Abstract
Oligomers of the β-amyloid peptide Aβ have emerged as important contributors to neurodegeneration in Alzheimer’s disease. Mounting evidence suggests that Aβ trimers and higher-order oligomers derived from trimers have special significance in the early stages of Alzheimer’s disease. Elucidating the structures of these trimers and higher-order oligomers is paramount for understanding their role in neurodegeneration. This paper describes the design, synthesis, X-ray crystallographic structures, and biophysical and biological properties of two stabilized trimers derived from the central and C-terminal regions of Aβ. These triangular trimers are stabilized through three disulfide cross-links between the monomer subunits. The X-ray crystallographic structures reveal that the stabilized trimers assemble hierarchically to form hexamers, dodecamers, and annular porelike structures. Solution-phase biophysical studies reveal that the stabilized trimers assemble in solution to form oligomers that recapitulate some of the higher-order assemblies observed crystallographically. The stabilized trimers share many of the biological characteristics of oligomers of full-length Aβ, including toxicity toward a neuronally derived human cell line, activation of caspase-3 mediated apoptosis, and reactivity with the oligomer-specific antibody A11. These studies support the biological significance of the triangular trimer assembly of Aβ β-hairpins and may offer a deeper understanding of the molecular basis of Alzheimer’s disease.
Introduction
In Alzheimer’s disease, the β-amyloid peptide Aβ assembles to form a multitude of soluble oligomers as well as insoluble fibrils.1,2 The Aβ oligomers have emerged as neurotoxic agents that lead to neurodegeneration in Alzheimer’s disease. The heterogeneity and metastability of the Aβ oligomers presents a tremendous challenge in understanding the molecular basis of Alzheimer’s disease. Specifically, the lack of homogeneous oligomers precludes detailed correlation of the biological properties of Aβ oligomers with their structural and biophysical properties.
To reduce the heterogeneity among assemblies of the Aβ peptide, researchers have prepared and studied Aβ oligomers that consist of Aβ monomers linked by chemical cross-links.3−9 These studies have helped determine the importance of different residues in Aβ oligomerization and have demonstrated that Aβ40 and Aβ42 form different types of oligomers. Cross-linked oligomers have been found to be toxic toward rat pheochromocytoma (PC12) cells and to inhibit long-term potentiation in rats, providing evidence for the role of Aβ oligomers in neurodegeneration in Alzheimer’s disease. Although cross-linking Aβ decreases the heterogeneity of Aβ oligomers, cross-linking has not yet produced structurally homogeneous oligomers. The high-resolution structures of the cross-linked oligomers that have been generated thus far, through either a single disulfide bond or through photoinduced cross-linking of unmodified proteins (PICUP), remain unknown.
In the past couple of years, our laboratory has identified and elucidated hitherto undiscovered modes of supramolecular assembly of macrocyclic β-sheet peptides derived from amyloidogenic peptides and proteins.10−13 We previously reported the X-ray crystallographic structures of two homologous trimers formed by two macrocyclic β-sheet peptides derived from Aβ17–36.10,14 These peptides contain Aβ17–23 and Aβ30–36 β-strands covalently linked by two δ-linked ornithine (δOrn) turn mimics and are designed to mimic an Aβ17–36 β-hairpin.15Figure 1 illustrates these peptides, 1 and 2, and shows their relationship to an Aβ17–36 β-hairpin. The δOrn that connects residues D23 and A30 replaces the Aβ24–29 loop; the δOrn that connects residues L17 and V36 reinforces β-sheet structure.16 We incorporated ornithine (α-linked) as a hydrophilic isostere of methionine at position 35 to improve the solubility of the peptides.14 Peptides 1 and 2 both contain an N-methyl group to block uncontrolled aggregation: peptide 1 contains an N-methyl group on G33; peptide 2 contains an N-methyl group on F20.
Figure 1.
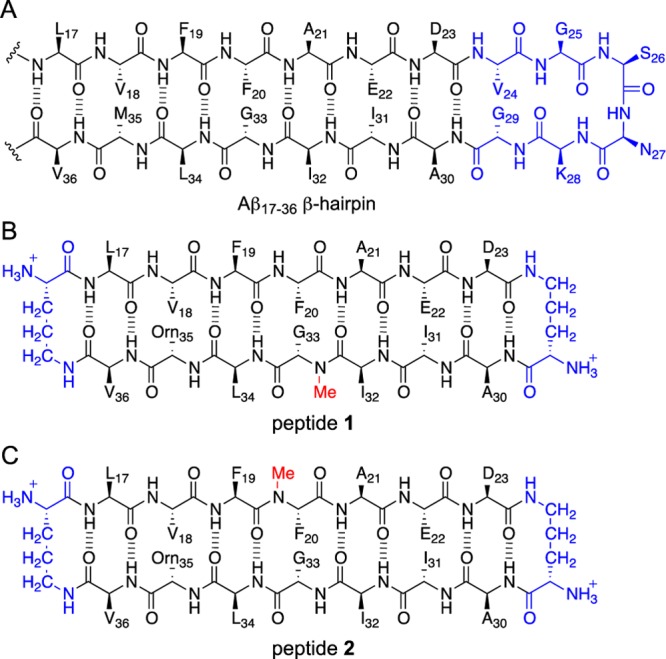
Chemical structures of an Aβ17–36 β-hairpin and peptides 1 and 2. The Aβ24–29 loop region of the Aβ17–36 β-hairpin is shown in blue to illustrate its relationship to the δOrn that connects D23 and A30 in peptides 1 and 2.
X-ray crystallography revealed that peptides 1 and 2 fold to form β-hairpins that assemble to form oligomers. In the X-ray crystallographic structures of peptides 1 and 2, three β-hairpins assemble in a triangular fashion to form trimers, which are stabilized by hydrogen bonding and hydrophobic interactions between monomers (Figure 2). At the three corners of each trimer, the main chain of residue V18 on one macrocyclic β-sheet hydrogen bonds with the main chain of residue E22 on the adjacent macrocyclic β-sheet. Clustering between hydrophobic residues at the corners of each trimer provides additional stability. In the crystal lattice, the trimers further assemble to form hexamers and dodecamers. The trimers, hexamers, and dodecamers formed by peptide 1 are morphologically identical to the trimers, hexamers, and dodecamers formed by peptide 2.
Figure 2.
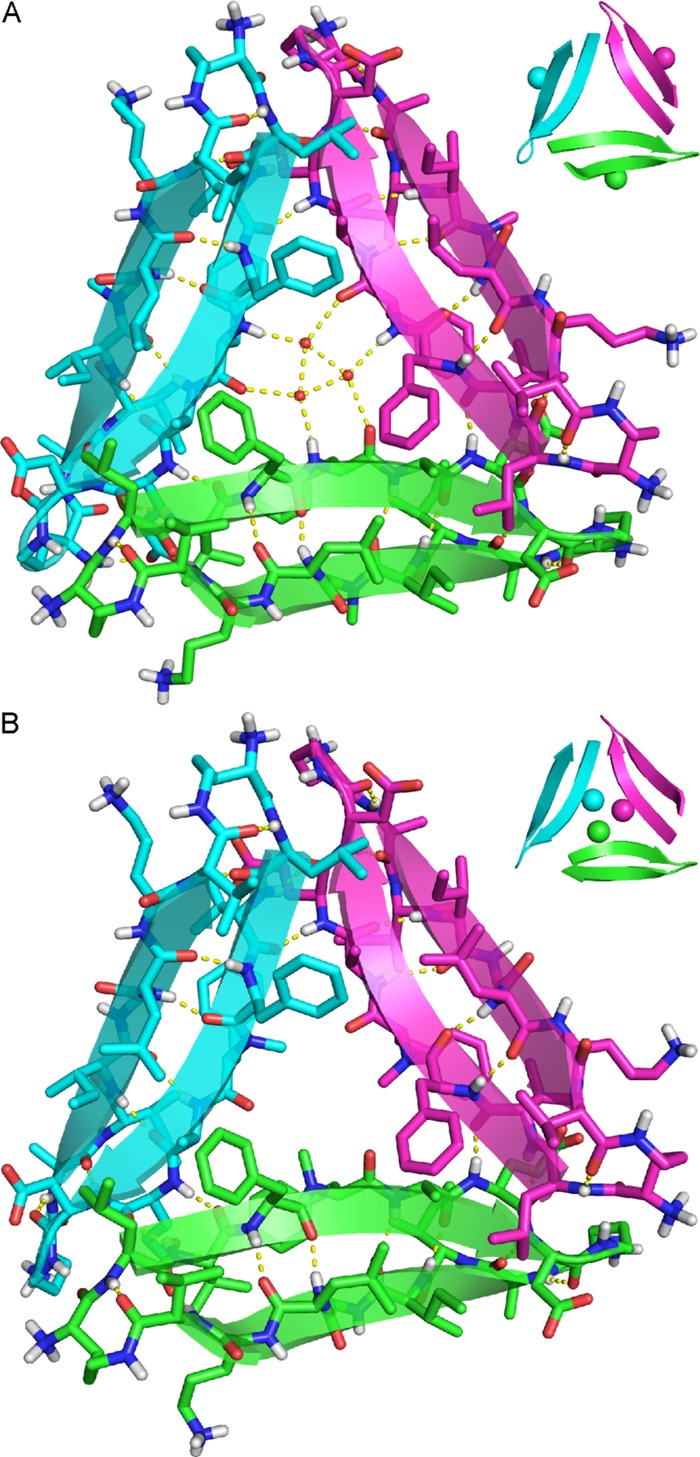
(A) X-ray crystallographic structure of the triangular trimer formed by peptide 1 (PDB 4NTR). The three ordered water molecules in the center of the trimer that form hydrogen bonds with the main chain of F20 are shown as small red spheres. In the inset, the N-methyl groups on G33 are shown as spheres. (B) X-ray crystallographic structure of the triangular trimer formed by peptide 2 (PDB 4NW9). In the inset, the N-methyl groups on F20 are shown as spheres.
The oligomers formed by peptides 1 and 2 are labile and dynamic in aqueous solution, making it difficult to correlate their biological and biophysical properties with their X-ray crystallographic structures. In the current study, we aimed to covalently stabilize the trimers formed by peptides 1 and 2 through chemical cross-linking, with the goal of investigating the biological significance of this triangular assembly (Chart 1). This paper describes the design, synthesis, and study of cross-linked trimers 5 and 6 (Figure 3). Peptides 3 and 4 are generated as cysteine-containing homologues of peptides 1 and 2 and are cross-linked to form trimers 5 and 6. The X-ray crystallographic structures of trimers 5 and 6 are determined and the higher-order assemblies that they form are elucidated at high resolution. Trimers 5 and 6 are also shown to form higher-order oligomers in aqueous solution that are toxic toward the human neuroblastoma cell line SH-SY5Y.
Chart 1. Design of Trimers 5 and 6.
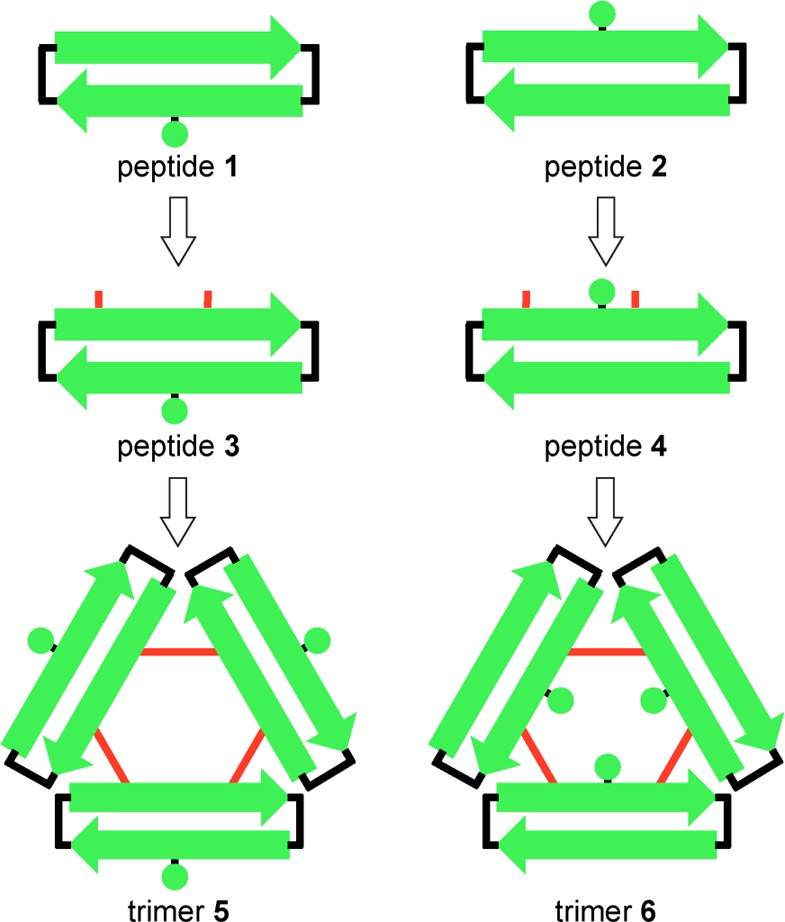
Figure 3.
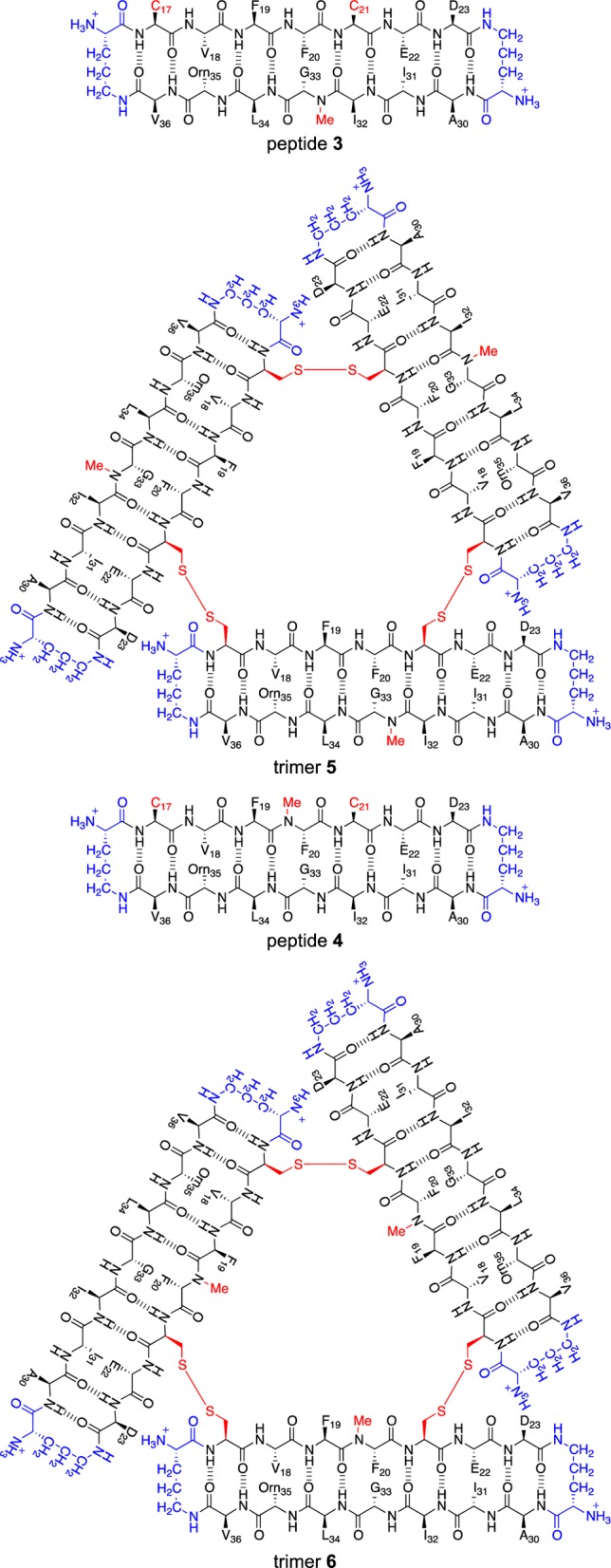
Chemical structures of peptides 3 and 4 and trimers 5 and 6.
Results
Design and Synthesis of Peptides 3 and 4 and Trimers 5 and 6
The X-ray crystallographic structures of the trimers formed by peptides 1 and 2 revealed a strategy for cross-linking these peptides into stable trimers. At the three corners of the triangular trimers, the side chain of residue L17 of one monomer subunit packs against the side chain of residue A21 of another monomer subunit. We hypothesized that mutating both L17 and A21 to cysteine would allow cross-linking the peptides to form covalent trimers containing three disulfide linkages. The resulting C17–C21 cross-links would be almost isosteric with L17 and A21, maintaining a similar level of hydrophobicity and not altering the charge of the trimer.
We synthesized peptides 3 and 4 by similar procedures to those we have developed for other macrocyclic β-sheet peptides: synthesis of the corresponding linear peptide on 2-chlorotrityl resin, followed by cleavage of the protected linear peptide from the resin, solution-phase macrocyclization, and global deprotection of the resulting macrocyclic peptide.10−13,17 We purified peptides 3 and 4 by reverse-phase HPLC (RP-HPLC) followed by lyophilization of pure fractions. Typical syntheses on a 0.1 mmol scale afforded ∼55 mg of peptides 3 and 4 in ≥95% purity. We rigorously purified peptides 3 and 4 to minimize off-target products in the subsequent oxidation reactions.
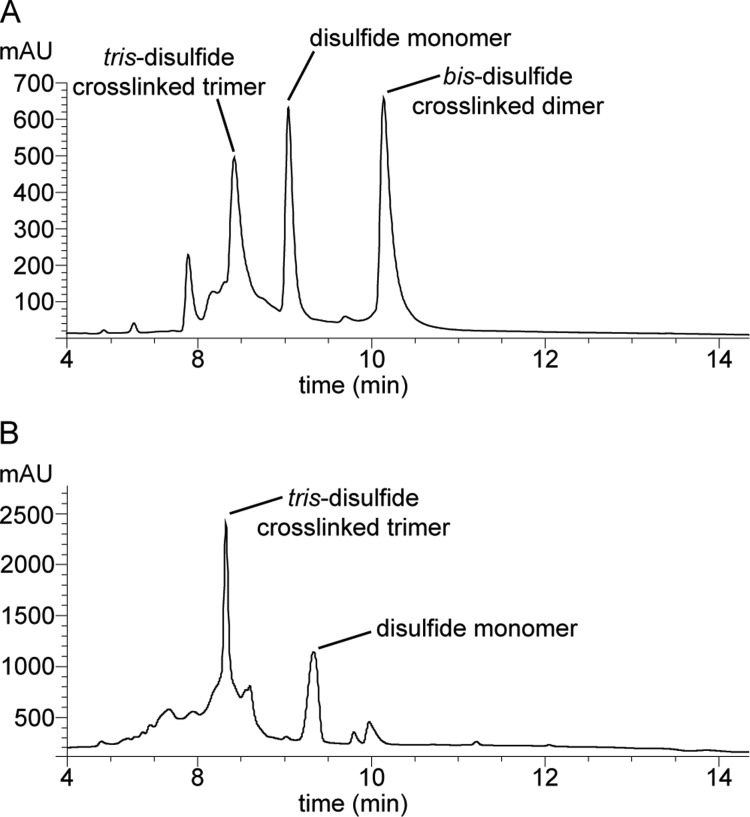
We anticipated that oxidation of peptides 3 and 4 to form trimers would be challenging. The peptides have the potential to form complex mixtures of monomeric, dimeric, trimeric, and higher oligomeric oxidation products. Five different oxidation products of trimer size or smaller are possible in the oxidation reactions of peptides 3 and 4: (1) a monomer that contains an intramolecular disulfide bond between C17 and C21, (2) an antiparallel bis-disulfide cross-linked dimer, (3) a parallel bis-disulfide cross-linked dimer, (4) an asymmetric tris-disulfide cross-linked trimer, and (5) a symmetric tris-disulfide cross-linked trimer (Figure 4). The desired trimers 5 and 6 are symmetric tris-disulfide cross-linked trimers.
Figure 4.
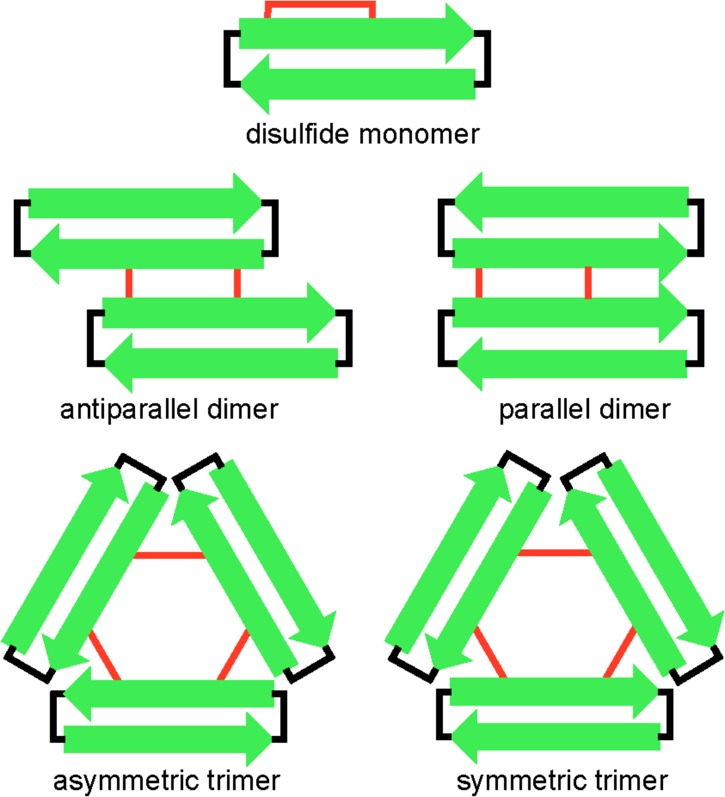
Cartoon illustrating the anticipated products of trimer size or smaller in the oxidation reactions of peptides 3 and 4.
We developed a two-step procedure for preparing trimers 5 and 6 from peptides 3 and 4. In the first step, we allow peptides 3 and 4 to oxidize at relatively high concentration of peptide (6 mM) in 20% (v/v) aqueous DMSO for 48 h.18,19 In the second step, we dilute the reaction mixture with water to a low concentration (∼250 μM) and allow the oxidized peptides to equilibrate over 48 h. Through this procedure, peptides 3 and 4 cross-link to form substantial amounts of the desired symmetric cross-linked trimers 5 and 6. In the oxidation reaction of peptide 3, we observe three major products: trimer 5, a cross-linked dimer, and the disulfide monomer (Figure 5A).20 In the oxidation reaction of peptide 4, we observe two major products: trimer 6 and the disulfide monomer; we do not observe appreciable amounts of either possible cross-linked dimer (Figure 5B). We purified trimers 5 and 6 by RP-HPLC followed by lyophilization of pure fractions to yield ∼15 mg of trimer 5 and ∼20 mg of trimer 6, each with ≥95% purity, from a 0.1 mmol scale synthesis of peptides 3 and 4.
Figure 5.
Analytical RP-HPLC traces of the mixture of products formed upon oxidation of peptide 3 (A) and peptide 4 (B). Analytical RP-HPLC was performed on a C18 column with an elution gradient of 5–95% CH3CN over 20 min.
X-ray Crystallographic Structure Determination of Trimers 5 and 6
We elucidated the structures of trimers 5 and 6 by X-ray crystallography. One of the challenges in X-ray crystallography is determining the X-ray crystallographic phases. Doing so often requires incorporation of a heavy atom, such as selenium, bromine, or iodine, through covalent modification.21 In previously solving the X-ray crystallographic structures of peptides 1 and 2, we prepared homologues containing p-iodophenylalanine. In solving the X-ray crystallographic structures of trimers 5 and 6, we employed two techniques for X-ray crystallographic phase determination that have not been widely used for peptides: sulfur single-wavelength anomalous diffraction (S-SAD) and postcrystallization incorporation of iodide ions into the crystal lattice.
We used S-SAD to determine the X-ray crystallographic structure of trimer 6. The intrinsic anomalous scattering of the sulfur atoms in the asymmetric unit provided sufficient data to determine the X-ray crystallographic phases. We collected five data sets from a single crystal of trimer 6 using an X-ray diffractometer equipped with a rotating copper anode, and we merged the data sets to increase the strength of the anomalous signal from sulfur.22,23 We then used the X-ray crystallographic structure generated by S-SAD (PDB 5SUS) as a search model for molecular replacement to solve the X-ray crystallographic phases of a higher resolution data set collected using a synchrotron radiation source (PDB 5SUR).
We used iodide ion incorporation and conventional SAD phasing to determine the X-ray crystallographic structure of trimer 5. To incorporate the iodide ions into the crystal lattice we soaked a crystal of trimer 5 in a mixture of crystallization buffer and aqueous potassium iodide (KI).24 The X-ray crystallographic structure of the KI-soaked trimer 5 (PDB 5SUU) was used as a search model for molecular replacement to determine the X-ray crystallographic phases of a higher resolution data set of unsoaked trimer 5 collected using a synchrotron radiation source (PDB 5SUT).
X-ray Crystallographic Structure and Supramolecular Assembly of Trimer 5
The X-ray crystallographic structure of trimer 5 reveals the hypothesized trimer, with three disulfide linkages between the monomeric subunits (Figure 6).25 As we envisioned, replacement of L17 and A21 with cysteine does not perturb the triangular trimer structure. Trimer 5 is composed of three folded macrocyclic β-sheets and is virtually identical to the trimers formed by peptides 1 and 2. Trimer 5 maintains the intersheet hydrogen bonds and hydrophobic clustering of amino acid side chains previously described for the trimers formed by peptides 1 and 2.10 At each corner of trimer 5, the main chain of residue V18 on one monomeric subunit hydrogen bonds with the main chain of residue E22 on the adjacent monomeric subunit (Figure 9A).
Figure 6.
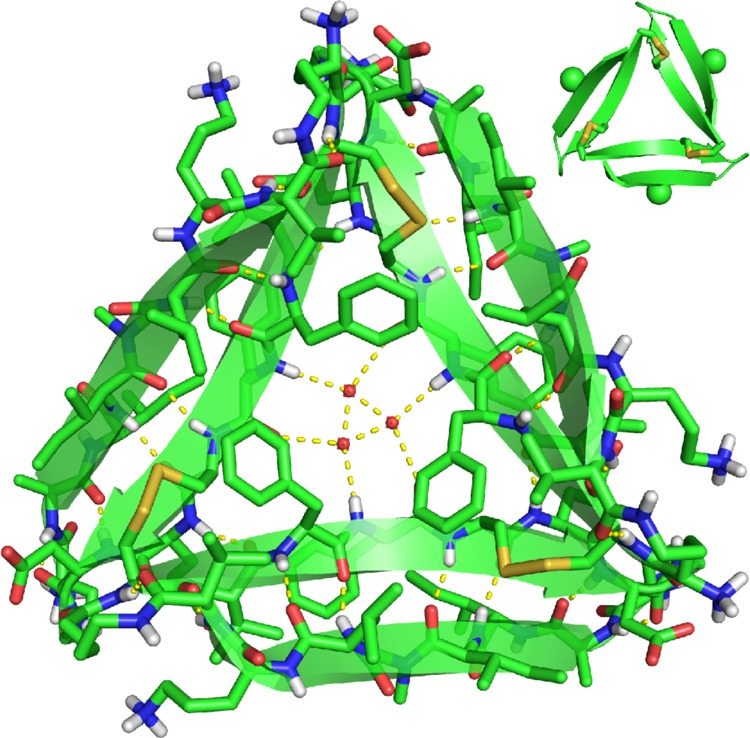
X-ray crystallographic structure of trimer 5 (PDB 5SUT). In the inset, the N-methyl groups on G33 are shown as spheres.
Figure 9.
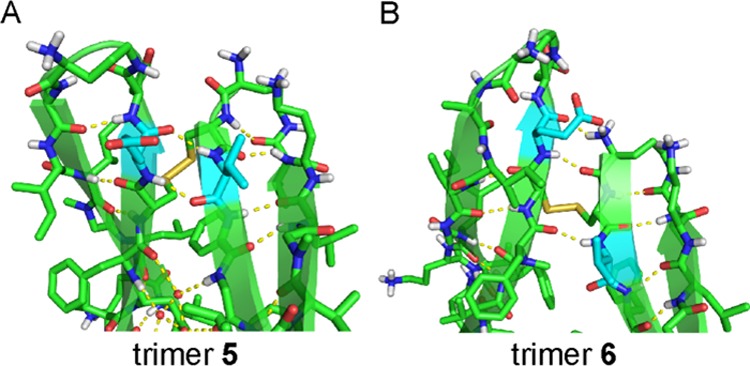
Contacts between the monomer subunits in trimer 5 and trimer 6. (A) Trimer 5. Residues V18 and E22 (highlighted in cyan) are aligned. (B) Trimer 6. Residues V18 and E22 (highlighted in cyan) are shifted out of alignment by two residues. The side chain of one F20 residue on trimer 5 is omitted for clarity.
The N-methyl groups in trimer 5 are located on the outer hydrogen-bonding edges of the trimer. These N-methyl groups block the outer hydrogen-bonding edges of the trimer from hydrogen bonding with other trimers in the crystal lattice. Three ordered water molecules fill the hole in the center of trimer 5, hydrogen bonding with each other and with the main chain of residue F20.
Clusters of hydrophobic residues in trimer 5 create two hydrophobic surfaces (Figure S1). The front surface displays the side chains of residues F19, I32, L34, and V36, as well as the C17–C21 disulfide linkages. We term this surface the “F19 face”. The back surface displays the side chains of residues V18, F20, and I31. We term this face the “F20 face”. Trimer 5 packs on both the F19 face and the F20 face to form higher-order assemblies in the crystal lattice.
In the X-ray crystallographic structure of trimer 5, two trimers pack to form a sandwich-like hexamer (Figure 7). In the hexamer, the F20 face of one trimer packs against the F20 face of another trimer (Figure 7B). The hexamers further assemble to form columns by stacking on their F19 faces (Figure 7C). The columns are arranged in a hexagonal fashion in the crystal lattice (Figure S2). The hexamer formed by trimer 5 is morphologically identical to the hexamers formed by peptides 1 and 2.
Figure 7.
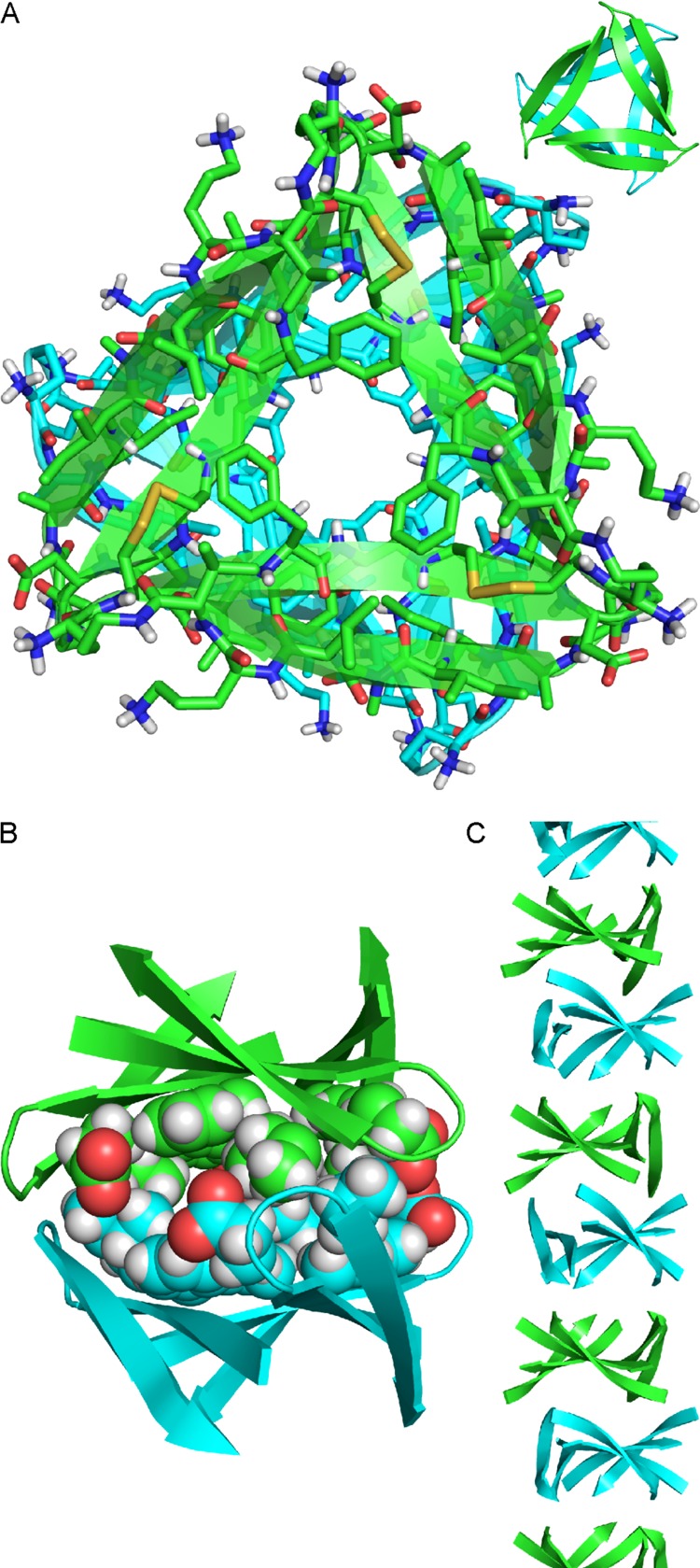
X-ray crystallographic structure of the sandwich-like hexamer formed by trimer 5. (A) Top view. (B) Side view. The side chains of residues F20, I31, and E22 are shown as spheres to illustrate the hydrophobic packing that occurs at the interface between the two trimers. (C) Column of stacked hexamers in the crystal lattice.
This mode of assembly, in which the hydrophobic faces displayed on triangular trimers pack together to form hexamers, appears to be characteristic of triangular trimers formed by amyloid-derived macrocyclic β-sheets and β-hairpins. Our laboratory has also observed this mode of assembly by a larger peptide derived from Aβ17–36 and by a macrocyclic β-sheet peptide derived from β2-microglobulin.11,16
X-ray Crystallographic Structure and Supramolecular Assembly of Trimer 6
The X-ray crystallographic structure of trimer 6 reveals a symmetric trimer that is cross-linked through disulfide linkages between C17 of one monomeric subunit and C21 of the adjacent monomeric subunit (Figure 8).26 Although trimer 6 is composed of three folded macrocyclic β-sheets, it differs in conformation from the trimers formed by peptides 1 and 2, and also differs in conformation from trimer 5. In the three other trimers, the main chains of residues V18 and E22 are hydrogen bonded at the corners of the trimer. In trimer 6 residues V18 and E22 shift out of alignment by two residues, such that residue V18 is across from residue F20 and residue E22 is across from δOrn (Figure 9B). In further contrast to trimer 5, the N-methyl groups in trimer 6 are sequestered in the center hole of the trimer, exposing the outer hydrogen-bonding edges and allowing trimer 6 to hydrogen bond with other trimers in the crystal lattice.
Figure 8.
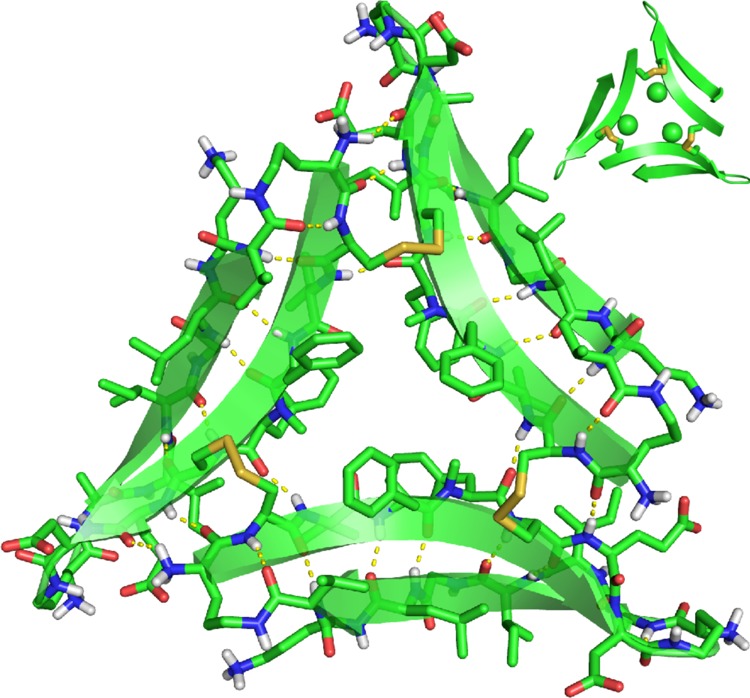
X-ray crystallographic structure of trimer 6 (PDB 5SUR). In the inset, the N-methyl groups on F20 are shown as spheres.
Clusters of hydrophobic residues in trimer 6 create two hydrophobic surfaces, which we term the “F19 face” and the “F20 face” (Figure S1). The F19 face displays the hydrophobic side chains of residues F19, I32, L34, and V36, as well as the C17–C21 disulfide linkages. The F20 face displays the hydrophobic side chains of residues V18, F20, and I31.
In the X-ray crystallographic structure of trimer 6, four trimers assemble in a tetrahedral fashion to form a ball-shaped dodecamer (Figure 10). The dodecamer is stabilized by a network of hydrogen bonds among the outer edges of the four trimers: the main chains of residues G33 and Orn35 on one trimer hydrogen bond with the main chains of residues I31 and δOrn on the adjacent trimers. The hydrophobic residues on the F20 faces of the four trimers line the inside of the dodecamer, creating a hydrophobic cavity approximately 2 nm in diameter.27
Figure 10.
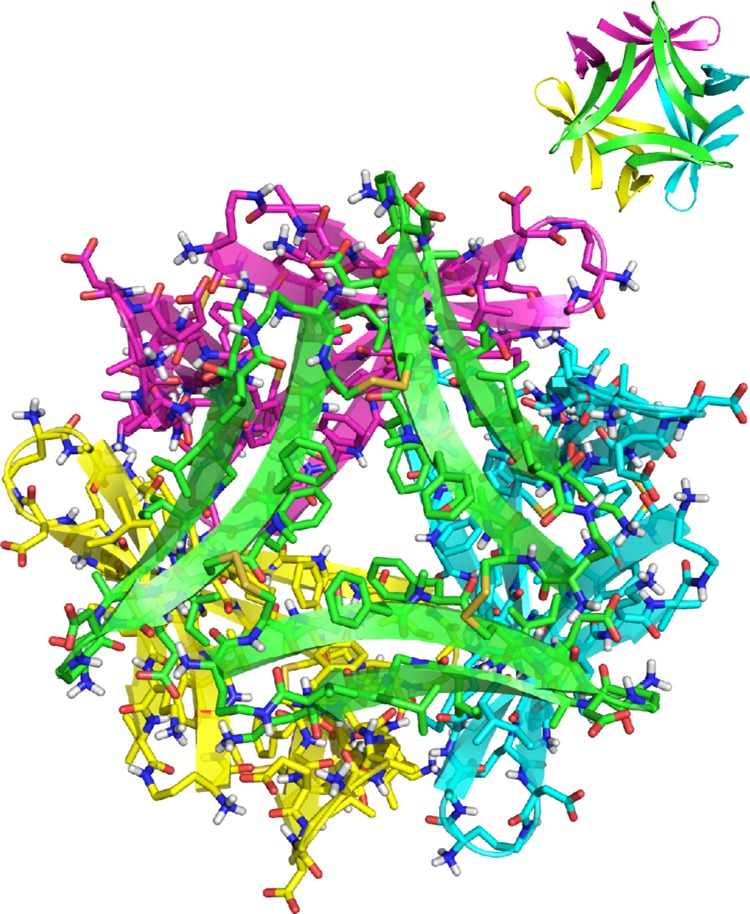
X-ray crystallographic structure of the ball-shaped dodecamer formed by trimer 6.
The ball-shaped dodecamers pack to form the crystal lattice. Within the crystal lattice, six dodecamers assemble to form annular porelike structures (Figure 11A). Hydrophobic packing between the F19 faces displayed on the exterior of each dodecamer stabilizes these annular porelike structures. At the interfaces between the dodecamers in the annular pore, the trimers pack to form sandwich-like hexamers (Figure 11B,C). The interfaces are stabilized by hydrophobic packing between the side chains of residues on the F19 faces of each trimer.
Figure 11.
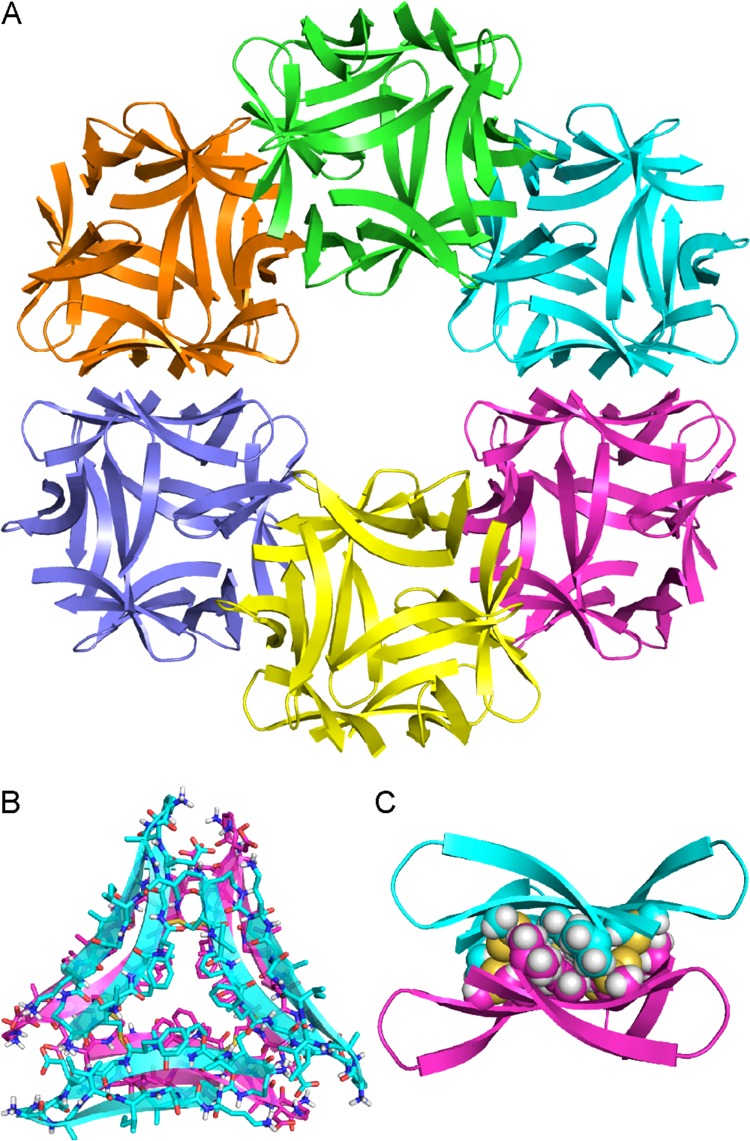
(A) X-ray crystallographic structure of an annular pore formed by trimer 6. (B) Sandwich-like hexamer formed by the trimers at the interface between two dodecamers in the annular pore (top view). (C) Side view of a sandwich-like hexamer. The side chains of residues on the F19 faces of the trimers are shown as spheres to illustrate the hydrophobic packing at the interface.
As illustrated above, trimer 5 and trimer 6 form different higher-order assemblies within the crystal lattice. Trimer 5 packs to form sandwich-like hexamers; trimer 6 assembles to form ball-shaped dodecamers that pack to form annular pores. The difference in the position of the N-methyl groups on the two trimers may explain the differences in the assemblies that form. In trimer 6, the N-methyl group on residue F20 is sequestered in the center hole of the trimer, exposing the outer hydrogen-bonding edges and allowing trimer 6 to hydrogen bond with the three other trimer 6 subunits that comprise the ball-shaped dodecamer. In trimer 5, the N-methyl group on residue G33 inhibits dodecamer formation by blocking hydrogen bonding with other trimers. Instead, trimer 5 forms a sandwich-like hexamer that is primarily stabilized by packing between the hydrophobic surfaces of the two trimers.
Biological Studies of Trimers 5 and 6
Trimers 5 and 6 provide tools to investigate the biological significance of the triangular assembly. We compared trimers 5 and 6 and peptides 1 and 2 in a series of biological and biophysical experiments to evaluate the effect of covalent stabilization of the trimers and to correlate differences in biological and solution-phase behavior with differences in structure.
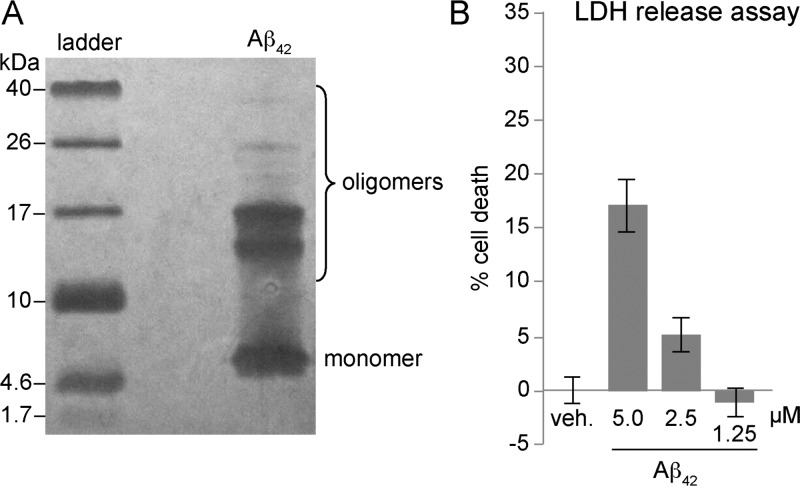
Aβ is known to be toxic toward neurons and neuronally derived cells.1,28 To corroborate the toxicity of Aβ, we prepared oligomers of Aβ42 and studied their toxicity toward the human neuroblastoma cell line SH-SY5Y. Aβ oligomers were prepared according to the procedure developed by Teplow and co-workers using recombinantly expressed Aβ42 pretreated with NH4OH (purchased from rPeptide).29,30 Under the conditions of the oligomer preparation, Aβ42 appears as a mixture of oligomers as assessed by SDS-PAGE (Figure 12A). We treated SH-SY5Y cells with varying concentrations of the mixture of Aβ42 oligomers and evaluated toxicity using a lactate dehydrogenase (LDH) release assay. The Aβ42 increased LDH release in a dose-dependent manner at concentrations as low as 2.5 μM, corroborating the toxicity of Aβ42 observed by other laboratories (Figure 12B).
Figure 12.
Aβ42 forms a mixture of oligomers and is toxic toward SH-SY5Y cells. (A) Silver-stained SDS-PAGE gel. Aβ42 was run at 250 μM. (B) Lactate dehydrogenase (LDH) release assay. Data represent the mean of five replicate wells ± s.d. Deionized water (vehicle, veh.) was used as a negative control.
LDH Release Assay
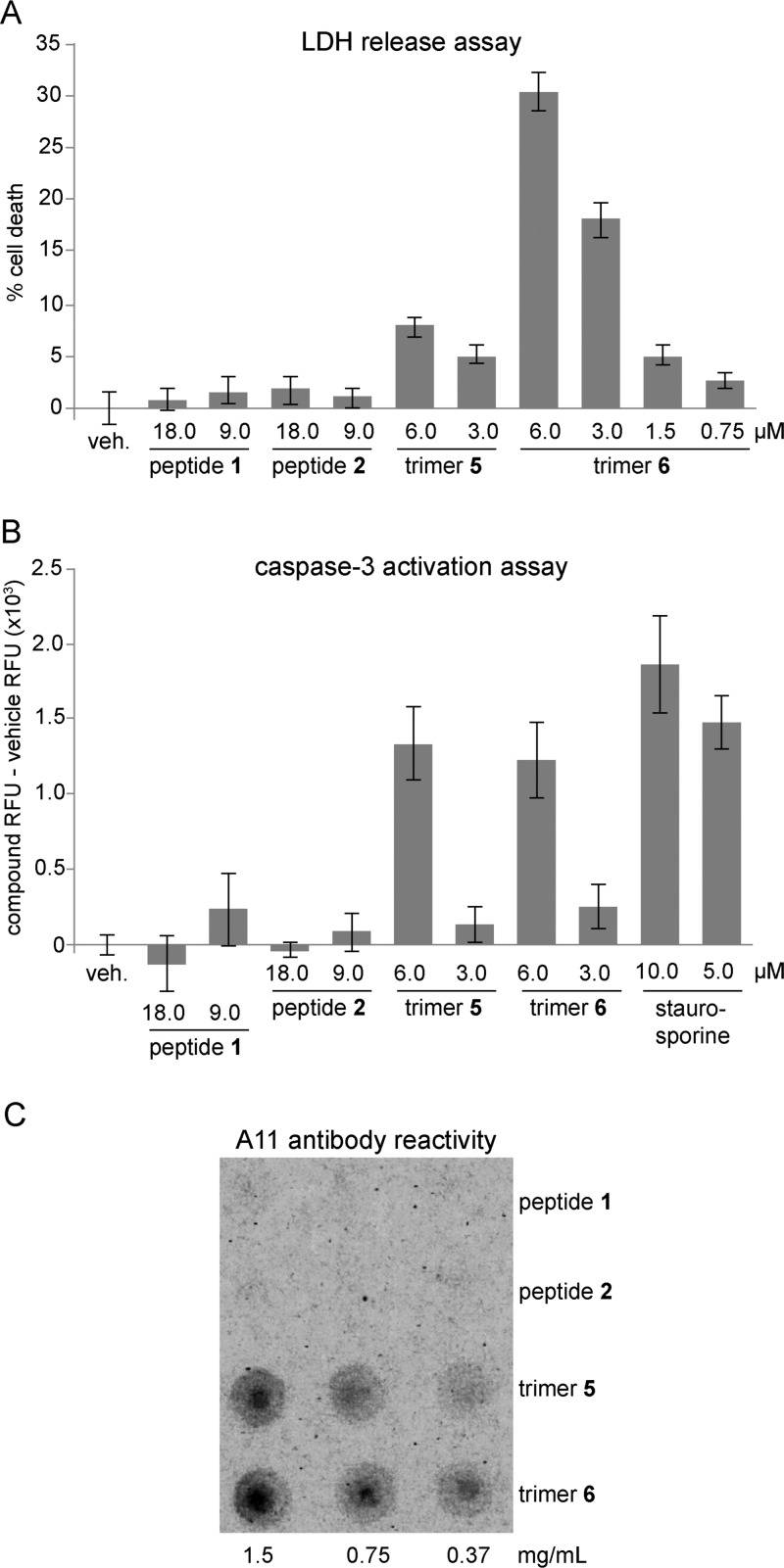
To test whether trimers 5 and 6 elicit toxicity similar to Aβ42, we evaluated the toxicity of the trimers toward SH-SY5Y cells using an LDH release assay. Deionized water (vehicle) and peptides 1 and 2 were used as controls. Trimer 6 increased LDH release in a dose-dependent manner at concentrations as low as 1.5 μM, indicating toxicity toward SH-SY5Y cells (Figure 13A). LDH release was observed as early as 48 h after addition to the cells and reached a maximum after 72 h (Figure S3). The toxicity of trimer 6 does not arise from in situ reduction to peptide 4, as peptide 4 showed no toxicity in LDH release assays (Figure S4). At equivalent concentrations, trimer 5 exhibited less toxicity than trimer 6, eliciting LDH release at concentrations as low as 3 μM. In contrast, monomeric peptides 1 and 2 showed little or no LDH release.
Figure 13.
Biological studies of trimers 5 and 6 and peptides 1 and 2. (A) LDH release assay. Data represent the mean of five replicate wells ± s.d. Deionized water (vehicle, veh.) was used as a negative control. (B) Caspase-3 activation assay. Data represent the mean of five replicate wells ± s.d. Staurosporine was used as a positive control. (C) Dot blot analysis of A11 antibody reactivity of trimers 5 and 6 and peptides 1 and 2.
Caspase-3 Activation Assay
One way in which Aβ oligomers elicit toxicity is by inducing caspase-3 mediated apoptosis.31,32 We used a rhodamine-based caspase-3 activity assay to evaluate whether trimers 5 and 6 also induce caspase-3 mediated apoptosis. At 6 μM, both trimer 5 and trimer 6 induced apoptosis within 72 h after addition to SH-SY5Y cells, whereas peptides 1 and 2 showed little or no effect (Figure 13B). Caspase-3 activity levels after treatment with trimer 5 or trimer 6 were comparable to that of the known caspase-3 activator staurosporine. These results suggest that trimers 5 and 6 may elicit toxicity by activating apoptosis.
A11 Antibody Reactivity
The LDH release and caspase-3 activation studies indicate that trimers 5 and 6 behave like oligomers of full-length Aβ and provide evidence for the biological significance of the triangular assembly. To evaluate further how the biological properties of trimers 5 and 6 compare to those of full-length Aβ, we examined the reactivity of the trimers with the oligomer-specific antibody A11 by dot blot analysis. Trimers 5 and 6 react with the A11 antibody, but peptides 1 and 2 do not (Figure 13C).
Reactivity with the A11 antibody is a hallmark of certain types of Aβ oligomers.33,34 The A11 antibody specifically recognizes oligomeric assemblies of Aβ, but does not recognize Aβ monomers or fibrils. The structures of the Aβ oligomers recognized by the A11 antibody are not known. The results from the dot blot experiment show that the A11 antibody recognizes trimers 5 and 6 as Aβ oligomers and suggest that oligomers of full-length Aβ may also contain triangular trimers.
Solution-Phase Biophysical Studies of Trimers 5 and 6
The differences in LDH release, caspase-3 activation, and A11 antibody reactivity between trimers 5 and 6 and peptides 1 and 2, suggest that covalent stabilization of the triangular trimer is necessary for these small peptides to mimic the oligomers of full-length Aβ at micromolar concentrations. Although peptides 1 and 2 assemble to form triangular trimers at the millimolar concentrations of crystallography experiments, they may be too small to assemble at the micromolar concentrations of biological and biophysical experiments. We turned to SDS-PAGE, size exclusion chromatography (SEC), and circular dichroism (CD) spectroscopy to probe the solution-phase behavior of trimers 5 and 6 and peptides 1 and 2, and thus explore these hypotheses.
SDS-PAGE
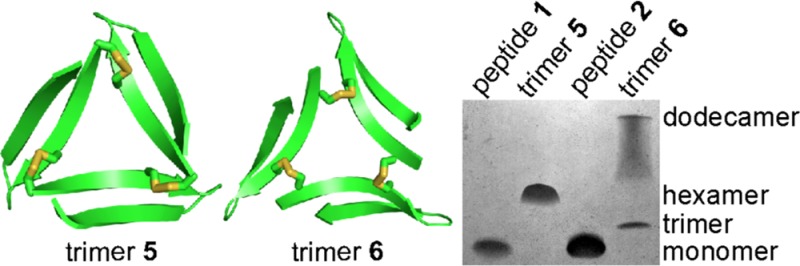
Tricine SDS-PAGE followed by silver staining reveals that trimers 5 and 6 assemble to form SDS-stable oligomers (Figure 14A).35,36 Trimer 5 migrates as a single band at a molecular weight consistent with a hexamer. Trimer 6 migrates as two bands: one consistent with the molecular weight of a dodecamer, the other consistent with the molecular weight of a trimer. The dodecamer band shows pronounced streaking, suggesting equilibria with lower molecular weight oligomers, such as nonamers and hexamers. Peptides 1 and 2 migrate as broad bands at molecular weights consistent with monomer or dimer.
Figure 14.
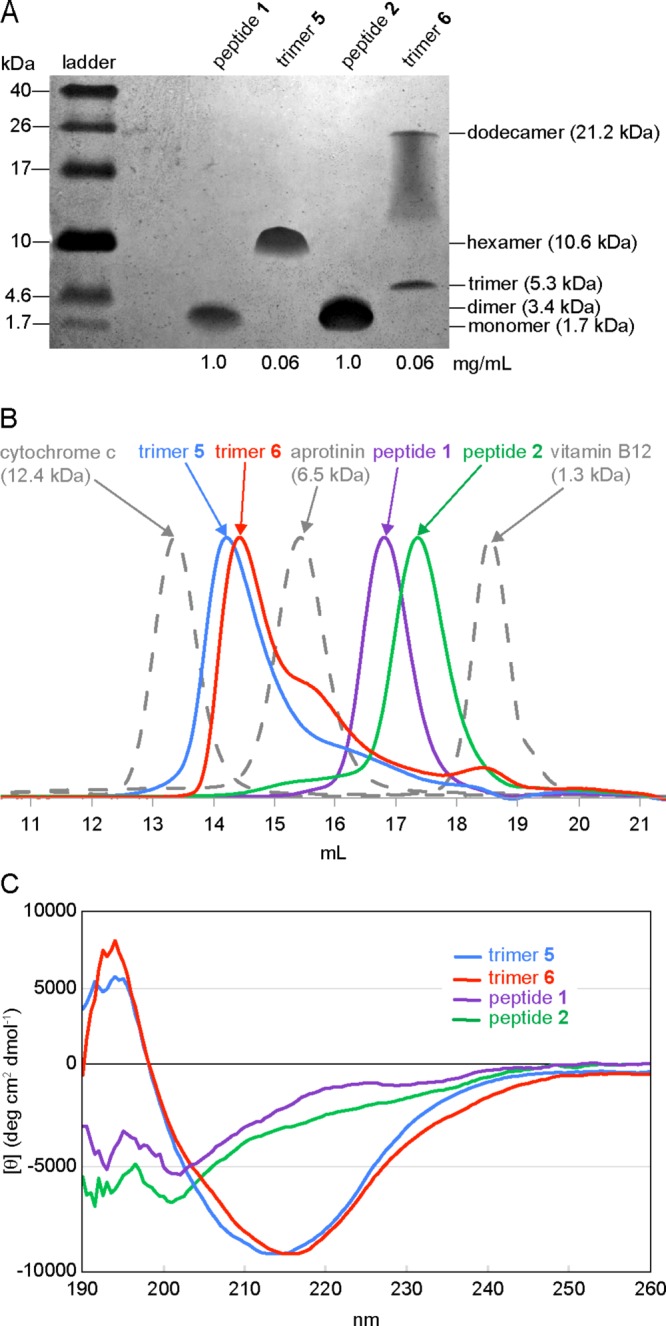
Solution-phase biophysical studies of trimers 5 and 6 and peptides 1 and 2. (A) Silver stained SDS-PAGE gel. SDS-PAGE was performed in Tris buffer at pH 6.8 with 2% (w/v) SDS. Molecular weights calculated for the monomer, dimer, trimer, hexamer, and dodecamer are listed in parentheses. (B) Size exclusion chromatography chromatograms. SEC was performed on 1.0-mg/mL solutions of trimers 5 and 6 and peptides 1 and 2 in 50 mM sodium acetate/50 mM acetic acid (pH 4.7) with a Superdex 75 10/300 column.37 (C) Circular dichroism spectra. Spectra were acquired at 0.3 mg/mL (50 μM trimers 5 and 6; 150 μM peptides 1 and 2) in 10 mM potassium phosphate buffer at pH 7.4.
Size Exclusion Chromatography
SEC reveals that trimers 5 and 6 also assemble to form higher-order oligomers in acetate buffer (Figure 14B).37 The elution profiles of trimers 5 and 6 were compared to those of size standards and peptides 1 and 2. The size standards vitamin B12 (1.3 kDa), aprotinin (6.5 kDa), and cytochrome c (12.4 kDa) eluted at 18.6, 15.4, and 13.4 mL, respectively. Trimer 5 elutes at 14.3 mL; trimer 6 elutes at 14.5 mL. These elution volumes fall between the elution volumes of the 6.5 and 12.4 kDa standards and are thus consistent with the molecular weight of a hexamer (10.6 kDa). The peaks for trimers 5 and 6 tail slightly, which may reflect a trimer-hexamer equilibrium in which the hexamer predominates. The tail of trimer 6 shows a distinct hump at 15.6 mL, suggesting a slow equilibrium between the trimer and the hexamer.
Under the conditions of the SEC experiments, peptides 1 and 2 do not assemble to form trimers. Peptide 1 elutes at 16.8 mL; peptide 2 elutes at 17.3 mL. These volumes are lower than would be expected for a 1.7 kDa monomer and higher than would be expected for a 5.3 kDa trimer, suggesting that peptides 1 and 2 may form dimers in solution.
Circular Dichroism
Circular dichroism spectra reflect the cooperative folding and assembly of macrocyclic β-sheet peptides (Figure 14C). The CD spectra of trimers 5 and 6 exhibit typical β-hairpin character as evidenced by negative bands at ∼215 nm and positive bands at ∼195 nm.38−40 In contrast, the CD spectra of peptides 1 and 2 show little β-hairpin structure. These results indicate that covalent stabilization not only locks in conformation but also promotes folding of the monomeric subunits into β-hairpins. Table 1 summarizes the results of the structural and biological studies described above.
Table 1. Structures, Stoichiometries, and Biological Activities of Trimers 5 and 6 and Peptides 1 and 2.
| oligomer size by |
|||||||
|---|---|---|---|---|---|---|---|
| compound | PDB ID | crystallography | SEC | SDS-PAGE | A11 reactivity | LDH release | caspase-3 activation |
| trimer 5 | 5SUT | 6 | 6 | 6 | yes | some | yes |
| trimer 6 | 5SUR | 6 and 12a | 6 | 3 and 12 | yes | yes | yes |
| peptide 1(10) | 4NTR | 3, 6, and 12 | 1–2 | 1–2 | no | no | no |
| peptide 2(10) | 4NW9 | 3, 6, and 12 | 1–2 | 1–2 | no | no | no |
In the X-ray crystallographic structure of trimer 6, the dodecamers further assemble to form annular porelike structures.
Discussion
X-ray crystallography provides a facile means to probe the structures of oligomers formed by β-hairpin peptides derived from amyloidogenic peptides and proteins. The solution-phase studies of trimers 5 and 6 provide evidence that their crystallographically observed assemblies are meaningful, and are not simply artifacts of the trimers packing to form a lattice. The hexamer formed by trimer 5 in the SDS-PAGE and SEC studies likely resembles the sandwich-like hexamer observed crystallographically, in which two trimers pack on their hydrophobic surfaces. The dodecamer formed by trimer 6 in the SDS-PAGE study likely resembles the ball-shaped dodecamer observed crystallographically, in which four trimers assemble in a tetrahedral fashion. Furthermore, the hexamer formed by trimer 6 in the SEC study may resemble the hexamer formed at the interface of the dodecamers in the annular pore.
The differences in solution-phase assembly between trimer 5 and trimer 6 may explain the greater toxicity of trimer 6 in the LDH release assay. The increased LDH release from cells treated with trimer 6 may reflect the propensity of trimer 6 to form dodecamers in a lipophilic environment, such as SDS micelles or cell membranes. In cell membranes, the dodecamers may further assemble to form annular pores and induce LDH leakage. The greater LDH release induced by trimer 6, in spite of comparable caspase-3 activation, suggests that LDH release and apoptosis might occur through different mechanisms.
β-Hairpins are thought to be the building blocks of some Aβ oligomers.41−44 The crystallographic and solution-phase assembly of trimers 5 and 6 support a model in which full-length Aβ folds into β-hairpins that come together to form triangular trimers that further assemble to form ball-shaped dodecamers.10,16 Dodecamers that are composed of triangular trimers arranged in a tetrahedral fashion are special because they display four hydrophobic faces that can pack with the hydrophobic faces of other dodecamers to form larger assemblies. The hierarchical assembly of triangular trimers into dodecamers that further assemble to form annular porelike structures is an emergent property of the triangular trimers observed by our laboratory. This mode of assembly may explain some of the large oligomeric assemblies observed for Aβ and other amyloidogenic peptides and proteins.
One type of large oligomeric assembly formed by Aβ has been termed annular protofibrils (APFs).45−47 APFs share a common donut-shaped morphology and appear to be composed of smaller spherical oligomers. APFs have also been observed for other amyloidogenic peptides and proteins, such as α-synuclein, islet amyloid polypeptide, and tau.48,49 The annular porelike assembly formed by trimer 6 could serve as a structural model for an APF formed by Aβ. Furthermore, the hierarchical assembly of trimers into dodecamers, which further assemble to form annular porelike structures, might be a common mode of hierarchical assembly for other amyloidogenic peptides and proteins.
Trimers are especially important among the various oligomers formed by full-length Aβ. Concentrations of Aβ trimers are elevated in cognitively normal adults who are at risk for Alzheimer’s disease.2,50,51 Trimers also appear to be the building blocks of the putative dodecamer of Aβ, termed Aβ*56, which was isolated from the brains of Tg2576 transgenic mice and shown to impair memory in healthy rats.52 Furthermore, Aβ trimers, but not monomers or dimers, have been shown to promote aggregation of the microtubule associated protein tau, which is also involved in the progression Alzheimer’s disease.53 Although the significance of triangular assemblies of Aβ β-hairpins in Alzheimer’s disease remains to be determined, the results described in this paper further support a model in which trimers are a central feature of Aβ oligomers.
Conclusion
The studies described in this paper embody our laboratory’s strategy for studying well-defined oligomers derived from Aβ. Stabilizing fragments of the Aβ peptide in a macrocyclic β-sheet peptide and blocking uncontrolled aggregation with an N-methyl group permits crystallization and elucidation of higher-order assemblies the peptide can form. The X-ray crystallographic structures of the higher-order assemblies can be used to develop strategies to cross-link the peptide and thus stabilize oligomers. The cross-linked oligomers provide a tool to investigate the biological significance of the crystallographically observed oligomers.
Trimers 5 and 6 constitute the first cross-linked oligomers of an Aβ-derived peptide in which the X-ray crystallographic structures are known. The results presented in this paper support the triangular trimer, as well as sandwich-like hexamers and ball-shaped dodecamers as biologically significant assemblies of the Aβ peptide. Trimers 5 and 6 assemble to form stable oligomers in solution and recapitulate the toxicity and A11 antibody reactivity of Aβ oligomers.
Trimers 5 and 6 offer the promise of relating Aβ oligomer structure with biological activity. The X-ray crystallographic structures of trimers 5 and 6 and the trimers formed by peptides 1 and 2 can serve as starting points for rationally designing small molecules that bind Aβ oligomers. The X-ray crystallographic structure of the trimer formed by peptide 1 has already been used in docking studies to explain the fluorescence of probes that bind Aβ oligomers.54,55 Trimers 5 and 6 provide stable targets that can be used to evaluate further binding of probes such as these. Trimers 5 and 6 may also serve as a starting point for discovering small molecules that inhibit the toxicity of Aβ oligomers. In addition, trimers 5 and 6 may serve as antigens for generating antibodies as probes for amyloid oligomers or as therapies for Alzheimer’s disease. We are currently investigating these applications and will report our findings in due course.
Acknowledgments
We thank Dr. Huiying Li and Mr. Ricardo Albay for helpful advice and assistance, Dr. Charles G. Glabe for a gift of A11 antibody, the Laser Spectroscopy Facility at the University of California, Irvine for assistance with circular dichroism measurements, the National Institutes of Health (NIH) National Institute of General Medical Sciences (NIGMS) for funding (Grant GM097562), and the Stanford Synchrotron Radiation Lightsource (SSRL) and the Berkeley Center for Structural Biology (BCSB) of the Advanced Light Source (ALS) for synchrotron data collection. Use of the Stanford Synchrotron Radiation Lightsource (SSRL) is supported by the Department of Energy and the NIGMS. The BCSB is supported in part by the NIH, NIGMS, and the Howard Hughes Medical Institute. The ALS is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.6b11748.
Procedures for the synthesis and crystallization of trimers 5 and 6, LDH release and caspase-3 assays, dot blot analysis, SDS-PAGE and silver staining, size exclusion chromatography, and circular dichroism spectroscopy; characterization data for peptides 1 and 2, and trimers 5 and 6; details of X-ray crystallographic data collection, processing, and refinement (PDF)
Data for 5SUR (CIF)
Data for 5SUS (CIF)
Data for 5SUT (CIF)
Data for 5SUU (CIF)
Crystallographic coordinates of trimers 5 and 6 were deposited into the Protein Data Bank (PDB) with codes 5SUR(PDB) and 5SUT (PDB) (data collected on a synchrotron at 1.00 Å wavelength) and 5SUS (PDB) and 5SUU (PDB) (data collected on an X-ray diffractometer at 1.54 Å wavelength).
The authors declare no competing financial interest.
Supplementary Material
References
- Benilova I.; Karran E.; De Strooper B. Nat. Neurosci. 2012, 15, 349–357. 10.1038/nn.3028. [DOI] [PubMed] [Google Scholar]
- Larson M. E.; Lesné S. E. J. Neurochem. 2012, 120, 125–139. 10.1111/j.1471-4159.2011.07478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G.; Lomakin A.; Teplow D. B. J. Biol. Chem. 2001, 276, 35176–35184. 10.1074/jbc.M102223200. [DOI] [PubMed] [Google Scholar]
- Bitan G.; Kirkitadze M. D.; Lomakin A.; Vollers S. S.; Benedek G. B.; Teplow D. B. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 330–335. 10.1073/pnas.222681699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitan G.; Vollers S. S.; Teplow D. B. J. Biol. Chem. 2003, 278, 34882–34889. 10.1074/jbc.M300825200. [DOI] [PubMed] [Google Scholar]
- Ono K.; Condron M. M.; Teplow D. B. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 14745–14750. 10.1073/pnas.0905127106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar G. M.; Li S.; Mehta T. H.; Garcia-Munoz A.; Shepardson N. E.; Smith I.; Brett F. M.; Farrell M. A.; Rowan M. J.; Lemere C. A.; Regan C. M.; Walsh D. M.; Sabatini B. L.; Selkoe D. J. Nat. Med. 2008, 14, 837–842. 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Nuallain B.; Freir D. B.; Nicoll A. J.; Risse E.; Ferguson N.; Herron C. E.; Collinge J.; Walsh D. M. J. Neurosci. 2010, 30, 14411–14419. 10.1523/JNEUROSCI.3537-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Malley T. T.; Oktaviani N. A.; Zhang D.; Lomakin A.; O’Nuallain B.; Linse S.; Benedek G. B.; Rowan M. J.; Mulder F. A.; Walsh D. M. Biochem. J. 2014, 461, 413–426. 10.1042/BJ20140219. [DOI] [PubMed] [Google Scholar]
- Spencer R. K.; Li H.; Nowick J. S. J. Am. Chem. Soc. 2014, 136, 5595–5598. 10.1021/ja5017409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer R. K.; Kreutzer A. G.; Salveson P. J.; Li H.; Nowick J. S. J. Am. Chem. Soc. 2015, 137, 6304–6311. 10.1021/jacs.5b01673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salveson P. J.; Spencer R. K.; Nowick J. S. J. Am. Chem. Soc. 2016, 138, 4458–4467. 10.1021/jacs.5b13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.; Kreutzer A. G.; Truex N. L.; Nowick J. S. Chem. Sci. 2016, 7, 6946–6951. 10.1039/C6SC01927G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- We have subsequently solved the X-ray crystallographic structure of a homologue of peptide 1 with methionine at position 35 and have observed an identical triangular trimer (unpublished results).
- Nowick J. S.; Brower J. O. J. Am. Chem. Soc. 2003, 125, 876–877. 10.1021/ja028938a. [DOI] [PubMed] [Google Scholar]
- We recently published the X-ray crystallographic structure of an Aβ17–36 β-hairpin containing the Aβ24–29 loop. This peptide forms the same type of triangular trimer as peptides 1 and 2.Kreutzer A. G.; Hamza I. H.; Spencer R. K.; Nowick J. S. J. Am. Chem. Soc. 2016, 138, 4634–4642. 10.1021/jacs.6b01332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer R.; Chen K. H.; Manuel G.; Nowick J. S. Eur. J. Org. Chem. 2013, 2013, 3523–3528. 10.1002/ejoc.201300221. [DOI] [Google Scholar]
- Tam J. P.; Wu C. R.; Liu W.; Zhang J. W. J. Am. Chem. Soc. 1991, 113, 6657–6662. 10.1021/ja00017a044. [DOI] [Google Scholar]
- Khakshoor O.; Nowick J. S. Org. Lett. 2009, 11, 3000–3003. 10.1021/ol901015a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- We confirmed the identity of each product in the oxidation reactions of peptides 3 and 4 using electrospray ionization mass spectrometry (ESI-MS). Although the monomers, dimers, and trimers share common mass-to-charge ratios that depend on their ionization states, each product has a unique isotope pattern, allowing us to easily identify monomer, dimer, and trimer.
- Spencer R. K.; Nowick J. S. Isr. J. Chem. 2015, 55, 698–710. 10.1002/ijch.201400179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q.; Dahmane T.; Zhang Z.; Assur Z.; Brasch J.; Shapiro L.; Mancia F.; Hendrickson W. A. Science 2012, 336, 1033–1037. 10.1126/science.1218753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma G. N.; Karplus P. A. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2006, 62, 707–716. 10.1107/S0907444906014946. [DOI] [PubMed] [Google Scholar]
- Dauter Z.; Dauter M.; Rajashankar K. R. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2000, 56, 232–237. 10.1107/S0907444999016352. [DOI] [PubMed] [Google Scholar]
- Trimer 5 does not appear as a discrete entity in the asymmetric unit of the X-ray crystallographic structure. Instead, the asymmetric unit contains two monomeric macrocyclic β-sheet subunits, which form two crystallographically distinct trimers through symmetry operations. The two crystallographically distinct trimers are virtually identical, differing primarily in rotamers of I31, I32, and Orn35.
- Trimer 6 does not appear as a discrete entity in the asymmetric unit of the X-ray crystallographic structure. Instead, the asymmetric unit contains four monomeric macrocyclic β-sheet subunits, which form two crystallographically distinct trimers. The two crystallographically distinct trimers are virtually identical, differing primarily in rotamers of E22, D23, and I32.
- We modeled four molecules of the crystallization cosolvent 1,6-hexanediol in the asymmetric unit of trimer 6. All four molecules reside within the hydrophobic cavity of the ball-shaped dodecamer, making contacts with the F20 faces of the trimer subunits and further stabilizing the dodecamer.
- Li Y. P.; Bushnell A. F.; Lee C. M.; Perlmutter L. S.; Wong S. K. Brain Res. 1996, 738, 196–204. 10.1016/S0006-8993(96)00733-0. [DOI] [PubMed] [Google Scholar]
- Teplow D. B. Methods Enzymol. 2006, 413, 20–33. 10.1016/S0076-6879(06)13002-5. [DOI] [PubMed] [Google Scholar]
- Fezoui Y.; Hartley D. M.; Harper J. D.; Khurana R.; Walsh D. M.; Condron M. M.; Selkoe D. J.; Lansbury P. T. Jr.; Fink A. L.; Teplow D. B. Amyloid 2000, 7, 166–178. 10.3109/13506120009146831. [DOI] [PubMed] [Google Scholar]
- Harada J.; Sugimoto M. Brain Res. 1999, 842, 311–323. 10.1016/S0006-8993(99)01808-9. [DOI] [PubMed] [Google Scholar]
- Jo J.; Whitcomb D. J.; Olsen K. M.; Kerrigan T. L.; Lo S. C.; Bru-Mercier G.; Dickinson B.; Scullion S.; Sheng M.; Collingridge G.; Cho K. Nat. Neurosci. 2011, 14, 545–547. 10.1038/nn.2785. [DOI] [PubMed] [Google Scholar]
- Kayed R.; Head E.; Thompson J. L.; McIntire T. M.; Milton S. C.; Cotman C. W.; Glabe C. G. Science 2003, 300, 486–489. 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- Glabe C. G. J. Biol. Chem. 2008, 283, 29639–29643. 10.1074/jbc.R800016200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H. Nat. Protoc. 2006, 1, 16–22. 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- Simpson R. J. CSH Protoc 2007, 2007, pdb.prot4727. 10.1101/pdb.prot4727. [DOI] [PubMed] [Google Scholar]
- A buffer of 50 mM sodium acetate and 50 mM acetic acid (pH 4.7) was used in the SEC studies. We initially attempted to perform the SEC studies in 0.1 M potassium phosphate buffer at pH 7.4 and in 50 mM Tris buffer at pH 6.8 with 150 mM NaCl. We observed prolonged retention under both conditions suggesting nonspecific interaction with the column matrix.
- Ramírez-Alvarado M.; Blanco F. J.; Serrano L. Nat. Struct. Biol. 1996, 3, 604–612. 10.1038/nsb0796-604. [DOI] [PubMed] [Google Scholar]
- Maynard A. J.; Sharman G. J.; Searle M. S. J. Am. Chem. Soc. 1998, 120, 1996–2007. 10.1021/ja9726769. [DOI] [Google Scholar]
- Anderson J. M.; Jurban B.; Huggins K. N.; Shcherbakov A. A.; Shu I.; Kier B.; Andersen N. H. Biochemistry 2016, 55, 5537–5553. 10.1021/acs.biochem.6b00732. [DOI] [PubMed] [Google Scholar]
- Yu L.; Edalji R.; Harlan J. E.; Holzman T. F.; Lopez A. P.; Labkovsky B.; Hillen H.; Barghorn S.; Ebert U.; Richardson P. L.; Miesbauer L.; Solomon L.; Bartley D.; Walter K.; Johnson R. W.; Hajduk P. J.; Olejniczak E. T. Biochemistry 2009, 48, 1870–1877. 10.1021/bi802046n. [DOI] [PubMed] [Google Scholar]
- Scheidt H. A.; Morgado I.; Huster D. J. Biol. Chem. 2012, 287, 22822–22826. 10.1074/jbc.M112.367474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi T.; Masuda Y.; Irie K.; Akagi K.; Monobe Y.; Imazawa T.; Takegoshi K. Biochem. Biophys. Res. Commun. 2012, 428, 458–462. 10.1016/j.bbrc.2012.10.096. [DOI] [PubMed] [Google Scholar]
- Tay W. M.; Huang D.; Rosenberry T. L.; Paravastu A. K. J. Mol. Biol. 2013, 425, 2494–2508. 10.1016/j.jmb.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner J. H.; Cheung C. L.; Woolley A. T.; Lieber C. M. Prog. Biophys. Mol. Biol. 2001, 77, 73–110. 10.1016/B978-008044031-6/50037-9. [DOI] [PubMed] [Google Scholar]
- Lin H.; Bhatia R.; Lal R. FASEB J. 2001, 15, 2433–2444. 10.1096/fj.01-0377com. [DOI] [PubMed] [Google Scholar]
- Lashuel H. A.; Hartley D.; Petre B. M.; Walz T.; Lansbury P. T. Jr. Nature 2002, 418, 291–292. 10.1038/418291a. [DOI] [PubMed] [Google Scholar]
- Quist A.; Doudevski I.; Lin H.; Azimova R.; Ng D.; Frangione B.; Kagan B.; Ghiso J.; Lal R. Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 10427–10432. 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayed R.; Pensalfini A.; Margol L.; Sokolov Y.; Sarsoza F.; Head E.; Hall J.; Glabe C. J. Biol. Chem. 2009, 284, 4230–4237. 10.1074/jbc.M808591200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesné S. E.; Sherman M. A.; Grant M.; Kuskowski M.; Schneider J. A.; Bennett D. A.; Ashe K. H. Brain 2013, 136, 1383–1398. 10.1093/brain/awt062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handoko M.; Grant M.; Kuskowski M.; Zahs K. R.; Wallin A.; Blennow K.; Ashe K. H. JAMA Neurol. 2013, 70, 594–599. 10.1001/jamaneurol.2013.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesné S.; Koh M. T.; Kotilinek L.; Kayed R.; Glabe C. G.; Yang A.; Gallagher M.; Ashe K. H. Nature 2006, 440, 352–357. 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- Sherman M. A.; LaCroix M.; Amar F.; Larson M. E.; Forster C.; Aguzzi A.; Bennett D. A.; Ramsden M.; Lesné S. E. J. Neurosci. 2016, 36, 9647–9658. 10.1523/JNEUROSCI.1899-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teoh C. L.; Su D.; Sahu S.; Yun S. W.; Drummond E.; Prelli F.; Lim S.; Cho S.; Ham S.; Wisniewski T.; Chang Y. T. J. Am. Chem. Soc. 2015, 137, 13503–13509. 10.1021/jacs.5b06190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv G.; Sun A.; Wei P.; Zhang N.; Lan H.; Yi T. Chem. Commun. (Cambridge, U. K.) 2016, 52, 8865–8868. 10.1039/C6CC02741E. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.