Abstract
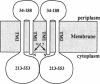
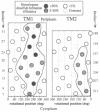
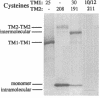
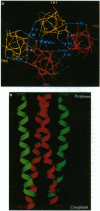
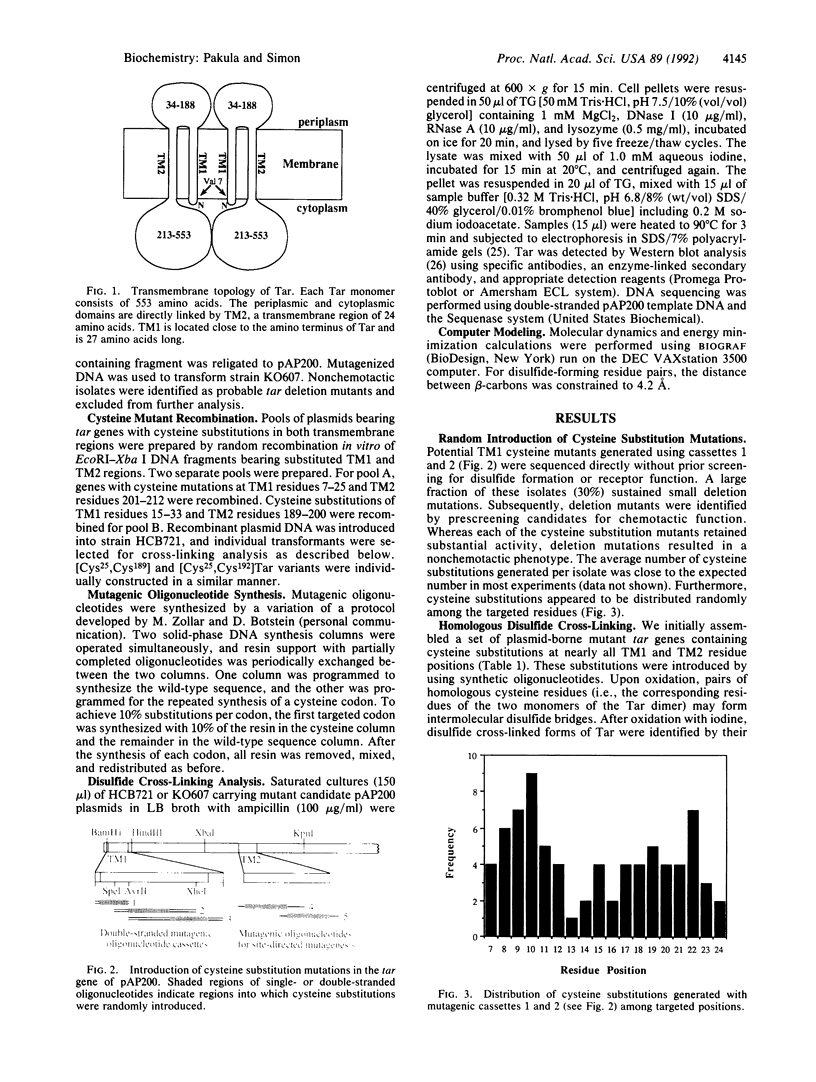
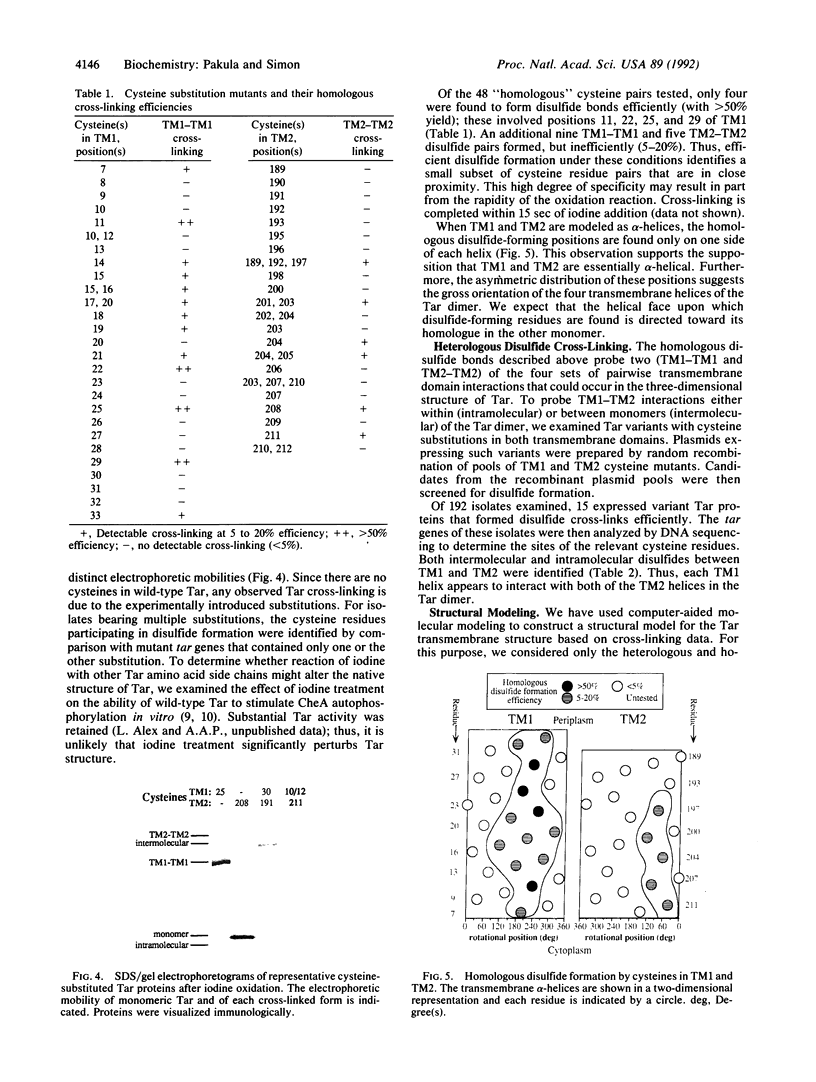
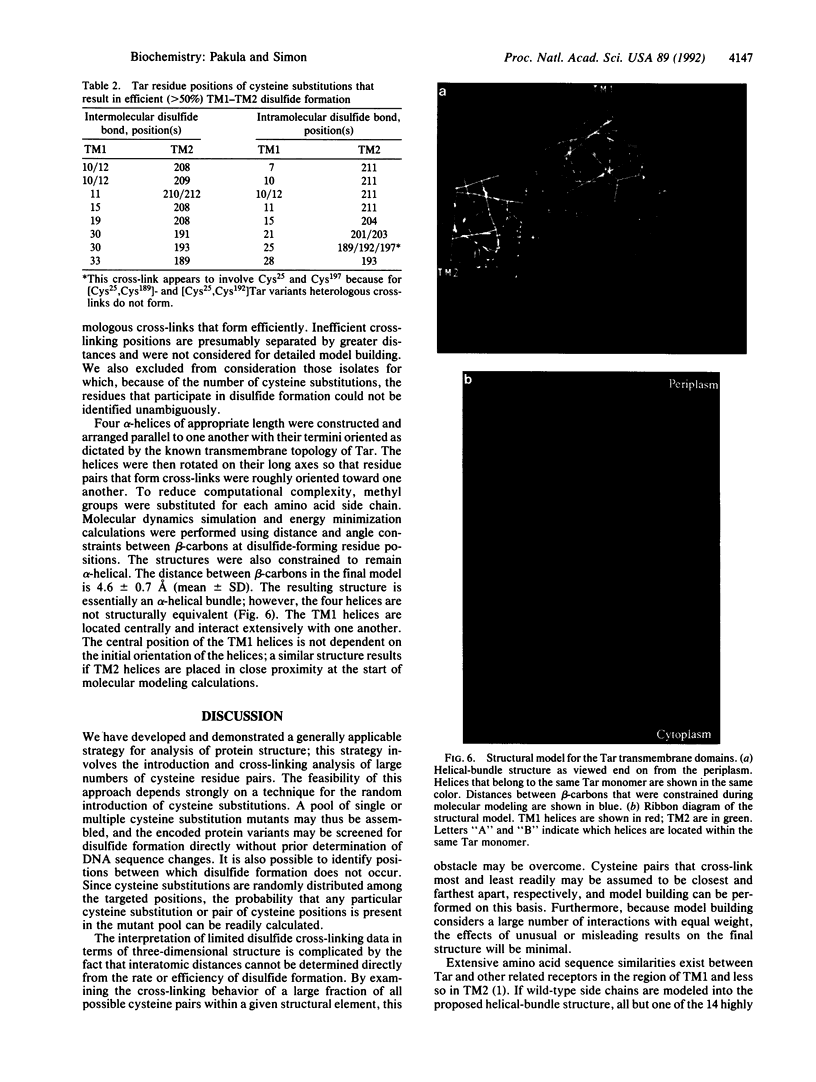
We have devised a generally applicable strategy for analysis of protein structure and have applied it to examine the structure of the transmembrane portion of the Tar receptor of Escherichia coli. The basis of our approach is the use of disulfide cross-linking to identify residues that are within close proximity. To generate and test large numbers of cysteine pairs, we used an unusual method of mutagenesis by which cysteine substitutions can be created randomly at a number of targeted codons. Cysteine-substituted proteins encoded by mutagenized genes may be screened directly for disulfide formation within oligomers or, alternatively, different pools of genes may be randomly recombined to generate gene populations with substitutions in multiple regions. Thus, it is possible to detect a variety of disulfide cross-links between and within individual protein molecules. Interactions between the four membrane-spanning stretches of the Tar dimer were probed by measuring the tendency of 48 cysteine substitutions throughout this region to form disulfide cross-links with one another. We have interpreted these data to suggest a helical-bundle structure for the transmembrane region. The four helices of this bundle are not structurally equivalent: the two TM1 helices interact closely, whereas the TM2 helices are more peripherally located.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Simon M. I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990 Dec 21;63(6):1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- Boyd A., Kendall K., Simon M. I. Structure of the serine chemoreceptor in Escherichia coli. Nature. 1983 Feb 17;301(5901):623–626. doi: 10.1038/301623a0. [DOI] [PubMed] [Google Scholar]
- Bubis J., Khorana H. G. Sites of interaction in the complex between beta- and gamma-subunits of transducin. J Biol Chem. 1990 Aug 5;265(22):12995–12999. [PubMed] [Google Scholar]
- Dahl M. K., Boos W., Manson M. D. Evolution of chemotactic-signal transducers in enteric bacteria. J Bacteriol. 1989 May;171(5):2361–2371. doi: 10.1128/jb.171.5.2361-2371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falke J. J., Dernburg A. F., Sternberg D. A., Zalkin N., Milligan D. L., Koshland D. E., Jr Structure of a bacterial sensory receptor. A site-directed sulfhydryl study. J Biol Chem. 1988 Oct 15;263(29):14850–14858. [PubMed] [Google Scholar]
- First E. A., Taylor S. S. Induced interchain disulfide bonding in cAMP-dependent protein kinase II. J Biol Chem. 1984 Apr 10;259(7):4011–4014. [PubMed] [Google Scholar]
- Karlin A., Holtzman E., Yodh N., Lobel P., Wall J., Hainfeld J. The arrangement of the subunits of the acetylcholine receptor of Torpedo californica. J Biol Chem. 1983 Jun 10;258(11):6678–6681. [PubMed] [Google Scholar]
- Karnik S. S., Khorana H. G. Assembly of functional rhodopsin requires a disulfide bond between cysteine residues 110 and 187. J Biol Chem. 1990 Oct 15;265(29):17520–17524. [PubMed] [Google Scholar]
- Krikos A., Conley M. P., Boyd A., Berg H. C., Simon M. I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Mutoh N., Boyd A., Simon M. I. Sensory transducers of E. coli are composed of discrete structural and functional domains. Cell. 1983 Jun;33(2):615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtenbach E., Curtis C. A., Pedder E. K., Aitken A., Harris A. C., Hulme E. C. Muscarinic acetylcholine receptors. Peptide sequencing identifies residues involved in antagonist binding and disulfide bond formation. J Biol Chem. 1990 Aug 15;265(23):13702–13708. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lynch B. A., Koshland D. E., Jr Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Dec 1;88(23):10402–10406. doi: 10.1073/pnas.88.23.10402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Milburn M. V., Privé G. G., Milligan D. L., Scott W. G., Yeh J., Jancarik J., Koshland D. E., Jr, Kim S. H. Three-dimensional structures of the ligand-binding domain of the bacterial aspartate receptor with and without a ligand. Science. 1991 Nov 29;254(5036):1342–1347. doi: 10.1126/science.1660187. [DOI] [PubMed] [Google Scholar]
- Milligan D. L., Koshland D. E., Jr Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J Biol Chem. 1988 May 5;263(13):6268–6275. [PubMed] [Google Scholar]
- Oosawa K., Mutoh N., Simon M. I. Cloning of the C-terminal cytoplasmic fragment of the tar protein and effects of the fragment on chemotaxis of Escherichia coli. J Bacteriol. 1988 Jun;170(6):2521–2526. doi: 10.1128/jb.170.6.2521-2526.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo A. F., Koshland D. E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983 Jun 3;220(4601):1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- Singer S. J. The structure and insertion of integral proteins in membranes. Annu Rev Cell Biol. 1990;6:247–296. doi: 10.1146/annurev.cb.06.110190.001335. [DOI] [PubMed] [Google Scholar]
- Springer M. S., Goy M. F., Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang V. C., Peralta J. M., Simons A. R. Enzyme-linked immunoelectrotransfer blot techniques (EITB) for studying the specificities of antigens and antibodies separated by gel electrophoresis. Methods Enzymol. 1983;92:377–391. doi: 10.1016/0076-6879(83)92032-3. [DOI] [PubMed] [Google Scholar]
- Wolfe A. J., Conley M. P., Berg H. C. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6711–6715. doi: 10.1073/pnas.85.18.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zagursky R. J., Berman M. L. Cloning vectors that yield high levels of single-stranded DNA for rapid DNA sequencing. Gene. 1984 Feb;27(2):183–191. doi: 10.1016/0378-1119(84)90139-2. [DOI] [PubMed] [Google Scholar]