Abstract
We report the complete sequence of the 4,274,642-bp genome of Haloarcula marismortui, a halophilic archaeal isolate from the Dead Sea. The genome is organized into nine circular replicons of varying G+C compositions ranging from 54% to 62%. Comparison of the genome architectures of Halobacterium sp. NRC-1 and H. marismortui suggests a common ancestor for the two organisms and a genome of significantly reduced size in the former. Both of these halophilic archaea use the same strategy of high surface negative charge of folded proteins as means to circumvent the salting-out phenomenon in a hypersaline cytoplasm. A multitiered annotation approach, including primary sequence similarities, protein family signatures, structure prediction, and a protein function association network, has assigned putative functions for at least 58% of the 4242 predicted proteins, a far larger number than is usually achieved in most newly sequenced microorganisms. Among these assigned functions were genes encoding six opsins, 19 MCP and/or HAMP domain signal transducers, and an unusually large number of environmental response regulators—nearly five times as many as those encoded in Halobacterium sp. NRC-1—suggesting H. marismortui is significantly more physiologically capable of exploiting diverse environments. In comparing the physiologies of the two halophilic archaea, in addition to the expected extensive similarity, we discovered several differences in their metabolic strategies and physiological responses such as distinct pathways for arginine breakdown in each halophile. Finally, as expected from the larger genome, H. marismortui encodes many more functions and seems to have fewer nutritional requirements for survival than does Halobacterium sp. NRC-1.
Halophilic archaeal organisms thrive in extreme environments of ∼4.5 molar salt such as solar salterns, the Great Salt Lake, and the Dead Sea. About four years ago, we sequenced the genome of the first halophilic archaeon, that of Halobacterium sp. NRC-1, because we were fascinated about how protein structures as well as metabolic strategies and physiologic responses might be altered to meet this extreme high-salt environment. Indeed, our expectations about fascinating biology turned out to be correct (Ng et al. 2000). The genome sequence of Halobacterium sp. NRC-1 revealed several interesting physical adaptations to a high-salt environment, including an acidic proteome of average pI of ∼4.5, believed to be essential for circumventing the salting-out of proteins in the hypersaline cytoplasm (Ng et al. 2000; Kennedy et al. 2001). It was also evident from the genome that Halobacterium sp. NRC-1 has complex requirements for growth: For example, it lacks enzymes for synthesis of at least eight amino acids; thus, it must live in an organic-rich environment. A third observation was that this halophile encodes an array of sensors, signal transducers, and transcriptional regulators, including multiple general transcription factors, that enable it to sense and tailor its physiology to perturbations in a diverse set of environmental factors characteristic of its extreme environment—high or low oxygen concentrations, harsh radiation, and the potential for desiccation. Finally, the genome sequence has catalyzed systems approaches to the study of halophilic archaeal biology. These systems-level studies have provided fascinating insights into gene regulatory control of energy transduction under anaerobic conditions and mechanisms for a robust stress response to extraordinary levels of UV irradiation (Baliga et al. 2002, 2004). This rapid progress in the systems-level study of halophilic archaea will further benefit from comparative genomic analyses of related halophiles (Goo et al. 2004).
We have long been aware of the power of comparative genomics to reveal fascinating evolutionary, structural, regulatory, and functional aspects of the biology of the corresponding organisms (Koop and Hood 1994), and accordingly, we decided to sequence the genome of a second halophile, Haloarcula marismortui (ATCC 43049). The primary goal here was to use comparative genomics as a means to add to our understanding of both the shared and unique features of halophiles. We demonstrate that the H. marismortui genome is 4275 kb in size and is composed of nine replicons and 4242 protein-coding genes. It encodes many more functions than does the Halobacterium sp. NRC-1 genome. Moreover, the multitiered approach we have used for functional annotation of the proteome has provided insights into functions for >58% of all encoded proteins—a higher percentage than is usually characterized in most prokaryotic genome sequencing efforts. Analysis of the H. marismortui genome sequence has also provided further support for proposed general characteristics of halophilic archaea such as an acidic proteome, multiple replicons (including a high G+C content large chromosome and multiple lower G+C content mini-replicons), multiple insertion sequence (ISH) elements throughout the genome, and the presence of multiple copies of general transcription factors.
Results and Discussion
Genome organization and evolutionary implications
Halophilic archaea share a common genome organizational theme: Multiple replicons falling into two correlated G+C content and size classes
The 4275-kb genome of H. marismortui is divided into relatively high and low G+C content replicons, including the large chromosome I, a 3132-kb replicon with a 62.36% G+C content, and eight smaller replicons ranging from 33 to 410 kb with a G+C contents ranging from 54.25% to 60.02% (average = 57%), respectively (Table 1). A comparison of halophilic archaeal genomes suggests that this bipartite genome-content organization in the form of a large high G+C content chromosome and multiple smaller replicons of lower G+C content is a general characteristic of all members of this group. For instance, the partially sequenced 4104-kb genome of Haloferax volcanii consists of two types of chromosomes that differ in their G+C content, a large 2920-kb chromosome with 65% G+C content and four smaller plasmids or mini-chromosomes, ranging from 6.4 to 690 kb, with an average G+C content of 55% (Charlebois et al. 1991). Likewise, the 2571-kb genome of Halobacterium sp. NRC-1 is organized as a 2014-kb chromosome of 67.9% G+C content and two smaller replicons pNRC100 (191 kb) and pNRC200 (365 kb) with an average G+C content of 58%. The implication of this genome-content organization remains a mystery, although the lower G+C content replicons seem to harbor a relatively higher density of repeat elements (see below).
Table 1.
Architecture of the 4,274,642 bp H. marismortui genome
| Replicons | Size (bp) | % G+C | RNA | IS H elementsa | Transposases |
|---|---|---|---|---|---|
| Chromosome I | 3,131,724 | 62.36 | 48 tRNAs, rrnA, rrnC | 13 | 15 |
| Chromosome II | 288,050 | 57.23 | 1 tRNA and rrnB | 7 | 7 |
| pNG700 | 410,554 | 59.12 | 5S rRNA | 2 | 2 |
| pNG600 | 155,300 | 58.33 | 1 tRNA | none | none |
| pNG500 | 132,678 | 54.48 | none | 15 | 22 |
| pNG400 | 50,060 | 57.35 | none | 3 | 3 |
| pNG300 | 39,521 | 60.02 | none | none | none |
| pNG200 | 33,452 | 55.63 | none | none | none |
| pNG100 | 33,303 | 54.25 | none | none | none |
The three ribosomal RNA operons each containing the 16S, 23S, and 5S rRNAs are designated as rrnA, rrnB, and rrnC.
Number of complete insertion sequences' elements containing terminal inverted repeats. The incomplete ISH elements are not included in this table
In addition to the large 3132-kb large chromosome, at least three other replicons in H. marismortui are considered chromosomes because they encode functions essential for survival
As mentioned above, the relevance of multiple replicons to the biology of halophilic archaea remains puzzling. However, it has been observed that the smaller replicons in Halobacterium sp. NRC-1 encode functions essential for survival (Ng et al. 1998, 2000). We make a similar observation in at least three mini-replicons in H. marismortui, and in some cases, we also observe that these replicons may be involved in functional segregation. For example, the replicon pNG600 in H. marismortui encodes the lone copy of the gene for aconitase, an important TCA cycle enzyme; a DNA polymerase B family protein, again the only copy in the H. marismortui genome; the large and small subunits of exonuclease VII; and two transcription factor B (TFB) orthologs. With respect to functional segregation, we find on this replicon nearly a dozen cation transport proteins of seemingly different specificities (copper, zinc, mercury, cadmium), a mercuric reductase, and several metal-ion dependent transcription regulators— suggesting this mini-chromosome is also essential for appropriately handling heavy metal stress.
Likewise, the pNG700 replicon encodes four enzymes, all of which are essential for folate metabolism, viz. methylenetetrahydrofolate dehydrogenase, 5, 10-methylenetetrahydrofolate reductase, formyltetrahydrofolate synthetase, and formimidoyltetrahydrofolate cyclodeaminase. The genes for urease, agmatinase, creatininase, and a spermidine/putrescine transport system are also encoded on the pNG700 replicon; suggesting this replicon encodes important functions that act downstream to arginine breakdown (see below). Finally, in addition to encoding one of three rRNA operons in H. marismortui, chromosome II also encodes several enzymes such as carbamoyl phosphate synthase, succinate-semialdehyde dehydrogenase, pyruvate dehydrogenase, acetyl-CoA acetyltransferase, citrate lyase, β chain, and GMP synthase, all of which are essential for core metabolic processes. Accordingly, it appears that haloarchaea have evolved a genome organization that requires multiple replicons—but still retain a single major chromosome. The interesting questions are whether there are essential genes that we do not recognize on some of the other smaller replicons, whether the other mini-chromosomes are on their way to capturing vital genes or whether they will not capture essential genes and thus remain dispensable. This can be evaluated by comparative genomic analysis as and when genome sequences are determined for other closely related halophilic archaea.
The lower G+C content chromosomes in halophilic archaeal genomes act as reservoirs for IS elements
IS elements are segments of DNA that can physically transpose from one region of the genome to another, often resulting in gene disruptions as well as large-scale genome reorganizations. The Halobacterium sp. NRC-1 genome contains a total of 91 ISH (for halobacterial IS elements) elements, of which 69 or ∼75% are located in the lower G+C content mini-chromosomes pNRC100 and pNRC200, which together constitute a fifth of the total genome content, suggesting that these replicons may function as reservoirs of ISH elements (Ng et al. 2000). In H. marismortui, >40 copies of intact ISH elements from 14 families are unevenly distributed among the replicons (Table 1). As in Halobacterium sp. NRC-1, 27 (about two-thirds) of the total 40 ISH elements in H. marismortui are also present in the small, lower G+C content replicons, which together represent less than a third of the full genome. The replicon with the highest density of ISH elements, pNG500 (G+C content: 54.5%), has 15 copies of intact IS from 10 families and >10 partial ISH elements. These observations provide further support to the hypothesis that the lower G+C chromosomes in halophilic archaea serve as reservoirs of ISH elements (Table 1). Furthermore, it has also been suggested for Halobacterium sp. NRC-1 that these AT rich islands of high ISH density in the mini-chromosomes harbor proteins of viral origin and potentially represent islands of prophage coding remnants (Bonneau et al. 2004). It has also been suggested that these insertions elements play a vital role in the reorganization of the chromosomal architecture of the Halobacterium sp. NRC-1 genome (Ng et al. 1998).
Comparisons of genome compositions and architectures suggest that Halobacterium sp. NRC-1 has evolved a streamlined or smaller genome arising from a common ancestor to H. marismortui
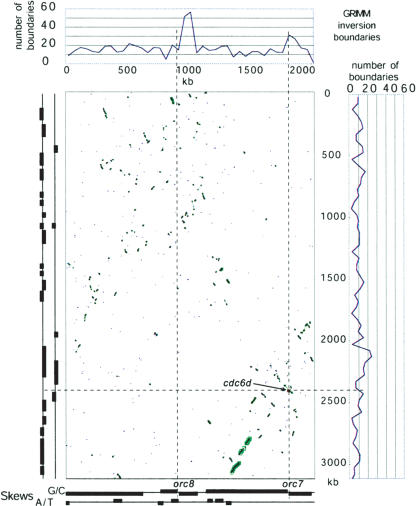
We compared the genome structures for the two halophilic archaea (Halobacterium sp. NRC-1 and H. marismortui) (Supplemental Fig. A) to understand the evolutionary and functional constraints that shaped both genomes (for details, see Supplemental material). The two large chromosomes of both species are clearly orthologous: A DNA level comparison demonstrated that the large chromosome in H. marismortui is 65%–70% identical to the large chromosome in Halobacterium sp. NRC-1. Moreover, threefourths of the genes in large Halobacterium sp. NRC-1 chromosomes have apparent orthologs in H. marismortui chromosome I, and these orthologous pairs amount to almost half the genes in the latter. We also see a few clear orthologs of genes encoded in the main chromosome of each halophile in the smaller replicons of the other species. These orthologs were detected at a higher frequency than would be expected if they were distributed randomly by sequence length. If the latter were true, then it would be simplest to assume that some genes translocated at random to previously unrelated chromosomes. Therefore, we conclude that the sequences in each genome encoding these orthologous pairs have a common origin and the similarity (conserved orthologous pairs) has been “diluted” over time. On the other hand, a much smaller fraction of the genes in the smaller H. marismortui replicons reveal potential orthologs to their counterparts in the mini-replicons of Halobacterium sp. NRC-1. This suggests one of two possibilities: Either the smaller replicons are entirely unrelated or their similarity has faded away to an extent that they appear unrelated. Finally, a dot plot depicting the location of the putative orthologous gene pairs in the large chromosomes shows that they are not colinear (Fig. 1). This implies that the chromosomal architectures have changed significantly, probably mediated at least in part by the ISs.
Figure 1.
Evolutionary comparison between the main chromosomes of H. marismortui (vertical axis) and Halobacterium sp. NRC-1 (horizontal axis). Scales are equal and in kilobasepairs (kb). Blue points in the dot-plot indicate the location of putative orthologous genes. Green boxes and lines indicate membership in ADHoRe clusters; larger boxes correspond to more populous clusters. The location of the cdc6d/orc7 orthologous pair is indicated in red. The graphs above and to the right depict the distribution of inversion boundaries as identified by GRIMM. Regions of significant nucleotide skews (absolute Z-score higher than three) are depicted in the graphs below and to the left of the dot-plot. Dotted lines are included as an aid for visualization.
We used the ADHoRe software (Vandepoele et al. 2002) to study a higher-confidence subset of 1459 orthologous pairs to identify regions in the two large halophilic archaeal chromosomes with conserved gene order and orientation (for a description of the ADHoRe software, see Supplemental material). The regions of micro-colinearity, depicted in green in Figure 1, reveal a large-scale (but fuzzy) “diagonal” of conservation, presumably reminiscent of the organization of the last common ancestor prior to divergence between these two species. Regions of micro-colinearity were also observed when comparing chromosome II and the pNG600 replicon in H. marismortui to the pNRC200 replicon in Halobacterium sp. NRC-1. The suggestion is that these two chromosomes were shared by the last common ancestor—the smaller replicons may have been added after the divergence of these two species.
The H. marismortui genome is larger than that of Halobacterium sp. NRC-1: in total sequence length, in the number of replicons, and in the number of genes. These differences could arise by the insertion (or deletion) of a limited number of large chromosomal segments, by a more widespread “stretching” (or compression) by way of many small insertion/deletion events (indels) or by both mechanisms. Interestingly, the distribution of regions of micro-colinearity shows that the length differences between H. marismortui chromosome I and Halobacterium sp. NRC-1 chromosomes are distributed throughout the entire length of the chromosomes, suggesting that since divergence there were no major additions (or removals) but rather scattered smaller indels. The H. marismortui chromosome I does not show signs of having acquired one or more major contiguous regions, for example, by horizontal transfer from another species. Furthermore, comparisons of intergenic distances shows very similar profiles between the two species (Supplemental Fig. B), with most intergenic distances in both species being 30 to 300 bp in length. This indicates that variation of intergenic segments also does not contribute significantly to the difference in genomic size, which, therefore, can be largely attributed to variation in gene number.
The conservation of over all gene order between these two organisms has been lost to an extent that makes it impossible to determine simple reconstructions of their evolutionary divergences. We therefore performed a detailed analysis of the rearrangements required for reconstructing the H. marismortui chromosome I from the Halobacterium sp. NRC-1 chromosome, using the GRIMM software (Tesler 2002) (for details, see Supplemental material). A parsimonious scenario produced by this analysis suggests a complex evolutionary history requiring at least 606 inversion events of different sizes, most frequently involving 30–200 genes, as well as single-gene inversions (this is in addition to the multiplicity of indels mentioned above). We mapped the inversion boundaries onto the sequence for the large chromosome in each genome, and found them to be concentrated in two “hotspots” in the Halobacterium sp. NRC-1 sequence. These two hotspots are in the vicinity of the orc8 and orc7 genes with locations that correspond to the origins of replication (see below). In the H. marismortui chromosome I, one single hotspot was apparent, corresponding to an origin of replication site orthologous to orc7 in Halobacterium sp. NRC-1.
We hypothesize that the frequent exposure to DNA damaging agents such as UV radiation and the desiccation-rehydration cycles in the natural environment of H. marismortui result in frequent double-stranded breaks and stalled replication forks, which together may facilitate extensive homologous recombinations and large-scale genomic rearrangements, mostly inversions (Bishop and Schiestl 2000). Furthermore, the systems-level study of the Halobacterium sp. NRC-1 response to UV irradiation demonstrated stress-induced up regulation of transposases, which may also mediate these large-scale genomic rearrangements (Baliga et al. 2004). The data also suggest that the initiation of the replication process in H. marismortui may also be involved in a similar manner with these genomic rearrangements. If so, consistent with our observations, such inversions would have one end at or near the origin of replication, leading to the hotspots pattern observed, and the sizes of these inversions would be highly variable. These evolutionary comparisons present a snapshot into the dynamic processes of evolutionary divergence between these two species, at a point at which most of the long-range colinearity has already been lost, yet enough of it remains locally to reveal the mechanistic and functional constraints acting on the sequence.
Comparative genomic analysis of the two halophiles does not provide convincing evidence for the hypothesis that the H. marismortui genome is chimeric
The suggestion that the H. marismortui genome is chimeric arose from the observation that there were two distinct rRNA operons in this organism (Mylvaganam and Dennis 1992; Dennis et al. 1998). In addition to the two previously reported copies of rRNA operons (rrnA and rrnB) in H. marismortui, we identified a third complete rRNA operon (rrnC) and a fourth copy of a 5S rRNA gene. Interestingly, the three ribosomal RNA operons each encoding the 5S, 16S, and 23S rRNA genes were identified on two different replicons: Operons rrnA and rrnC are encoded on chromosome I, and the rrnB operon is encoded on chromosome II. The fourth copy of the 5S rRNA is encoded on pNG700. As has been reported previously, we identified numerous pair-wise single nucleotide sequence differences among the rRNA paralogs with up to 83 differences in the three 16S rRNA copies, up to 45 differences among the three 23S rRNAs, and up to seven differences among the four 5S rRNA copies. The two rRNA operons encoded on chromosome I, rrnA and rrnC, share a high degree of similarity with most of these differences being in rrnB, (Mylvaganam and Dennis 1992; Dennis et al. 1998), which as indicated above, is encoded on a different replicon altogether.
A hypothesis depicting H. marismortui as a chimera between two related organisms has been advanced to account for the relatively extensive differences between the 16S rRNA genes. We evaluated three explanations for the presence of the 16S rRNA variants in H. marismortui: (1) because the rrnB operon is encoded on chromosome II, which appears to be weakly related in its organization to the three replicons in Halobacterium sp. NRC-1, this entire replicon might have been acquired later during evolution; (2) the 16S rRNA gene in rrnB, which is host to most of the sequence differences, may alone have been acquired through lateral gene transfer; and (3) the rrnB operon arose through duplication of and subsequent divergence from the rrnA or rrnC operon.
In considering explanations 1 and 2, for survival of two rRNA gene variants in a chimeric genome, two protein synthesis machineries should exist to differentially translate the proteins encoded by each of the two component genomes (Mylvaganam and Dennis 1992). However, the differences in the rRNA variants in H. marismortui lie in paired complementary sites, making it unlikely that they alter the structures of the ribosomal machinery. Therefore, the presence of two protein synthesis machineries is unlikely. Furthermore, we see no support for this hypothesis from the evolutionary comparison of chromosome I to that of Halobacterium sp. NRC-1 chromosome, which did not reveal any large-scale genomic insertions in the former (see above). Therefore, the first two explanations are unlikely, and the genome sequence analysis favors the third explanation.
H. marismortui genome may encode multiple origins of replication
Although archaea have circular genomes and their genes are organized into operons as in bacteria, their DNA replication proteins are more closely related to those in the eukaryotic machinery. The initiation of replication occurs at the origin through loading of the minichromosome maintenance protein Mcm, which activates the origin recognition complex (Orc1; also known as the cell division cycle protein [Cdc6]) (Bohlke et al. 2002). Whereas most archaea including Halobacterium sp. NRC-1 encode a single Mcm protein required for the initiation of DNA replication, H. marismortui, similar to Methanococcus jannaschii (Bernander 1998), encodes at least three Mcm proteins, two on chromosome I and the one on pNG300. Similarly, in contrast to the nine Orc1/Cdc6-like replication proteins encoded in the Halobacterium sp. NRC-1 genome, at least 17 Orc1/Cdc6 homologs were identified in H. marismortui.In H. marismortui the cdc6 genes are distributed among the nine replicons as follows: chromosome I, cdc6c-j; chromosome II, cdc6a-b; pNG700, cdc6k; pNG600, cdc6l-m; pNG500, cdc6n-o; pNG300, cdc6p; and pNG100, cdc6q. In most archaea the cdc6 gene is located adjacent to the origin of replication (Bernander 2000), which also coincides with sharp changes from positive to negative value in nucleotide compositional skews, for example, GC skew (i.e., strand-specific overrepresentation of guanine versus cytosine) (Grigoriev 1998; Kennedy et al. 2001; Berquist and DasSarma 2003).
In chromosome I of H. marismortui, we observed significant but, relative to Halobacterium sp. NRC-1, much less extensive regions of GC skew (Supplemental Fig. C, panel A). A single sharp boundary is evident, from a region with excess guanines (2.09–2.37 Mb) to a region with excess cytosines (2.42–2.50 Mb). The H. marismortui orc7 ortholog is located within this region (2.415 Mb) (Berquist and DasSarma 2003). Independent of G+C content and GC skew, a sequence may also be biased in adenine versus thymine (AT skew). We measured the AT skew in both halophilic species and found the AT skew to be more extensive and significant in H. marismortui, which, not surprisingly, has a lower G+C content than does Halobacterium sp. NRC-1 (Supplemental Fig. C, panel B). Interestingly, the boundary between several of the significant transitions from high to low AT skew in chromosome I of H. marismortui coincide with the location of the cdc6 genes.
Given these observations, it is tempting to speculate that H. marismortui has multiple origins of replications. However, computational (Kennedy et al. 2001; Zhang and Zhang 2003) as well as experimental analysis in Halobacterium sp. NRC-1 (Berquist and DasSarma 2003) has demonstrated only two origins of replication, both of which coincide with the location of two of the nine cdc6 gene orthologs (orc7 and orc8). Therefore, the implication of multiple copies of cdc6 in the genome of these halophiles remains unclear. (For a description of other proteins involved in DNA replication, repair, and recombination, see Supplemental material).
Proteome analysis: Structure and function
An annotation procedure comprised of four steps—the identification of primary sequence similarities, protein family signatures, and three-dimensional structural similarities, as well as positioning in a functional association network—provides functional insights for a greater number of proteins than obtained by any single procedure alone.
Gene prediction and function assignment
Analysis of the H. marismortui genome with the gene prediction program Glimmer predicted 4242 putative protein coding genes, which together in total account for ∼84.5% of the genome sequence. In an attempt to functionally annotate each of the 4242 proteins, we have applied a multi-tiered strategy that included BLASTP and additional software providing, protein family signatures, transmembrane region predictions (TMHMM), and ab initio structure predictions (e.g., Rosetta; for details, see Methods) (Chivian et al. 2003). We have also used the organization of genes into operons and applied comparative genomics approaches to construct a functional association network (see below) to provide contextual information for each protein.
Proteins were separated into functional domains, when domain boundaries were detectable, by using Ginzu (Chivian et al. 2003). Up to 3319 of the 4242 predicted proteins in H. marismortui analyzed by this approach appeared to contain a single domain and 923 contained multiple domains. Because of limitations in detecting domain boundaries for proteins with few or no homologs, a significant fraction of these single domain proteins may in fact have multiple domains. Of the 5607 H. marismortui protein domains analyzed, 4338 domains (∼77.4%) had significant matches by at least one annotation method to entries in public sequence and structure databases. Accordingly, if the match had a defined function, then the H. marismortui protein could be putatively assigned a similar function.
Functional association network
Comparative genomic approaches provide powerful means for understanding gene function through the assignment of an unknown protein to a known, protein interaction, or gene regulatory networks. To facilitate this functional annotation and help determine possible networks of function and gene regulation in the H. marismortui genome, we constructed a functional association network (Supplemental Fig. D; Supplemental Table A) among the 4242 proteins encoded in the H. marismortui genome. Briefly, we compared, using BLASTP, the primary sequences for all of these proteins to themselves as well as to 350,111 proteins from 89 organisms whose complete genome sequences have been determined (65 eubacteria, 16 archaea, and eight eukaryotes). Altogether 6589 functional associations were identified; these functional associations were of the following four major types: domain fusion association (genes separate in some genomes and fused in others often share related functions), phylogenetic pattern association (genes that are present together in many genomes often share related functions), operons (genes that are tightly linked can be coordinately expressed to carry out related functions), and protein families (paralogs often carry out distinct but related functions) (Marcotte et al. 1999; Overbeek et al. 1999; Pellegrini et al. 1999; Moreno-Hagelsieb and Collado-Vides 2002) (for detailed descriptions on each of these functional associations, see Supplemental material).
In this functional association network, we have identified putative functions for 231 of the 1223 proteins that did not have a significant match to any characterized protein sequence, family signature or structure (Supplemental Fig. D, panel B). This was done by virtue of the fact that these proteins were linked to known functional proteins by an operon relationship, gene fusion, or phylogenetic pattern association(s). This represents an additional 5.4% of all H. marismortui proteins and ∼19% of the unknown function category proteins, for which we now have some functional insights. Although this association network does not identify the exact function of a protein, it may nevertheless help suggest a general functional role for a protein with little or no sequence similarity to characterized proteins by virtue of the fact that it is associated with genes of known function (for an illustration of this approach, which implicates at least 40 genes in the denitrification process, see below) (Marcotte et al. 1999; Bonneau et al. 2004).
In summary, BLASTP identified orthologs for 3099 proteins in the GenBank non-redundant protein, SWISS-PROT, and EMBL-EBI proteome analysis databases. Of these, 1667 (∼39.3%) matched characterized proteins and were assigned putative functions, 1432 (∼33.8%) matched proteins of unknown function, and the remaining 1143 proteins did not match any extant entry in the databases. Protein family signatures and similarities of predicted three-dimensional protein structures assigned putative functions to an additional 555 (∼13%) proteins that escaped annotation by BLASTP (Supplemental Table B). Finally, the functional association network provided insight into putative functions for an additional 231 proteins (∼5.4%). Therefore altogether, these annotation approaches provided putative functional information for at least 2453 (∼58%) of the total 4242 proteins.
The integrated annotation and network discovery approach implicates at least 40 genes in the denitrification process
We illustrate the power of the multi-tiered strategy to annotating genome sequences with an example of its application to annotating genes directly or indirectly associated with the denitrification process in H. marismortui. Reduction of nitrate into nitrogen and nitrous oxide is an important process in the global nitrogen cycle. Organisms from all three domains of life are capable of carrying out this process. In addition to the experimentally characterized nitrite reductase (NirK) and nitrate reductase (NarGH) in H. marismortui (Yoshimatsu et al. 2000, 2002; Ichiki et al. 2001), the genome analysis identified at least four additional proteins with functions important for denitrification, viz. a nitrilase, a carbon-nitrogen hydrolase involved in reduction of organic nitrogen compounds and ammonia production; an ammonia-dependent NAD synthetase; a dissimilatory nitrate reductase; and an ammonium transporter.
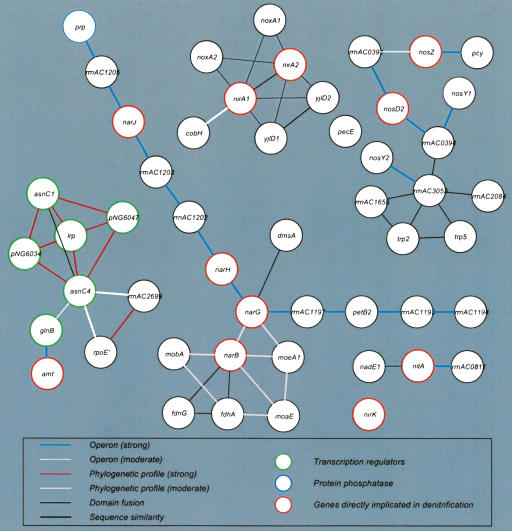
The genome organization and function associations of the denitrification proteins provide additional insights into the roles that other proteins might be playing in this process (Fig. 2). The narG and narH genes are in an operon with genes encoding cytochrome b6, a Rieske domain Fe-S protein; NarJ a chaperone believed to be necessary for molybdenum cofactor assembly; a protein phosphatase, a putative ABC-type phosphate transport system protein; and four proteins of unknown function. NarG also has phylogenetic associations with a third nitrate reductase (NarB), and a molybdenum cofactor biosynthesis protein (MoeA). Likewise, NarB has similar phylogeny as two MoeA proteins, and a molybdenum-guanine dinucleotide biosynthesis protein. This evolutionary functional association of denitrification genes with molybdenum cofactor biogenesis genes is indicative of the fact that nitrate reductase function requires molybdenum cofactor. Similarly, the operon-like organization of the genes encoding Amt, an ammonium transporter; GlnB, a regulator of glutamine synthetase activity; and Lrp, a leucine-responsive transcription regulator, suggests a regulatory link between ammonium uptake and its assimilation by glutamine synthetase.
Figure 2.
Functional associations among proteins implicated in the denitrification process in H. marismortui. (Inset) Key indicates functional relevance of node and edge color attributes (for other details, see text). A detailed list of dentrification proteins is included in Supplemental Table C.
Altogether, the integrated annotation and network discovery approach has identified at least 40 genes in H. marismortui that are potentially associated with the denitrification process (Fig. 2; Supplemental Table C; Stouthamer 1991). There is an obvious relationship to the denitrification process for some of these functions such as requirement of molybdenum cofactor biogenesis for nitrate reductase function. Although the functional association network does not provide precise roles in the denitrification process for other proteins, such as the protein phosphatase and cytochrome b6 as well as the unknown function proteins, it does, however, provide a clear path to their experimental analysis.
The unique physiochemical properties of H. marismortui proteins may facilitate structural genomics
As in Halobacterium sp. NRC-1, the H. marismortui proteome is highly acidic with an average isoelectric point of 5.0. The acidic residues in these proteins are predominantly present on the surface of the folded proteins. This unusual property is believed to be necessary to maintain the structure and function of these proteins in the highly salt saturated cytoplasm (Kennedy et al. 2001). In addition, this high negative surface charge probably makes many haloarchaeal proteins soluble that are insoluble under similar conditions in organisms displaying less protein surface charge. Furthermore, archaeal proteins in general have been shown to have higher stability owing to the shorter loops and, in some cases, for example, in hyperthermophiles, a higher density of cysteine residues paired in disulfide linkages (Mallick et al. 2002). These unusual properties of haloarcheal proteins directly address several technical hurdles encountered in structural biology, that is, insolubility of majority of proteins upon overexpression or during crystallization. Moreover, these physicochemical properties provide important constraints for the correct folding of proteins, which in turn should facilitate higher quality ab initio three-dimensional structure predictions by programs such as Rosetta (Bonneau et al. 2004).
H. marismortui physiology
Overview of the semi-automated approach to metabolic reconstruction
The annotation procedures that we have developed assigned, in an automated manner, Enzyme Commission (EC) designations to all proteins that matched a characterized protein sequence, three-dimensional structure, or family signature (for details, see Methods). Subsequently, by using a Web-based intermediary software tool (wbi), we searched all of these EC numbers against biochemical pathways in the Kyoto Encyclopedia of Genes and Genome (KEGG) (Kanehisa 2002). The EC numbers that matched biochemical steps were highlighted (by KEGG) in the pathway maps and hyperlinked (by the wbi tool) to the corresponding annotation entries in the Systems Biology Experiment, Analysis and Management System (SBEAMS) database, which is routinely used at ISB for storing, managing, and exploring diverse data types, such as genome sequence, annotation, microarray data, and proteomics data. The following insights into H. marismortui physiology were realized by manually verifying functional annotations for proteins assigned to specific enzymatic steps in KEGG biochemical pathways. We also made use of the KEGG pathway display to simultaneously visualize and compare biochemical pathways' memberships of Halobacterium sp. NRC-1 and H. marismortui proteomes (Fig. 2).
Overview of central metabolism
Glycolysis and the modified Entner-Doudoroff (ED) pathways are the major mechanisms of sugar breakdown in H. marismortui. The products of the oxidized sugars are acted upon by the TCA cycle enzymes, all of which have also been identified, for energy production as well as for synthesis of important precursors for amino acid biosynthesis. Enzymes for synthesis of up to 16 amino acids are encoded in the H. marismortui genome sequence. Halobacterium sp. NRC-1 on the other hand is unable to synthesize at least eight amino acids. H. marismortui may also catabolize some amino acids as sources of energy and/or use them to provide metabolic carbon and nitrogen. Finally, the reverse TCA cycle (reductive carboxylate cycle) may work together with the phosphoenol pyruvate (PEP) carboxylase and enzymes of gluconeogensis to fix inorganic carbon into sugars (for a description of oxidative phosphorylation, lipid metabolism, cell envelope components, and transporters in H. marismortui, see Supplemental material).
In addition to the expected extensive similarity in the physiologies of Halobacterium sp. NRC-1 and H. marismortui, there are striking differences such as one of two distinct arginine catabolism pathways in each organism
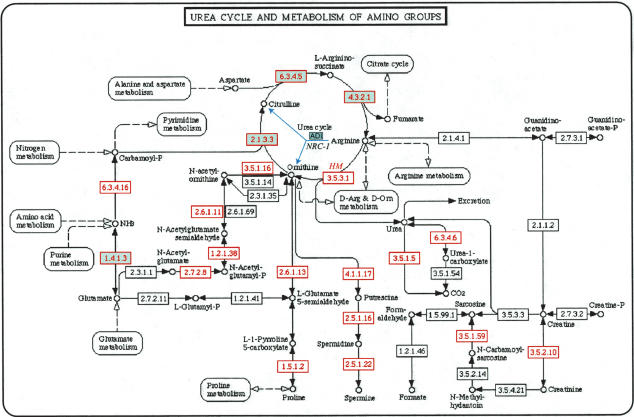
While growing under anaerobic conditions, fermentation of arginine through the arginine deiminase (ADI) pathway is a major source of energy for Halobacterium sp. NRC-1 (Ruepp and Soppa 1996). In striking contrast, H. marismortui degrades arginine via the arginase pathway to yield ornithine and urea, which is further degraded by a multi-subunit urease into ammonia and carbon dioxide (Supplemental Table D; Mizuki et al. 2004). Ornithine, on the other hand, may be converted into one of two amino acids, namely, back into arginine or glutamate; conversion of ornithine back to arginine results in conversion of aspartate to fumarate, and hence, the arginase pathway in H. marismortui may also interface with the TCA cycle (Fig. 3). However, unlike the ADI pathway, the breakdown of arginine by arginase does not yield energy. In fact a single round of the urea cycle utilizes three ATP molecules for every aspartate that is metabolized. Therefore, the main purpose of the arginase pathway and the urea cycle in H. marismortui might be to convert, through the TCA cycle, excess amino acids imported from the external environments into important metabolic intermediates.
Figure 3.
Comparative metabolic reconstruction in H. marismortui and Halobacterium sp. NRC-1. An example of metabolic reconstruction using the Web intermediary software and KEGG: arginine metabolism and the urea cycle in H. marismortui compared with Halobacterium sp. NRC-1. Enzymes identified in Halobacterium sp. NRC-1 are shaded in green, and those identified in H. marismortui are shown with a red border and red font. The ADI pathway of arginine fermentation in Halobacterium sp. NRC-1 is indicated with solid blue arrows. Each enzyme identified in H. marismortui is hyperlinked to its corresponding entry in the SBEAMS annotation database. This view of KEGG metabolic pathways for the two halophiles is available on http://halo.systemsbiology.net.
Analysis of gene organization provides insights into coordinate regulation of different aspects of metabolism, which in turn will enable the construction of hypothetical gene regulatory networks
The physical organization into operons of genes of related function is beneficial to an organism by allowing for coregulation and coexpression of genes of a selectable phenotype (Lawrence 1999; Omelchenko et al. 2003). This is evident in the operon-like organization and coordinate regulation of arginine metabolism genes in Halobacterium sp. NRC-1 (Ruepp and Soppa 1996; Ng et al. 2000) with those encoding other aspects of metabolism (for further description, see Supplemental material; Baliga et al. 2002). On the basis of these observations, we analyzed the organization of arginine metabolism genes in H. marismortui in an attempt to reconstruct their possible coordinated regulation with other aspects of metabolism. For example, the arginine synthesis gene arcB, in H. marismortui is in an operon with three of four genes encoding interconversion of glutamate and ornithine—an important intermediate of arginine metabolism. On the other hand, the arginine degradation gene arg2, which encodes an arginase, is in an operon with the glutamate dehydrogenase gene gdhA1, which encodes the interconversion of glutamate and oxoglutarate, a TCA cycle intermediate. Together these observations suggest coordinate regulation of genes encoding both arginine synthesis and breakdown with those encoding aspects of the TCA cycle and glutamate metabolism.
Similarly, the organization into an operon of one of two copies of the argB gene, which encodes interconversion of glutamate and ornithine, with an isoprenoid synthesis gene mvk, suggests coordinated regulation of both pathways (Baliga et al. 2002). Based on these observations, we speculate that the analysis of operon organization in prokaryotic genomes will play a key role in inferring their metabolic and gene regulatory networks.
The arginine synthesis and degradation functions in the two halophiles are segregated on different replicons
In Halobacterium sp. NRC-1, the genes encoding arginine synthesis are all encoded on the large chromosome, and the arginine fermentation genes are encoded on the smaller mini-chromosome pNRC200. Similarly, in H. marismortui all genes of arginine synthesis are encoded on the large chromosome. In contrast, most genes of arginine breakdown, with the exception of the two arginase genes on the large chromosome, are encoded on the mini-chromosome pNG700: These include an operon of seven genes for biogenesis of the urease enzyme complex; genes for agmatinase and creatininase, which similar to urease act downstream to the breakdown of arginine; and three other proteins of a spermidine/putrescine transport system. These observations lead us to conclude that notwithstanding alternate means for arginine breakdown in these halophiles, the genes of the synthesis and degradation pathways share common genomic organizational themes. The implication of this physical segregation on different replicons of synthesis and degradation functions, however, is not fully understood.
H. marismortui encodes a complex environmental response system
Photobiology: opsins, cryptochrome/photolyase, clock regulators, and transducers
Similar to Halobacterium sp. NRC-1, H. marismortui inhabits an extreme environment characterized by high salt concentration, low oxygen solubility, and high light intensity. An evolutionary adaptation common to these two organisms is a set of opsin proteins that use light energy to maintain physiological ion concentrations, facilitate phototaxis, and generate chemical energy in the form of a proton gradient. Halobacterium sp. NRC-1 encodes four archaeal opsins Sop1, Sop2, Hop, and Bop, which individually complex with a retinal chromophore to form the two sensory rhodopsins sRI and sRII and the ion pumps halorhodopsin (hR) and bacteriorhodopsin (bR), respectively. In contrast, six opsin genes are found in H. marismortui, including orthologs of all four opsin genes in Halobacterium sp. NRC-1. But the functions of the two remaining opsins (Xop1 and Xop2) can be only tentatively suggested at this time. Xop1 appears to contain most of the functionally important residues of bacteriorhodopsin. For example, it possesses an equivalent complement of carboxyl-containing residues, including the aspartic acid residue at position 96 (bR residue numbering is used for this discussion), which seems to be unique to bacteriorhodopsin and its orthologs. It also contains a tryptophan residue at position 138, which is known from high-resolution crystal structures of bacteriorhodopsin and its mutants to be a key component of the complex hydrogen-bond network on the extracellular side of the protein, which undergoes substantial rearrangements late in the photocycle (Supplemental Fig. E; Luecke et al. 2001; Rouhani et al. 2001; Facciotti et al. 2003). Thus, although it seems that Xop1 may function as a second bacteriorhodopsin, the significance of evolutionarily maintaining two such genes is difficult to explain unless there is some further differentiation of function. One possibility is that the two bacteriorhodopsins could be spectrally tuned to a slightly different λmax, thus helping to ensure a greater energy producing spectral range. Xop2, on the other hand, is more difficult to functionally annotate. Despite having some similarity to other known sensory opsins, particularly at positions 96(Y) and 138(F), the residue at position 211 (F or Y) important for the interface with the cognate Htr signal transducer is not present in Xop2. Instead, the residue 211(V) in Xop2 is reminiscent of the ion pumping opsins. This observation alone, however, does not preclude the possibility that Xop2 may function as a photoreceptor, and if Xop2 indeed functions as a photoreceptor, its cognate sensory transducer is not evident because, unlike other characterized sop (sensory-opsin) genes, xop2 is not found in an operon with any known htr (transducer) gene. The enzymes and regulation of retinal biosynthesis, a key component of the functional bacteriorhodopsin, are described in the Supplemental material.
In addition to the six opsins, two candidate sensory pigments (Phr1 and Phr2) that absorb maximally in the blue wavelength have been identified. Although highly similar at the primary sequence level, these two proteins may have different biological functions. For example, only one of these two proteins in many organisms including Halobacterium sp. NRC-1 (McCready and Marcello 2003; Baliga et al. 2004) has been implicated in DNA repair, in which it functions as a photolyase. On the other hand, the second protein may function as a cryptochrome that can mediate cellular responses to blue light (Cashmore et al. 1999; Hitomi et al. 2000).
Nine halocyanin precursor-like proteins were identified in H. marismortui genome; the physiological role of these plastocyanin-like proteins in halophilic archaea is not clear (Mattar et al. 1994). We also find proteins such as phycocyanobilins, which are major components of macromolecular complexes termed phycobilisomes that are involved with capture of energy from light (Glazer 1989). H. marismortui has at least 29 unique proteins containing one or more GAF domains, a light-responsive domain found in plant and cyanobacterial phytochromes, and invertebrate cGMP-stimulated phosphodiesterases (Aravind and Ponting 1997), of which several are likely to be phytochrome or phytochrome-like genes.
Finally, at least two kaiC circadian clock regulator-like genes were also detected. By contrast, the Halobacterium sp. NRC-1 genome encodes only a single kaiC ortholog. The presence of kaiC in the archaeal halophiles suggests that, as has been reported in cyanobacteria (Xu et al. 2003), they are capable of regulating their metabolism in response to the day/night cycle.
Transducers
During an environmental response, upon sensing a change in an environmental factor, a sensor such as sRI transmits signals to a bound transducer, which in case of sRI is HtrI. The transducer in turn communicates the signal to the chemotaxis apparatus and to other aspects of metabolism. The signal transducers characterized by the presence of at least one of two domains—MCP (PF00015) and HAMP (PF00672)—associated with methyl-accepting chemotaxis receptors that are found in abundance in both Halobacterium sp. NRC-1 and H. marismortui. Although Halobacterium sp. NRC-1 encodes 17 proteins containing the MCP and/or the HAMP domain(s), H. marismortui encodes 21 such proteins. Although often associated with transmembrane receptors, several of these proteins also appear to be soluble proteins. RrnAC0183, for instance, is believed to be a soluble heme-associated aerotaxis transducer.
H. marismortui encodes a large family of multidomain proteins that can act as both sensors and transcriptional or posttranscriptional regulators
H. marismortui has a well-developed sensory apparatus for sensing and responding to changes in physiologically available oxygen levels. We find 49 proteins with one or more PAS/PAC domains known to be associated with a variety of cofactors with redox states that are easily changed (Taylor and Zhulin 1999), many of which are also associated with signal-transduction histidine kinase domains. Those proteins with both kinase and sensory domains serve as both sensors and signal transducers.
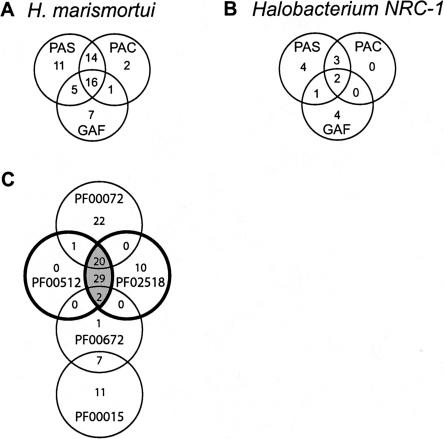
In addition to a wealth of sensory/receptor proteins, the H. marismortui genome contains a large number of sensory transduction proteins. At least 43 individual proteins containing the response regulator domain signature (PF00072), typically present in the sensor partner in a bacterial two-component system, were found. Many of these proteins are also transcription regulators that can directly regulate gene expression via N-terminal DNA binding domains. An alternative signal transduction mechanism using a histidine kinase (comprised of two domains: phosphoacceptor [PF00512] and ATPase [PF02518]) is also relatively abundant in H. marismortui. More than 62 individual proteins contain one or both of these his-kinase domains. Many of these proteins also contain PAS, PAC, GAF, or some combination of these sensory domains as well (Fig. 4; Supplemental Table E).
Figure 4.
Distribution in H. marismortui and Halobacterium sp. NRC-1of sensory domains among unique proteins having at least one such domain. The distribution of light-sensing (GAF [PF01590]) and redox-sensing domains (PAS [PF00989]), (PAC [PF00785]) in H. marismortui (A) and in Halobacterium sp.NRC-1 (B). (C) The distribution of methyl accepting chemotaxis (MCP–PF00015), HAMP (PF00672), histidine kinase (PF00512 and PF02518), and response regulator (PF00015) domains in H. marismortui. The blue circles represent the kinase domains. The red shaded area indicates the number of proteins that also contain PAS, PAC, or GAF domains.
Included among the response regulators is an intriguing group of large multidomain proteins (Oye and Oye1–3), containing multiple sensory related domains. The largest of these, Oye1, contains 13 PAC domains, 10 PAS domains, and two GAF domains connected to two signal transducing histidine kinase domains—27 domains total. The precise functional role of these large multidomain proteins in environmental sensing and response regulation is unclear. It is clear, however, that archaeal organisms have large multidomain sensory and signaling proteins capable of integrating a variety of different functions and stimuli. A comparison of the number and distributions of H. marismortui sensory proteins with those of similar proteins in Halobacterium sp. NRC-1 is enlightening and discussed further in the Supplemental material.
Relative to Halobacterium sp. NRC-1, the presence of nearly five times as many environmental response regulators in H. marismortui suggests that it has an enormously enhanced capacity to adapt to changes in a larger variety of distinct environmental factors.
The transcription regulatory networks in halophilic archaea are comprised of both transcription regulators and multiple copies of general transcription factors
The archaeal transcription apparatus resembles a stripped down version of the eukaryotic RNA polymerase II machinery. In all archaea, the preinitiation complex, which assembles prior to transcription initiation, is comprised of one TBP, one TFB, in some cases one TFE α subunit, and the multi-subunit RNA polymerase. None of the other factors such as TFIIF, TFIIH, and the β subunit of TFIIE necessary for transcription initiation by the corresponding RNA polymerase II in eukaryotes is present or essential in archaea (Bell and Jackson 1998).
Halophiles characteristically encode multiple TBP and/or TFB transcription factors. The multiplicity of these factors is hypothesized to play a role in transcription regulation through an assembly of pair-wise TBP-TFB combinations (Baliga et al. 2000). Unlike Halobacterium sp. NRC-1, which encodes at least six TBPs, H. marismortui genome encodes a single TBP located on chromosome I. In contrast to seven TFBs present in Halobacterium sp. NRC-1, H. marismortui genome encodes at least eight TFBs. In H. volcanii differential regulation of a TFB upon heat shock has been implicated in regulation of genes necessary for the appropriate response (Thompson et al. 1999). The presence of multiple TFBs in archaeal halophiles therefore suggests that this regulatory phenomenon may be common to many organisms in this group. Whereas two TFIIB signature repeats (PF00382) are present in all TFBs, a ninth protein in H. marismortui contains a single TFIIB repeat signature domain. Consistent with all heretofore genomically analyzed archaea, H. marismortui encodes a single TFE α subunit and lacks the β subunit. The lack of multiple TBPs was initially surprising because it suggests fewer combinatorial pairings with TFBs and therefore, perhaps, a simpler general transcription factor regulatory network for a genome that encodes nearly twice as many functions than does Halobacterium sp. NRC-1. This concern was subsequently addressed upon analyzing the transcription regulators encoded in H. marismortui (see below).
We identified, by using protein family signatures and Rosetta-predicted three-dimensional structures, at least 192 proteins with domains of DNA-binding function (Supplemental Table F). In comparison, at least 155 proteins in Halobacterium sp. NRC-1 contain domain(s) of DNA-binding function (Bonneau et al. 2004). Therefore, despite the likelihood of a simpler general transcription factor network, the presence of a larger complement of transcriptional regulators in H. marismortui points to a more complex gene regulatory network than is present in Halobacterium sp. NRC-1. In both halophilic archaea, the majority of these putative transcriptional regulators predicted through ab initio structure predictions are of the helix-turn-helix (HTH) “winged-helix” repressor DNA-binding domain type. Likewise, the predominant class of DNA-binding proteins identified through Pfam are also of the HTH type, with PF04937 being the most common HTH-type transcription factor family encoded in both halophilic genomes. It is interesting to note that with the completion of H. marismortui genome sequence, we have added 30 new members to the PF04937 domain family, which prior to the sequencing of this genome contained only 39 members, suggesting that several uncharacterized families of proteins await discovery in archaeal genomes yet to be sequenced.
Because the function of many DNA-binding proteins is to regulate gene expression via protein–DNA interactions, the precise biological functions of all predicted transcription factors and transcriptional regulators identified in this manner remain unknown until the cognate binding sites in the genome of the transcriptional factors and regulators are discovered and the correlation between regulator binding and gene expression is understood. Therefore, we await experimental data such as genome-wide transcription factor localization and genome-wide gene expression analyses to provide detailed functional information concerning these putative transcriptional regulators. Furthermore, the comparative analysis of the gene regulatory networks in the two halophiles will also provide insights into both unique and shared regulatory schema in each of these organisms (for details on RNA polymerase subunits and other aspects of the transcription process, see Supplemental material).
Coda
The genome sequence of H. marismortui has provided fascinating insights into the evolution of genome architecture of archaeal halophiles, viz.: (1) It has confirmed the speculation that haloarchaeal genomes are organized into a high G+C content large chromosome and multiple replicons of lower G+C composition; (2) many of the smaller replicons encode functions important for survival and therefore may represent the beginnings of genome organization into multiple chromosomes; (3) the ISs that can reconfigure genomic architecture are mostly encoded on the smaller replicons of lower G+C content; and (4) Halobacterium sp. NRC-1 and H. marismortui share a common ancestor, and the former has evolved an incrementally reduced genome. Furthermore, we have annotated the predicted proteins in the H. marismortui genome sequence using a multitiered approach, including analysis of primary sequence similarities, three-dimensional structural similarities, conserved phylogeny among genes, gene organization within and across genome(s), and gene fusion events. These approaches have permitted the functional annotation of a larger number of genes in the genome than is routinely achieved for most prokaryotic genome sequences. This effort has greatly facilitated the comparative genomics of metabolic pathways and protein networks to understand gene function, physiology and possible regulatory interactions among genes in H. marismortui and those encoded in Halobacterium sp. NRC-1.
Finally, upon comparing the environmental responses and regulatory functions encoded in the two halophiles, it was clear that relative to Halobacterium sp. NRC-1, H. marismortui is capable of adapting to many more diverse environmental conditions. This capacity of an organism to sense and respond to changes in environmental factors is encoded in its genome sequence in the form of genes and their cis-control elements that together specify signal transduction networks, metabolic networks, and gene regulatory circuits. In fact, the relatively small genome size of Halobacterium sp. NRC-1 coupled to availability of a multiplicity of advanced genomic and proteomic strategies for its study make it an attractive archaeal model organism in which to determine the complete gene regulatory network (Baliga et al. 2002, 2004; Weston 2004). Comparative genomic analyses of Halobacterium sp. NRC-1 and H. marismortui are expected to facilitate the discovery of these gene regulatory networks by providing insights into linkages such as protein-protein and protein-DNA interactions that are more likely to be better conserved among closely related species. A comprehensive knowledge of these biological networks will enable their modeling in a predictive framework such that new behaviors and processes can be engineered for future biotechnological applications.
Methods
Sequencing and assembly
Whole-genome shotgun sequencing method was applied to determine the H. marismortui (ATCC 43049) genome. Small- and large-insert shotgun libraries were constructed in the Escherichia coli vector pNG170 (Ng et al. 2000) with agarose gel-purified 1.6–2-kb and 4.5–5-kb genomic DNA fragments produced by sonication. Big-dye terminator chemistry was used to obtain paired sequence reads for both termini of 20,832 PCR amplicons from the 1.6-2-kb library as well as the inserts in 4224 plasmids from the 4.5–5-kb library (Ng et al. 1998). The phred, phrap, and consed software packages were used for sequence base calling and quality assessment, assembly, and editing (Ewing and Green 1998; Ewing et al. 1998; Gordon et al. 1998). The average sequencing success rate was 96%, and the high-quality nucleotide sequences (phred quality values, q ≥ 20) provided ∼7.2× coverage of the whole genome. By using the phredPhrap script with the “hide-and-seek” assembly approach (Ng et al. 2000), 81 contigs were obtained from the primary sequences. The sequence gaps and low-quality regions were then covered by sequencing the opposite-ends of ∼900 PCR-amplified genome fragments and 161 dye-primer sequencing reads of selected shotgun clones. The ribosomal RNA operon sequences were assembled separately by using an oligo-directed assembly approach (W.V. Ng, unpubl.) facilitated by the published rrnA and rrnB operon sequences (Dennis et al. 1998). The shotgun reads from the linked contigs without identical repetitive sequences were grouped and assembled without repeat masking to produce longer contig sequences. The final sequences were produced by merging the linked long contig sequences from the second assembly by using the phrap.longread script in the phrap software package.
Sequence annotation
The putative coding sequences (CDSs) were predicted by using the Glimmer program (Delcher et al. 1999). Primary functional assignment was performed by BLASTP searches of predicted protein sequences against GenBank non-redundant protein, SWISS-PROT, and EMBL-EBI proteome analysis databases. The BLASTP outputs were processed by using the MSPcrunch program (Sonnhammer and Durbin 1994). Functions were assigned to the predicted genes with significant matches. The tRNA genes were predicted using the tRNAScan-SE software (Lowe and Eddy 1997).
Domain parsing and domain annotation
To annotate predicted proteins, we used Ginzu (Chivian et al. 2003), a hierarchically organized combination of sequence-based methods (PSI-BLAST and HMMER) to separate proteins into domains and, subsequently, to annotate the protein domains as previously described (Altschul et al. 1997; Bateman et al. 2000). Briefly, regions of the query sequences with a significant match by PSI-BLAST to the Protein Data Bank (PDB) are masked and the remaining protein sequence is searched against Pfam using HMMER. Subsequently, domains with Pfam and/or PDB matches are masked and the sequence is searched using PSI-BLAST against the National Institute of Biotechnology Information non-redundant (nr) sequence database. Multiple sequence alignments resulting from this final PSI-BLAST run are then parsed into domains by using the chilli-eye-ball algorithm (Chivian et al. 2003). The domain parsing, Pfam, PSI-BLAST, and PDB-BLAST results for the H. marismortui proteome were stored in SBEAMS (see below). Transmembrane segments within proteins were detected with the TMHMM algorithm (Sonnhammer et al. 1998). By using a Python script, EC numbers were parsed from Pfam description Web pages and designated to all matching H. marismortui proteins.
Annotation of proteins using InterProScan and KEGG
In an automated process and by using a local installation of InterProScan (see http://www.ebi.ac.uk/interpro/README1.html), InterPro annotations were assigned to 422 proteins and TIGRFAM annoations were assigned to 495 proteins. Of these, 135 had EC numbers. In another automated process using the KEGG (http://www.genome.ad.jp/kegg-bin/srch_orth_html) database, more than 1700 COG (http://ncbi.nih.gov/COG) assignments were made to 1327 proteins (in some cases more than one COG assignment was made to one protein).
Rosetta structure prediction and structure–structure searches
Rosetta is a software tool that can predict ab initio from a primary sequence the three-dimensional structure of a protein. For each query sequence, 9000 independent simulations were carried out, each one resulting in a unique low-energy conformation. This ensemble of conformations was then clustered and ranked as previously described (Bonneau et al. 2002), resulting in 20 models for each query. The cluster threshold indicates the tightness of the cluster, whereas the mammoth Z-score provides a measure of similarity between the predicted structure to its orthologous structure in the PDB (Ortiz et al. 2002). EC number designations were automatically made for the proteins with matches to characterized enzymes in the PDB.
Biological network construction
Operon prediction
Two genes in the H. marismortui genome were considered to be in an operon if they were encoded on the same strand and within 40 bp of each other (Moreno-Hagelsieb and Collado-Vides 2002a).
Phylogenetic profile and domain fusion
To reconstruct phylogenetic distribution and domain fusion associations among proteins encoded in the H. marismortui, the protein sequences were compared with each other and with 350,111 proteins from 89 complete genomes (65 eubacteria, 16 archaea, and 8 eukaryotes) by using the program BLASTP under default settings. Phylogenetic profiles were constructed from the BLASTP results, summarizing the distribution in different organisms of homologs for each H. marismortui protein; subsequently, pairs of nonhomologous (E-value > 10-4) proteins with similar phylogenetic profiles were identified by measuring the mutual information (MI) between the profiles (Date and Marcotte 2003). Protein pairs with MI >0.85 were indicated in the association network as strong phylogenetic profile edges, whereas pairs of proteins with MI >0.70 but <0.85 were indicated as moderate phylogenetic profile edges.
Domain fusion linkages were inferred for each pair of nonhomologous H. marismortui proteins that showed significant sequence similarity (E-value < 10-6) to distinct, non-overlapping regions of the same protein in the database (Verjovsky Marcotte and Marcotte 2002).
Data visualization and integration using Cytoscape and SBEAMS
Cytoscape is a network visualization and exploration tool that enables simultaneous visualization of microarray and proteomics data in the context of a protein interaction/functional association network. Cytoscape can also incorporate several external sources of annotation such as KEGG and GO and provide in the form of plug-ins an array of integrated data analysis programs such as biomodule calculation, simulation of regulatory dynamics, network aware promoter, and protein motif recognition (Shannon et al. 2003).
SBEAMS
SBEAMS is a software and database framework developed at ISB that provides convenient HTML display of data tables with seamless access to external databases through a Web interface. SBEAMS was used as the primary database for the H. marismortui genome sequencing project, wherein the multitude of data types described in this work were stored, maintained, and accessed throughout the annotation process. In addition, external users accessing the database can add comments to any annotation in the table, thus facilitating our efforts to curate the annotation after initial release.
SBEAMS Interface with KEGG
We created a “Web intermediary” to search for KEGG metabolic pathways for all of the H. marismortui enzymes. EC numbers were attributed in an automated manner to each protein that matched a characterized protein sequence, family signature, or structure and subsequently stored in SBEAMS. At the user's request, by pressing a button on a Web page, all of these EC numbers were submitted to KEGG, and their individual mapping to metabolic pathways were retrieved. This mapping is returned by KEGG in the form of a long list in HTML, with all participating enzymes listed below each pathway. Each pathway in the list is hyperlinked to the HTML for the particular pathway map, which upon request is appropriately reprocessed by the Web intermediary so that EC numbers in that pathway corresponding to a given H. marismortui gene are highlighted and hyperlinked to the SBEAMS annotation table.
Evolutionary comparisons of the H. marismortui and the Halobacterium sp. NRC-1 genomes
Establishment of orthology relationships
We compared the 4242 predicted protein sequences from H. marismortui to the 2627 annotated protein sequences from Halobacterium sp. NRC-1 by using the BLAST2 implementation in Biofacet (Gene-IT), with an E-value cutoff of 0.0001. We selected those alignments with at least 30% identity over at least 80 amino acids (total, 7251 alignments) and used a mutual best match procedure to identify 1708 putative orthologous pairs. We similarly defined a reduced, higher-confidence subset of orthologous pairs by using 40% as identity cutoff over at least 200 amino acids. ADHoRe analysis (Vandepoele et al. 2002) was performed on the high-confidence subset of 1459 putative orthologous pairs, with maximum gap size of 25 and default quality parameter of 0.9. GRIMM analysis (Tesler 2002) was performed on the set of 1300 putative orthologous pairs between Halobacterium sp. NRC-1 chromosome and H. marismortui chromosome I, modeling evolution of a circular chromosome of signed genes.
Compositional skew analysis
We divided the genomic sequences into non-overlapping 10-kb windows and calculated for each window the G/C skew, defined as (G-C)/(G+C), where G and C represent the number of guanines and cytosines, respectively, within that window. The results were filtered by averaging over nine consecutive windows (90 kb) and normalized to the average and standard deviation of values observed in shuffled sequences. The resulting values are expressed as Z-scores (number of standard deviations away from the average). A/T skew was similarly measured.
Acknowledgments
We acknowledge Kerry Deutsch and Erik Schweighofer for assistance in setting up computing infrastructure and data submission, Glenn Tesler for help running the GRIMM software, and Gene-IT for providing the BioFacet software. This work was supported by National Science Foundation (NSF) grants 0220153 and 0313754 to N.S.B. and L.H., Department of Defense grant DAAD13-03-0-005 to N.S.B. and L.H., and NSC92-2314-B-010-055 from the National Science Council in Taiwan to W.V.N. E.M. and S.D. acknowledge grant support from the Welch Foundation, NSF, and Packard Foundation. We acknowledge Shiladitya Das-Sarma's help in obtaining initial funding for this project in the form of a collaborative NSF grant (0135595) with L.H.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2700304.
Footnotes
[Supplemental material is available online at www.genome.org and http://halo.systemsbiology.net. The sequence data from this study have been submitted to GenBank under accession nos. AY59290–AY59298].
References
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind, L. and Ponting, C.P. 1997. The GAF domain: An evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22: 458-459. [DOI] [PubMed] [Google Scholar]
- Baliga, N.S., Goo, Y.A., Ng, W.V., Hood, L., Daniels, C.J., and DasSarma, S. 2000. Is gene expression in Halobacterium NRC-1 regulated by multiple TBP and TFB transcription factors? Mol. Microbiol. 36: 1184-1185. [DOI] [PubMed] [Google Scholar]
- Baliga, N.S., Pan, M., Goo, Y.A., Yi, E.C., Goodlett, D.R., Dimitrov, K., Shannon, P., Aebersold, R., Ng, W.V., and Hood, L. 2002. Coordinate regulation of energy transduction modules in Halobacterium sp. analyzed by a global systems approach. Proc. Natl. Acad. Sci. 99: 14913-14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baliga, N.S., Bjork, S.J., Bonneau, R., Pan, M., Iloanusi, C., Kottemann, M.C.H., Hood, L., and DiRuggiero, J. 2004. Systems level insights into the stress response to UV radiation in the halophilic archaeon Halobacterium NRC-1. Genome Res. 14: 1025-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman, A., Birney, E., Durbin, R., Eddy, S.R., Howe, K.L., and Sonnhammer, E.L. 2000. The Pfam protein families database. Nucleic Acids Res. 28: 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell, S.D. and Jackson, S.P. 1998. Transcription in archaea. Cold Spring Harb. Symp. Quant. Biol. 63: 41-51. [DOI] [PubMed] [Google Scholar]
- Bernander, R. 1998. Archaea and the cell cycle. Mol. Microbiol. 29: 955-961. [DOI] [PubMed] [Google Scholar]
- ———. 2000. Chromosome replication, nucleoid segregation and cell division in archaea. Trends Microbiol. 8: 278-283. [DOI] [PubMed] [Google Scholar]
- Berquist, B.R. and DasSarma, S. 2003. An archaeal chromosomal autonomously replicating sequence element from an extreme halophile, Halobacterium sp. strain NRC-1. J. Bacteriol. 185: 5959-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop, A.J.R. and Schiestl, R.H. 2000. Homologous recombination as a mechanism for genome rearrangements: Environmental and genetic effects. Hum. Mol. Genet. 9: 2427-2434. [DOI] [PubMed] [Google Scholar]
- Bohlke, K., Pisani, F.M., Rossi, M., and Antranikian, G. 2002. Archaeal DNA replication: Spotlight on a rapidly moving field. Extremophiles 6: 1-14. [DOI] [PubMed] [Google Scholar]
- Bonneau, R., Strauss, C.E., Rohl, C.A., Chivian, D., Bradley, P., Malmstrom, L., Robertson, T., and Baker, D. 2002. De novo prediction of three-dimensional structures for major protein families. J. Mol. Biol. 322: 65-78. [DOI] [PubMed] [Google Scholar]
- Bonneau, R., Baliga, N.S., Deutsch, E.W., Shannon, P., and Hood, L. 2004. Comprehensive de novo structure prediction in a systems-biology context for the archaea Halobacterium sp. NRC-1. Genome Biol. 5: R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.J., and Liu, D. 1999. Cryptochromes: Blue light receptors for plants and animals. Science 284: 760-765. [DOI] [PubMed] [Google Scholar]
- Charlebois, R.L., Schalkwyk, L.C., Hofman, J.D., and Doolittle, W.F. 1991. Detailed physical map and set of overlapping clones covering the genome of the archaebacterium Haloferax volcanii DS2. J. Mol. Biol. 222: 509-524. [DOI] [PubMed] [Google Scholar]
- Chivian, D., Kim, D.E., Malmstrom, L., Bradley, P., Robertson, T., Murphy, P., Strauss, C.E., Bonneau, R., Rohl, C.A., and Baker, D. 2003. Automated prediction of CASP-5 structures using the Robetta server. Proteins 53(Suppl 6): 524-533. [DOI] [PubMed] [Google Scholar]
- Date, S.V. and Marcotte, E.M. 2003. Discovery of uncharacterized cellular systems by genome-wide analysis of functional linkages. Nat. Biotechnol. 21: 1055-1062. [DOI] [PubMed] [Google Scholar]
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27: 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis, P.P., Ziesche, S., and Mylvaganam, S. 1998. Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui. J. Bacteriol. 180: 4804-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using phred, II: Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using phred, I: Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Facciotti, M.T., Cheung, V.S., Nguyen, D., Rouhani, S., and Glaeser, R.M. 2003. Crystal structure of the bromide-bound D85S mutant of bacteriorhodopsin: Principles of ion pumping. Biophys. J. 85: 451-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer, A. 1989. Light guides: Directional energy transfer in a photosynthetic antenna. J. Biol. Chem. 264: 1-4. [PubMed] [Google Scholar]
- Goo, Y.A., Roach, J., Glusman, G., Baliga, N.S., Deutsch, K., Pan, M., Kennedy, S., DasSarma, S., Ng, W.V., and Hood, L. 2004. Low-pass sequencing for microbial comparative genomics. BMC Genomics 5: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Grigoriev, A. 1998. Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res. 26: 2286-2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitomi, K., Okamoto, K., Daiyasu, H., Miyashita, H., Iwai, S., Toh, H., Ishiura, M., and Todo, T. 2000. Bacterial cryptochrome and photolyase: Characterization of two photolyase-like genes of Synechocystis sp. PCC6803. Nucleic Acids Res. 28: 2353-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiki, H., Tanaka, Y., Mochizuki, K., Yoshimatsu, K., Sakurai, T., and Fujiwara, T. 2001. Purification, characterization, and genetic analysis of Cu-containing dissimilatory nitrite reductase from a denitrifying halophilic archaeon, Haloarcula marismortui. J. Bacteriol. 183: 4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa, M. 2002. The KEGG database. Novartis Found. Symp. 247: 91-101; discussion 101-103, 119-128, 244-252. [PubMed] [Google Scholar]
- Kennedy, S.P., Ng, W.V., Salzberg, S.L., Hood, L., and DasSarma, S. 2001. Understanding the adaptation of Halobacterium species NRC-1 to its extreme environment through computational analysis of its genome sequence. Genome Res. 11: 1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koop, B.F. and Hood, L. 1994. Striking sequence similarity over almost 100 kilobases of human and mouse T-cell receptor DNA. Nat. Genet. 7: 48-53. [DOI] [PubMed] [Google Scholar]
- Lawrence, J. 1999. Selfish operons: The evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 9: 642-648. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luecke, H., Schobert, B., Lanyi, J.K., Spudich, E.N., and Spudich, J.L. 2001. Crystal structure of sensory rhodopsin II at 2.4 angstroms: Insights into color tuning and transducer interaction. Science 293: 1499-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallick, P., Boutz, D.R., Eisenberg, D., and Yeates, T.O. 2002. Genomic evidence that the intracellular proteins of archaeal microbes contain disulfide bonds. Proc. Natl. Acad. Sci. 99: 9679-9684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotte, E.M., Pellegrini, M., Ng, H.L., Rice, D.W., Yeates, T.O., and Eisenberg, D. 1999. Detecting protein function and protein–protein interactions from genome sequences. Science 285: 751-753. [DOI] [PubMed] [Google Scholar]
- Mattar, S., Scharf, B., Kent, S., Rodewald, K., Oesterhelt, D., and Engelhard, M. 1994. The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J. Biol. Chem. 269: 14939-14945. [PubMed] [Google Scholar]
- McCready, S. and Marcello, L. 2003. Repair of UV damage in Halobacterium salinarum. Biochem. Soc. Trans. 31: 694-698. [DOI] [PubMed] [Google Scholar]
- Mizuki, T., Kamekura, M., Dassarma, S., Fukushima, T., Usami, R., Yoshida, Y., and Horikoshi, K. 2004. Ureases of extreme halophiles of the genus haloarcula with a unique structure of gene cluster. Biosci. Biotechnol. Biochem. 68: 397-406. [DOI] [PubMed] [Google Scholar]
- Moreno-Hagelsieb, G. and Collado-Vides, J. 2002. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics 18(Suppl 1): S329-S336. [DOI] [PubMed] [Google Scholar]
- Mylvaganam, S. and Dennis, P.P. 1992. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130: 399-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng, W.V., Ciufo, S.A., Smith, T.M., Bumgarner, R.E., Baskin, D., Faust, J., Hall, B., Loretz, C., Seto, J., Slagel, J., et al. 1998. Snapshot of a large dynamic replicon in a halophilic archaeon: Megaplasmid or minichromosome? Genome Res. 8: 1131-1141. [DOI] [PubMed] [Google Scholar]
- Ng, W.V., Kennedy, S.P., Mahairas, G.G., Berquist, B., Pan, M., Shukla, H.D., Lasky, S.R., Baliga, N.S., Thorsson, V., Sbrogna, J., et al. 2000. From the cover: Genome sequence of halobacterium species NRC-1. Proc. Natl. Acad. Sci. 97: 12176-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omelchenko, M.V., Makarova, K.S., Wolf, Y.I., Rogozin, I.B., and Koonin, E.V. 2003. Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome Biol. 4: R55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz, A.R., Strauss, C.E., and Olmea, O. 2002. MAMMOTH (matching molecular models obtained from theory): An automated method for model comparison. Protein Sci. 11: 2606-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek, R., Fonstein, M., D'Souza, M., Pusch, G.D., and Maltsev, N. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. 96: 2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini, M., Marcotte, E.M., Thompson, M.J., Eisenberg, D., and Yeates, T.O. 1999. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 96: 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhani, S., Cartailler, J.P., Facciotti, M.T., Walian, P., Needleman, R., Lanyi, J.K., Glaeser, R.M., and Luecke, H. 2001. Crystal structure of the D85S mutant of bacteriorhodopsin: Model of an O-like photocycle intermediate. J. Mol. Biol. 313: 615-628. [DOI] [PubMed] [Google Scholar]
- Ruepp, A. and Soppa, J. 1996. Fermentative arginine degradation in Halobacterium salinarium (formerly Halobacterium halobium): Genes, gene products, and transcripts of the arcRACB gene cluster. J. Bacteriol. 178: 4942-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon, P., Markiel, A., Ozier, O., Baliga, N.S., Wang, J.T., Ramage, D., Amin, N., Schwikowski, B., and Ideker, T. 2003. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13: 2498-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer, E.L. and Durbin, R. 1994. An expert system for processing sequence homology data. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2: 363-368. [PubMed] [Google Scholar]
- Sonnhammer, E.L., von Heijne, G., and Krogh, A. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6: 175-182. [PubMed] [Google Scholar]
- Stouthamer, A.H. 1991. Metabolic regulation including anaerobic metabolism in Paracoccus denitrificans. J. Bioenerg. Biomembr. 23: 163-185. [DOI] [PubMed] [Google Scholar]
- Taylor, B.L. and Zhulin, I.B. 1999. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63: 479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesler, G. 2002. GRIMM: Genome rearrangements web server. Bioinformatics 18: 492-493. [DOI] [PubMed] [Google Scholar]
- Thompson, D.K., Palmer, J.R., and Daniels, C.J. 1999. Expression and heat-responsive regulation of a TFIIB homologue from the archaeon Haloferax volcanii. Mol. Microbiol. 33: 1081-1092. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Saeys, Y., Simillion, C., Raes, J., and Van De Peer, Y. 2002. The automatic detection of homologous regions (ADHoRe) and its application to microcolinearity between Arabidopsis and rice. Genome Res. 12: 1792-1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verjovsky Marcotte, C.J. and Marcotte, E.M. 2002. Predicting functional linkages from gene fusions with confidence. Appl. Bioinformatics 1: 37-44. [PubMed] [Google Scholar]
- Weston, A.D., Baliga, N.S., Bonneau, R., and Hood, L. 2004. Systems approaches applied to the study of Saccharomyces cerevisiae and Halobacterium sp. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PubMed]
- Xu, Y., Mori, T., and Johnson, C.H. 2003. Cyanobacterial circadian clockwork: Roles of KaiA, KaiB and the kaiBC promoter in regulating KaiC. EMBO J. 22: 2117-2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimatsu, K., Sakurai, T., and Fujiwara, T. 2000. Purification and characterization of dissimilatory nitrate reductase from a denitrifying halophilic archaeon, Haloarcula marismortui. FEBS Lett. 470: 216-220. [DOI] [PubMed] [Google Scholar]
- Yoshimatsu, K., Iwasaki, T., and Fujiwara, T. 2002. Sequence and electron paramagnetic resonance analyses of nitrate reductase NarGH from a denitrifying halophilic euryarchaeote Haloarcula marismortui. FEBS Lett. 516: 145-150. [DOI] [PubMed] [Google Scholar]
- Zhang, R. and Zhang, C.T. 2003. Multiple replication origins of the archaeon Halobacterium species NRC-1. Biochem. Biophys. Res. Commun. 302: 728-734. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://halo.systemsbiology.net; Sequence, data, annotations, and analyses.
- http://www.ebi.ac.uk/interpro/README1.html); InterProScan.
- http://www.genome.ad.jp/kegg-bin/srch_orth_html; KEGG database.
- http://ncbi.nih.gov/COG; COG.