Sci 21: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]