Abstract
Although the cost of generating draft-quality genomic sequence continues to decline, refining that sequence by the process of “sequence finishing” remains expensive. Near-perfect finished sequence is an appropriate goal for the human genome and a small set of reference genomes; however, such a high-quality product cannot be cost-justified for large numbers of additional genomes, at least for the foreseeable future. Here we describe the generation and quality of an intermediate grade of finished genomic sequence (termed comparative-grade finished sequence), which is tailored for use in multispecies sequence comparisons. Our analyses indicate that this sequence is very high quality (with the residual gaps and errors mostly falling within repetitive elements) and reflects 99% of the total sequence. Importantly, comparative-grade sequence finishing requires ∼40-fold less reagents and ∼10-fold less personnel effort compared to the generation of near-perfect finished sequence, such as that produced for the human genome. Although applied here to finishing sequence derived from individual bacterial artificial chromosome (BAC) clones, one could envision establishing routines for refining sequences emanating from whole-genome shotgun sequencing projects to a similar quality level. Our experience to date demonstrates that comparative-grade sequence finishing represents a practical and affordable option for sequence refinement en route to comparative analyses.
The strategy of “shotgun sequencing” (Sanger et al. 1982; Wilson and Mardis 1997b; Green 2001) has emerged as the most cost-effective approach for the de novo generation of large amounts of genomic sequence data. Whether applied on individual large-insert clones (C. elegans Sequencing Consortium 1998; International Human Genome Sequencing Consortium 2001), whole genomes (Adams et al. 2000; Venter et al. 2001; Aparicio et al. 2002; Mouse Genome Sequencing Consortium 2002), or a combination of both (Rat Genome Sequencing Project Consortium 2004), shotgun-sequencing strategies are typically performed in two broad phases. In the initial “shotgun” phase, highly redundant sequence data are obtained by generating sequence reads from one or both insert ends of randomly selected subclones derived from the starting DNA (large-insert clone or whole genome). This phase involves high-throughput methodologies and is responsible for generating the great majority of the actual sequence. In the second “finishing” phase, the assembled sequence emanating from the shotgun phase is analyzed and refined, with additional sequence data typically generated to attain long-range continuity and to improve accuracy. Sequence finishing is a low-throughput, craftsman-like process that involves highly skilled personnel performing both computational and experimental procedures in a customized fashion; as a result, it is also relatively expensive.
For sequencing the human genome, the Human Genome Project appropriately set very high standards with respect to the quality of the finished sequence (Felsenfeld et al. 1999; International Human Genome Sequencing Consortium 2001; see www.genome.wustl.edu/Overview/finrulesname.php?G16=1). Specifically, there was a rigorous set of standards that ensured consistency among different sequencing centers and a well-defined quality specification that required a low error rate (less than one error per 10,000 bases), the absence of gaps, and confirmation of the final sequence by comparison with a restriction enzyme digest-based fingerprint of each clone. Implementation of these standards yielded a remarkably accurate human genome sequence (International Human Genome Sequencing Consortium 2004), which has provided a powerful foundation for subsequent annotation efforts (Stein 2001; Ashurst and Collins 2003), comparisons with other species' sequences (Aparicio et al. 2002; Mouse Genome Sequencing Consortium 2002; Rat Genome Sequencing Project Consortium 2004), and efforts to untangle complex genomic structures, such as segmental duplications (Bailey et al. 2002). However, achieving such high standards required a considerable investment in sequence finishing, estimated to have been 30%–40% of the total cost. At present and with the recent decline in the costs of producing shotgun-sequence data, the resources required to perform such high-quality sequence finishing now correspond to 40%–70% of the total cost (data not shown).
It is well recognized that the quality of the sequence generated for the human genome, which we refer to as human-grade finished sequence, is substantially better than that available at the end of the shotgun phase. The latter full-shotgun draft sequence is simply derived from the automated assembly of the full collection of shotgun sequence reads (e.g., that providing greater than eightfold average sequence coverage). It is important to point out that in the progression from full-shotgun to human-grade finished sequence, there is not a linear relationship between the associated additional costs and the enhancement in sequence quality. Indeed, early in this progression, significant gains in quality can be achieved with even small amounts of additional effort (Wilson and Mardis 1997b; Gordon et al. 2001), whereas in later stages, large amounts of effort are often required to accomplish even small quality improvements.
In contemplating the sequencing of additional vertebrate genomes beyond the first pair of high-quality reference sequences (i.e., those of the human [International Human Genome Sequencing Consortium 2001, 2004] and mouse [Mouse Genome Sequencing Consortium 2002] genomes), the relative value of sequence finishing is of great interest. Specifically, understanding the relationship between overall sequence quality and the ability to extract relevant information by comparative analyses becomes important, especially in the context of analyzing sequences from multiple species. Motivated to generate genomic sequence from multiple species suitable for comparative analyses (Margulies et al. 2003a,b; Thomas et al. 2003), we sought to investigate whether an intermediate grade of finished sequence could be produced that was both cost-effective and appropriate in terms of quality. Toward that end, we have established an approach for generating what we call comparative-grade finished sequence. Here we report details about comparative-grade finished sequence, as generated on a large scale for bacterial-artificial chromosome (BAC) clones (Shizuya et al. 1992; Birren et al. 1998). In addition, we assess the relative quality of this sequence and the effort and costs associated with producing it.
Results
Conceptualization of comparative-grade finished sequence
By studying large data sets of genomic sequence generated from multiple species (Thomas et al. 2003; see www.nisc.nih.gov), we have gained considerable insight about the quality of sequence needed to perform detailed comparative analyses. By using that knowledge, we sought to establish an experimental approach for producing substantially higher-quality sequence with a small amount of additional effort beyond the generation and assembly of highly redundant shotgun sequence reads. The resulting sequence product (comparative-grade finished sequence) would be an intermediate between full-shotgun draft sequence and human-grade finished sequence.
Our experience in comparative sequence analyses indicated the need for a minimal set of characteristics for such a sequence product. First, the shotgun sequence reads must provide sufficient redundancy to ensure a high-accuracy consensus sequence and long-range continuity following assembly; when applied to a diverse set of different species' genomes, this most reliably and cost-effectively can be accomplished with greater than eightfold average coverage, thereby comfortably exceeding established minimum thresholds (Bouck et al. 1998). Second, the sequence must be devoid of gross misassemblies and regions of notably poor quality (see Methods for details). Third, the assembled sequence contigs must be definitively ordered and oriented. It is important to emphasize that the above characteristics were conceptualized based on our initial efforts to use multispecies sequences to perform comparative studies of gene structure, genome dynamics, and sequence conservation (Thomas et al. 2003). Indeed, the lack of such characteristics greatly hampers long-range sequence assemblies (e.g., of multiple BACs) and multispecies sequence analyses, annotations, and comparisons. Such problems become exacerbated when comparative analyses include more distantly related sequences.
Based on the above insights and desired characteristics, we established a core set of specifications for comparative-grade finished sequence, albeit ones that could be obtained with a minimal amount of additional effort. Similar to the G16 standards established for generating the human genome sequence (see www.genome.wustl.edu/Overview/finrulesname.php?G16=1), the requisite features of comparative-grade finished sequence can be implemented in a routine fashion by technicians accustomed to sequence finishing (i.e., “finishing technicians”). The major specifications for comparative-grade finished sequence are (1) the underlying sequence assembly must be based on at least eightfold average coverage in high-quality (Phred Q20) bases; (2) detectable, major artifacts resulting from the sequence-assembly process (e.g., misassemblies) must be resolved; (3) suspect, low-quality consensus sequence must be removed from the ends of sequence contigs; and (4) all sequence contigs >2 kb in size must be ordered and oriented relative to one another, and this must be verified using independent data. The latter include sequence derived from overlapping BACs, orthologous sequences from other species, and the results of PCR-based studies (see Methods). Note that because the contigs are ordered and oriented, we submit comparative-grade finished sequence as “phase 2” genomic sequence to GenBank (see www.ncbi.nlm.nih.gov/HTGS); indeed, because the order and orientation of contigs is based on a strong compilation of evidence (including read-pair information, PCR verification, and independent supporting data), this really represents an enhanced type of phase 2 sequence.
Generation of comparative-grade finished sequence
By using the above-detailed specifications, we have now generated >350 Mb of comparative-grade finished sequence from >1900 BACs. Importantly, for ∼67% of the BACs, the comparative-grade finishing process was exclusively computational, requiring no additional experimental work. The small amount of experimental effort with the remaining BACs solely involved performing PCR to establish contig order and orientation. The latter is required when a nascent BAC sequence contains more than one “uncaptured gap” (i.e., a gap with zero or one spanning subclone; “captured gaps” are those with two or more spanning subclones with insert-end sequences that each reside within the adjacent contigs). Some uncaptured gaps require multiple attempts at PCR amplification to confirm contig adjacency, often using several sets of primers or a variety of reaction conditions to generate an authentic product (see Methods).
Quality of comparative-grade finished sequence
To investigate the relative quality of different types of generated sequence, we focused more intensely on a set of 116 BACs from 18 species (the complete list of clones is available at www.nisc.nih.gov/data). All 116 BACs, which together reflect ∼15 Mb of total sequence, were derived from the same orthologous region encompassing the CFTR gene (Thomas et al. 2003), which in the human genome is fairly average with respect to general genomic features (e.g., 38.4% GC content, 1.1% exonic sequence, and 40.3% repetitive sequence).
For each BAC, we generated and analyzed three types of sequence (see Methods): (1) full-shotgun draft sequence (in all cases, the assembled sequence provided greater than eightfold average coverage in high-quality bases); (2) comparative-grade finished sequence; and (3) human-grade finished sequence. Note that the two types of finished sequence were generated independently, in each case starting with the same full-shotgun assembled sequence for that BAC. Also, the human-grade finished sequence met the standards established for finishing the human genome sequence; specifically, there were no gaps and an overall Phrap-estimated error rate of ∼0.02 errors per 10,000 bases. The human-grade finished sequence was then annotated for repetitive and exonic sequence (Thomas et al. 2003), with a summary provided in Table 1.
Table 1.
General annotated features of the 116 BAC sequences from multiple nonhuman species used for investigating the quality of comparative-grade finished sequence
| Total sequence (from 116 BACs) | 14,948,004 bases |
| Total repeats | 4,989,823 bases (33.4%) |
| Simple repeats | 92,410 bases (0.6%) |
| Exons | 250,700 bases (1.7%) |
Comparison of the full-shotgun draft sequence to either type of finished sequence revealed the markedly inferior quality of a nonfinished product. The full-shotgun draft sequence of the 116 analyzed BACs consists of 507 contigs. Of these, 199 are at least partially ordered and oriented due to the presence of BAC insert-end sequences, whereas the remaining 308 contigs are ordered and oriented only in the comparative-grade finished sequence (but not in the full-shotgun draft sequence). Such a lack of contig order and orientation can hinder gene-annotation efforts. This is illustrated in Figure 1, which shows that Genscan (Burge and Karlin 1997) gene predictions are less accurate using full-shotgun draft sequence compared to using the corresponding comparative- or human-grade finished sequence. Manual gene-annotation efforts are also hindered by the lack of contig order and orientation. Indeed, of the 132 genes wholly or partially included within the 116 analyzed BACs, 78 are disrupted by one or more gaps (pre-dominantly within their introns). Annotation of the latter genes is profoundly challenging without knowing the spatial relationships of contigs. In addition, the presence of low-quality regions, such as those at the ends of nascent contigs, greatly reduces the overall accuracy of full-shotgun draft sequence. For example, the progression from full-shotgun draft to comparative-grade finished sequence resulted in the removal of 67,099 bases from contig ends (of the ∼15 Mb of total sequence). Of that removed sequence, only 26,489 bases can be aligned to the human-grade finished sequence, with 2052 of those bases being in error. The remaining 40,610 bases of non-aligned, trimmed sequence likely reflect chimeric, contaminant, or notably poor-quality sequence. Finally, it is important to stress that the assembly of long-range sequences by the pair-wise merging of BAC sequences, either manually or using automated tools, is very problematic in the absence of quality-trimmed, ordered, and oriented contigs (data not shown). These results thus confirm the expectation that assembled draft sequence represents a considerably lower-quality product than either type of finished sequence.
Figure 1.
Predicted gene structures using different grades of genomic sequence. The annotated relative positions of exons in the rat CAPZA2 (A), baboon CAV2 (B), and lemur GASZ (C) genes are indicated. In each case, the exon positions predicted by Genscan (Burge and Karlin 1997) using each of the different types of genomic sequence are shown below (generated for BAC clones RP31-188L2 [GenBank no. AC087041], RP41-479B1 [GenBank no. AC084730], and LB2-246N5 [GenBank no. AC123544], respectively). The positions of gaps in the full-shotgun draft and comparative-grade finished sequence are shown as grey boxes. Note that using full-shotgun draft sequence (with unordered contigs separated by stretches of 50 Ns), Genscan incorrectly predicts the positions of a number of exons whose positions are correctly predicted by using comparative-grade finished sequence (with ordered and oriented contigs separated by stretches of 50 Ns) or human-grade finished sequence. There are also cases in which Genscan incorrectly predicts exons using all three types of sequence.
We systematically compared the comparative-grade finished sequence to the corresponding human-grade finished sequence for the set of 116 BACs. Analysis of the regions missing in comparative-grade finished sequence (i.e., gaps) revealed the data summarized in Table 2. A total of 344 gaps averaging 458 bases in size are present in the comparative-grade finished sequence, for an average of three gaps per BAC (or 23 gaps per megabase). The 157,505 gap bases (i.e., bases missing from the comparative-grade finished sequence but present in the human-grade finished sequence) only reflect 1.1% of the total sequence. These gap bases correspond disproportionately to certain types of sequence. For example, ∼50% of the gap bases fall within repetitive sequences (total repeats) (Table 2), whereas only 33% of the sequence corresponds to total repeats (Table 1). Similarly, 3.7% of the gap bases fall within simple repeats (predominantly stretches of mono-, di-, tri-, and tetranucleotides) (Table 2), whereas only 0.6% of the sequence corresponds to simple repeats (Table 1). Finally, 2.4% of gap bases fall within annotated exons (Table 2), whereas exons constitute 1.7% of the sequence (Table 1). In 44 instances (among the 116 BACs), the comparative-grade finished sequence consisted of two overlapping contigs that were not joined during assembly. Since all of the underlying sequence was actually present, these “virtual gaps” were not included among the data in Table 2.
Table 2.
Characteristics of gaps within the ∼15 Mb of comparative-grade finished sequence
| Number of gaps | 344 |
| Number of gap bases | 157,505 bases (1.1% of total sequence) |
| Average gap size | 458 bases (range 3 to 4,763) |
| Gap bases in total repeats | 78,535 bases (49.9%) |
| Gap bases in simple repeats | 5,797 bases (3.7%) |
| Gap bases in exons | 3,777 bases (2.4%) |
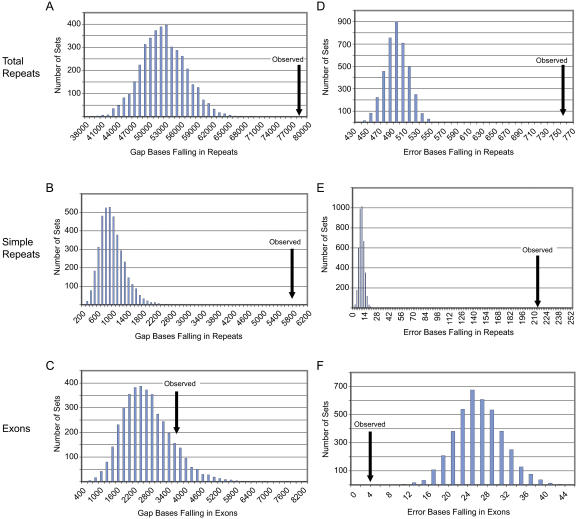
More rigorous analyses demonstrate the statistically significant enrichment of repetitive sequences and the lack of a statistically significant enrichment of exonic sequences within the gaps of comparative-grade finished sequence (Fig. 2). Specifically, a total of 4000 simulated data sets were generated by randomly placing the same number and size of gaps across the total sequence. The number of gap bases falling within total repeats, simple repeats, and exons were then counted after each simulation. The observed number of gap bases in comparative-grade finished sequence falling within total (Fig. 2A) and simple (Fig. 2B) repeats was considerably higher than that seen with the simulated data sets (P < 0.00025). These results indicate that gaps are more likely to occur in repetitive than nonrepetitive sequence. Finally, the observed number of gap bases residing within exons fell within the distribution generated with the simulated data sets (P = 0.20) (Fig. 2C).
Figure 2.
Analysis of gaps and errors in comparative-grade finished sequence by simulation studies. The histogram bar heights reflect the total gaps or errors falling within each class of annotated sequence (total repeats [A,D], simple repeats [B,E], and exons [C,F]) for the simulated data sets. The arrows point to the observed values with the generated comparative-grade finished sequence (for additional details, see Methods). Note that the observed low level of sequence errors falling in exons (F) likely reflects the fact that the generation of simulated data sets assumes a uniform distribution of errors across repetitive and nonrepetitive sequence (which in reality is not seen).
We investigated in greater detail the relative contribution of sequencing and cloning problems to the formation of different types of gaps. To begin with, we examined the lengths of the sequence reads assembling immediately adjacent to the 344 gaps in our data set. While all sequence reads in our assemblies average 546 Q20 bases in length (with an error of the mean of less than one base), reads that align to at least some portion of the 200 bases adjacent to a gap and are directed into that gap average 450 Q20 bases in length (with an error of the mean of 10 bases). Although this result indicates that shorter read lengths are associated with gaps, there does not seem to be a significant difference in the average lengths of reads directed into (1) the 63 simple-repeat-containing gaps (440 Q20 bases); (2) the 187 repeat-containing gaps devoid of simple repetitive elements (450 Q20 bases); and (3) the 94 gaps containing no repeats (460 Q20 bases). These results indicate that sequencing problems play a role in the formation of gaps in repetitive and nonrepetitive regions. A total of 68 (20%) of the gaps are not associated with adjacent sequence reads directed into the gap; these 68 gaps account for 14% of the simple-repeat–containing gaps, 26% of the repeat-containing gaps devoid of simple repetitive elements, and 11% of the gaps containing no repeats. Indeed, the majority of these gaps are uncaptured, most likely caused by cloning problems. Of note, we find a general enrichment of repetitive sequences in uncaptured versus captured gaps (data not shown). Together, these results are consistent with previous studies demonstrating the premature termination of sequencing reactions within repetitive regions (McMurray et al. 1998; Langan et al. 2002; Keith et al. 2004) and the known difficulty of cloning some types of repetitive sequences in common laboratory strains of Escherichia coli (Ishiura et al. 1989; Chissoe et al. 1997; Razin et al. 2001).
We also investigated the possibility that some gaps are a result of the Phrap-assembly process. Specifically, of the ∼102,000 nonassembled (singlet) reads from the 116 BAC assemblies, only 84 (averaging 152 Q20 bases in length) align to the human-grade finished sequence. Of these, only 38 align within a gap, and the overall short lengths of these reads (averaging 221 Q20 bases) make them unlikely to be capable of substantively improving the assemblies. The low-quality reads aligning within nongap sequences were presumably not assembled by Phrap because they lack an exact word match of significant length with other reads in the assembly.
Examination of the accuracy of comparative-grade finished sequence revealed no errors in the established order and orientation of sequence contigs among the 116 BACs and a modest 1466 bases (of the ∼15 Mb) that differ from the corresponding human-grade finished sequence (and are thus presumed to be errors) (Table 3). This reflects a combined error rate of less than one in 10,000 bases. As seen with the gaps, these errors occur disproportionately in certain types of sequence. For example, 51.5% and 14.6% of the errors fall within total repeats and simple repeats, respectively, yet these types of repeats constitute only 33.4% and 0.6% of the sequence, respectively (Table 1). Once again, this is not entirely unexpected, as the prokaryotic DNA polymerases used for DNA sequencing are known to have difficulty in faithfully replicating vertebrate repetitive sequences (Hite et al. 1996; Mytelka and Chamberlin 1996; McMurray et al. 1998). Finally, 0.3% of the errors fall within annotated exons (Table 3), which is less than the total percentage of exonic bases. Simulation studies again demonstrated the statistical significance of these findings (Fig. 2D–F), with errors being enriched in repetitive sequences but not in exons (P < 0.00025 for all three results).
Table 3.
Characteristics of errors within the ∼15 Mb of comparative-grade finished sequence
| Total number of sequence errors | 1,466 bases (0.99 errors per 10,000 bases) |
| Errors in total repeats | 755 bases (51.5%) |
| Errors in simple repeats | 214 bases (14.6%) |
| Errors in exons | 4 bases (0.3%) |
We also examined another set of 20 BACs (from six species) derived from a genomic region (DeSilva et al. 2002) that has different compositional properties (49.1% GC content, 1.0% exonic sequence, and 55.1% repetitive sequence in human) than the region analyzed above. These 20 BACs were specifically chosen because of their high GC content (ranging from 51.4% to 57.0%). Although the comparative-grade finished sequence generated for these clones had more gaps (∼47 per megabase, resulting in the absence of 2.7% of the sequence) and a slightly higher error rate (2.03 per 10,000 bases), a similar enrichment of gaps and errors in repetitive sequences was seen as above. Specifically, 46.4% of gap bases and 67.5% of sequence errors fall within annotated total repeats, whereas such repeats only constitute 38.0% of the sequence in these clones (which derive from multiple nonhuman species).
Relative costs of generating comparative-grade finished sequence
To roughly assess the relative costs associated with producing comparative-grade versus human-grade finished sequence, we systematically captured data about the experimental and computational efforts of our finishing technicians over a 100-d period. During this time, comparative-grade and human-grade finished sequence was generated for 167 and 12 BACs, respectively (both types of sequence finishing started with the full-shotgun draft sequence assemblies providing greater than eightfold average coverage). Details of the experimental (e.g., types of sequencing chemistries, number of custom oligonucleotide primers, and time working in the laboratory) and computational (e.g., time refining the sequence using various software tools) efforts required for each BAC were documented. The resulting information was then used to calculate the average direct time, elapsed time, and reagent costs associated with each type of sequence finishing (Fig. 3).
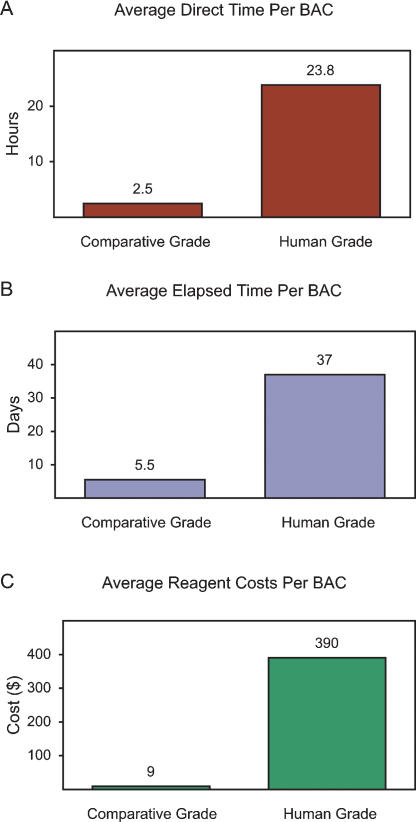
Figure 3.
Costs of generating comparative-grade versus human-grade finished sequence. The estimated average direct time (A; actual “handson” time that a finishing technician worked to finish a BAC sequence), elapsed time (B; interval of time from when a BAC was assigned to a finishing technician to when it was finished to a comparative-grade or human-grade stage), and reagent costs (C) required per BAC to perform comparative-grade and human-grade sequence finishing (starting with full-shotgun draft sequence) is indicated (for details, see text).
Comparative-grade sequence finishing required ∼10-fold less direct time compared to human-grade sequence finishing, involving an average of 2.5 h (median = 1.5; range = 0.2 to 17.3) versus 23.8 h (median = 21.5; range = 4.7 to >39) per BAC, respectively. A similar difference in elapsed time was encountered, averaging 5.5 d (median = 4; range = 1 to 41) versus 37 d (median = 30; range = 12 to 102), respectively. Note that the review of a larger set of BACs found a similar difference in average elapsed time, specifically 7.5 d (median = 5) versus 58 d (median = 41) for comparative-grade (751 BACs) and human-grade (128 BACs) finishing, respectively. A more dramatic (∼40-fold) difference was seen with the required reagent costs, averaging $9 (median = $0; range = $0 to $137) versus $390 (median = $292; range = $38 to $1230) per BAC for comparative-grade and human-grade finishing, respectively.
Application to whole-genome sequencing efforts
Our experience to date in generating and analyzing comparative-grade finished sequence has been limited to BAC-based sequencing projects. However, it will now be of interest to adapt our approach for use in whole-genome sequencing efforts, in particular those using a whole-genome shotgun sequencing strategy (Adams et al. 2000; Green 2001; Venter et al. 2001; Aparicio et al. 2002; Mouse Genome Sequencing Consortium 2002; Rat Genome Sequencing Project Consortium 2004).
To investigate the feasibility of refining whole-genome shotgun assemblies to the specifications established for comparative-grade finished sequence, we examined the sequence of the large genomic interval encompassing the CFTR gene (Thomas et al. 2003)—the same region from which virtually all of the above-analyzed BACs were derived—in four available whole-genome sequence assemblies (mouse, rat, chicken, and chimpanzee). In all four assemblies, the greater CFTR region resides within a single scaffold, with no evidence of gross misassemblies. Only in the case of the chimpanzee assembly are there significant sequence blocks unlinked to the primary scaffold: five contigs that are >2 kb in size, totaling ∼25 kb (out of 1.4 Mb). With very minor exceptions (involving a handful of insertions/deletions that are <2 kb in size), the sequence contigs in all assemblies are correct in their order and orientation. The amount of sequence missing within gaps is in most cases slightly larger for these assemblies compared to the BAC-derived full-shotgun or comparative-grade finished sequence (not surprising in light of the lower shotgun coverage associated with the whole-genome shotgun assemblies); specifically, in the analyzed region, the following amounts of sequence were missing in the whole-genome assemblies: (1) mouse, 2.4%; (2) rat, 2.9%; (3) chicken, 0.9%; and (4) chimpanzee, 4.9%. Consistent with the above findings, these gaps are enriched for repetitive sequences and rarely correspond to coding regions. These findings are encouraging, suggesting that carefully constructed whole-genome shotgun assemblies emerge at quality levels that are only slightly lower than the above-described comparative-grade finished sequence.
Discussion
Continual reductions in the cost of large-scale DNA sequencing, coupled with increasing enthusiasm for using comparative sequence analyses to unravel the complexities of vertebrate genomes (Cooper and Sidow 2003; Frazer et al. 2003; Pennacchio and Rubin 2003; Miller et al. 2004; Nobrega and Pennacchio 2004), has heightened interest to sequence many additional genomes. However, beyond the desire to generate near-perfect sequence for a handful of key reference genomes that will play central roles in myriad comparative studies, the quality standards for most future genome-sequencing efforts are not well established. Specifically, the extent of sequence finishing—the customized refinement of genomic sequence beyond the automated generation and assembly of shotgun sequence reads—that is desired or required for such projects is highly relevant and should be considered in conjunction with formulating plans for using the generated sequence (Palmer and McCombie 2002). Consideration must also be given to the fact that sequence finishing, especially to the standards appropriately set for the human genome sequence, remains an expensive endeavor.
As part of a larger program to generate and study sequences from the same targeted genomic regions in multiple vertebrates (Thomas et al. 2003), we have devised and investigated a new paradigm for sequence finishing that is tailored for comparative analyses. Based on first-hand experience in refining BAC-derived full-shotgun draft sequence to the point at which it is suitable for comparative studies, we established working specifications for comparative-grade finished sequence, a defined product with a quality somewhere between a Phrap assembly of full-shotgun sequence reads and human-grade finished sequence. In multiple studies performed to date (Margulies et al. 2003a,b; Thomas et al. 2003), we have found that comparative-grade finished sequence is well suited for rigorous comparisons of multispecies sequences; such findings are not surprising in light of the data reported here. Specifically, the quality of comparative-grade finished sequence is high with respect to both base accuracy (rivaling the standards established for human-grade finished sequence) and completeness (reflecting 99% of the total sequence).
Furthermore, the gaps and infrequent errors mostly correspond to repetitive sequences. Meanwhile, the cost associated with the comparative-grade finishing process is quite modest, reflecting a small fraction of the human-grade finishing process or even the resources required to generate the full-shotgun draft sequence itself (data not shown). This low cost primarily results from the heavy emphasis on computational-based sequence refinement, with the laboratory work limited to PCR-based studies, and the effective use of auxiliary data. Together, these findings indicate that comparative-grade finished sequence is an appropriate and cost-effective product for use in multispecies sequence comparisons.
It is important to distinguish between the sequence-finishing approach described here and sequence-improving routines that involve the medium-scale generation of additional sequence reads and initial rounds of computational-based sequence refinement (often referred to as prefinishing), such as those guided by the software tool Autofinish (Gordon et al. 2001). The generation of comparative-grade finished sequence is largely a computational endeavor, mostly involving rigorous review of sequence assemblies by a finishing technician. The associated laboratory work is limited to PCR-based studies for confirming contig-end relationships (required for ∼33% of BACs) and does not involve the generation of additional sequence reads. In contrast, most prefinishing routines involve the generation of additional sequence reads, with much less manual computational refinement of the sequence assemblies. Finally, in contrast to the comparative-grade finishing process described here, pre-finishing efforts are typically not associated with precise final quality specifications or a detailed verification process; in addition, they usually do not yield fully ordered and oriented contigs, a critical feature of comparative-grade finished sequence. Nonetheless, prefinishing routines are valuable components of the process en route to generating the highest quality sequence, and certainly something that we used for the generation of the human-grade finished sequence described here.
For the quality- and cost-assessment studies reported here, we did not stratify the analyzed BACs based on their species of origin. Rather, we compiled the data from all BACs, regardless of species, leading to general conclusions about generated vertebrate genomic sequence. However, it is interesting to note that there is considerable heterogeneity among the different species with respect to the difficulty in generating comparative-grade finished sequence (data not shown). For example, platypus and mouse sequences are often associated with more uncaptured gaps (compared to other species' sequences), requiring a larger number of PCR-based studies to establish contig order and orientation. Meanwhile, chimpanzee and baboon sequences are often associated with more misassemblies (compared to other species' sequences), requiring more computational manipulation to achieve an acceptable degree of refinement. Additional idiosyncrasies have been encountered with other species' sequences, often requiring slightly different approaches to generate comparative-grade finished sequence (in prep.). As such, the comparative-grade finishing process described here will inevitably be slightly refined when used in different projects, in some cases allowing further reductions in costs. For example, we recently found that restriction enzyme digest-based fingerprints generated in silico with the assembled sequence can be compared with the actual fingerprints generated from the starting clones, in many cases providing a cost-reducing alternative to PCR for verifying contig order and orientation (data not shown).
Indeed, the approach described here reflects a first-generation pipeline for producing comparative-grade finished sequence, with our studies to date emphasizing the sequence product and its specifications more than the process used to generate it. One can envision improving this process by using different sequence-assembly programs (e.g., ones that directly use read-pair information during sequence assembly), by further automating the process of trimming poor-quality sequence from contig ends, and by using mixtures of shotgun subclones with different insert sizes. For some of these developments, the data sets we have generated to date (including the primary sequence reads themselves) can be used for benchmarking more automated finishing pipelines. One can also imagine the design of variant approaches that involve generating lower-redundancy sequence coverage during the shotgun phase and then performing some directed prefinishing steps (including the acquisition of additional sequence reads) to enhance sequence quality as part of the comparative-grade sequence finishing process. In pursuing such refinements, it will be worthwhile to keep in mind that a lower-quality sequence product might be adequate for some comparative studies, and thus a lower sequence coverage and/or a simpler comparative-grade finishing process might suffice.
Comparisons of BAC-derived comparative-grade finished sequence and unrefined whole-genome shotgun sequence assemblies reveal encouraging similarities. As such, one could envision the adaptation of our nascent comparative-grade finishing routines for use in refining whole-genome assemblies (in their entirety or targeted portions therein) to the specifications reported here for the comparative-grade sequence finishing of BACs (or to some slightly modified specifications). To begin with, it appears that the whole-genome shotgun assembly processes generally yield few gross misassemblies, and interval-by-interval (perhaps in roughly BAC-sized segments) analyses could be used to correct the remaining ones (in a fashion similar to that used with individual BACs). Next, evidence for accurate long-range ordering and orienting of sequence contigs might naturally emerge from the broader whole-genome scaffolds, with any additional confirmation or refinement being performed, as needed. Of relevance, whole-genome shotgun sequencing projects typically include the generation of insert-end sequence reads from large-insert clones (e.g., BACs, fosmids, and large-insert plasmids) (Mouse Genome Sequencing Consortium 2002; Stein et al. 2003; Istrail et al. 2004); thus, following incorporation of such reads into a genome-wide sequence assembly, the corresponding clones could be used as substrates for PCR-based confirmation of contig order and orientation. Finally, the types of verification steps performed for BAC-derived sequences (e.g., comparisons to reference sequences and experimentally-derived restriction maps) could be scaled for use with whole-genome sequence assemblies, either globally or in a more targeted fashion. Of course, such refinement steps would require the development of robust software tools capable of handling the unique features of whole-genome shotgun assemblies.
Nonetheless, the systematic application of comparative-grade sequence finishing to whole-genome shotgun assemblies will face considerable obstacles, especially since the routines we have developed to date have been specifically tailored for BAC-based sequencing. For example, whole-genome shotgun assemblies are more vulnerable to problems relating to segmental duplications (Bailey et al. 2004), and these will inevitably present serious challenges to the finishing process that otherwise might be avoided in BAC-based sequencing. Furthermore, the overall scale of the effort will require robust adaptations of routines initially designed for BACs, likely including more automated tools for detecting and correcting misassemblies, for routine trimming of low-quality regions, for identifying notably poor-quality intervals that require more detailed study, and for systematic confirmation of contig order and orientation. Finally, an inherent advantage of BAC-based sequencing is the study of only a single haplotype within an individual clone; in the case of outbred species, comparative-grade sequence finishing of whole-genome shotgun assemblies will encounter serious challenges associated with the presence of two or more haplotypes.
In summary, the operational experience described here, coupled with a better understanding of the trade-offs associated with different grades of finished sequence, should provide a strong framework for the rational establishment of finishing standards for ongoing and future genome sequencing projects. It is also inevitable that as we gain a better understanding of the myriad uses of large collections of multispecies genomic sequences, approaches for additional refinement of the sequence-finishing process should readily emerge.
Methods
Generation of comparative-grade finished sequence
The initial shotgun phase of BAC sequencing was performed by using well-established methods (Wilson and Mardis 1997a,b; Ellsworth et al. 2000; DeSilva et al. 2002; Thomas et al. 2003). In brief, plasmid subclones containing 3- to 5-kb inserts (produced by physically shearing purified BAC DNA) were derived from each BAC, and sequence reads were generated from both ends of randomly selected subclones (forward and reverse “read pairs”) to provide at least eightfold average coverage in high-quality (Phred Q20) (Ewing and Green 1998; Ewing et al. 1998) bases. Sequence assemblies were performed by using Phrap (see www.phrap.org). The resulting full-shotgun draft sequence reflected the set of unmodified, assembled contigs >2 kb in size, and this was then subjected to sequence finishing, both to the standards established for the human genome (human-grade finished sequence; see above) and independently to the specifications detailed below for comparative-grade finished sequence. Note that different technicians performed the two types of sequence finishing starting with the same full-shotgun assemblies.
To generate comparative-grade finished sequence, a standardized set of analyses and refinements of the above-defined full-shotgun draft sequence is performed by finishing technicians in three stages. The entire process makes extensive use of key software tools, including the sequence-editing program Consed (Gordon et al. 1998), Phrapview (Gordon et al. 1998), Orchid (see www-shgc.stanford.edu/informatics/orchid.html), and PipMaker (Schwartz et al. 2000, 2003) (see bio.cse.psu.edu/pipmaker) to guide conservative refinements. Note that no additional sequence reads are generated to resolve ambiguities, to improve low-quality regions, or to derive missing sequence (i.e., to fill gaps). Additional details about the steps and procedures involved in comparative-grade sequence finishing are available at www.nisc.nih.gov/data.
Stage 1
As a first step, major misassemblies are untangled by breaking and rejoining sequence contigs, often using information about read pairs to guide decisions. Minor misassemblies (e.g., those caused by misplaced sequence reads that do not alter the consensus) are generally ignored, because these do not change the order or orientation of contigs. Second, BAC insert-ends are identified, and errors in the consensus sequence related to BAC vector-masking are edited. Third, regions within a contig suspected to contain a false join (e.g., stretches of ≥10 Q0 bases or regions linked by a chimeric sequence read, as tagged by Consed) are broken. Fourth, any readily detectable artifacts associated with the sequencing process (e.g., unbound fluorescence dye signals, incorrectly assigned consensus bases, and chimeric reads) that alter the consensus sequence are corrected or removed. Finally, all contig ends are trimmed of low-quality sequence by using the modified Mott algorithm with an error probability cutoff value of 0.05, as used by the base-calling program Phred (Ewing and Green 1998; Ewing et al. 1998). This final edited sequence forms the basis for the next stage, in which contig order and orientation is established. Note that edited assemblies consisting of a single contig bypass the second stage, because ordering/orienting is not required.
Stage 2
For edited assemblies consisting of two contigs, order and orientation is evident as long as both BAC insert-ends are identified within the sequence. For assemblies with more than two contigs or the absence of an identified BAC insert-end(s), further analyses are required. When possible, read-pair information is used first to establish the spatial relationships among contigs. Two contig ends are considered adjacent if (1) two or more read pairs connect the ends across the intervening gap, and (2) if the sum of the distances from the start of each read to its contig end is not too large relative to the distribution of subclone insert sizes (as calculated by Orchid or Consed). The programs Orchid, Phrapview, and Consed Assembly View are extensively used in this stage for visualizing relationships of read pairs (e.g., reviewing orientation and read-pair separation distances) and for listing potential contig relationships. By using these tools, contig-end adjacencies are established.
Stage 3
The verification process involves comparing the deduced spatial relationships of sequence contigs for each BAC to an independent data source. First, the refined sequence is compared to any independently derived, overlapping BAC sequence(s) to confirm colinearity by using the program Pal (S. Dear and G. Marth, unpubl.; see genome.wustl.edu/Overview/computerguid.php?commands=1). Second, the deduced order and orientation of sequence contigs is confirmed, when possible, by alignment to an orthologous reference sequence using the program PipMaker (Schwartz et al. 2000, 2003). For the BACs described here, high-quality human or mouse genomic sequence was typically used as the reference. Third, when the spatial relationship between two or more contigs cannot be established (or is ambiguous), PCR is performed with custom-designed oligonucleotides that prime near each contig end and point toward each other across the purported gap. Generation of a robust amplicon from the cognate BAC template is considered supportive evidence for contigend adjacency; with all such PCR studies, a suitable negative control is required (e.g., involving the use of the same oligonucleotides in a combinatorial fashion with oligonucleotides designed from other contig ends). Finally, the computational and experimental steps yielding the comparative-grade finished sequence for each BAC are subjected to independent scrutiny by a second finishing technician and a bioinformatician prior to completion.
Quality-assessment studies
Key to assessing the quality of comparative-grade finished sequence was the availability of human-grade finished sequence for the same set of analyzed BACs. Such data were available for the individual BACs and for the compiled assemblies of each genomic region for each species (see www.nisc.nih.gov/data). The methods used to assemble and annotate these sequences have been described (Thomas et al. 2003).
For each BAC, the comparative-grade finished sequence was compared to the human-grade finished sequence by using the program “cross_match” (version 0.990319, default parameters) (see www.phrap.org). For these comparisons, the fasta files of the comparative-grade finished sequence were first trimmed to include only the portion of the BAC sequence used to compile the multi-BAC assembly of the region (thereby ensuring that each segment of the sequence was only included once in the analysis, such as the data reported in Tables 1, 2, 3). Alignments were examined to assess whether the deduced order and orientation of the comparative-grade finished sequence contigs was correct. All mismatches, insertions, and deletions within the alignments were considered errors, and all regions of the human-grade finished sequence not included in the alignments were considered gaps. Note that in some cases, full-shotgun draft and comparative-grade finished sequences contain a gap even though the two adjacent contigs actually overlap; these were not counted as gaps in our analysis, because none of the human-grade finished sequence is actually missing.
Detected errors associated with a Phrap-assigned quality score of greater than or equal to Q40 (in the comparative-grade finished sequence) were scrutinized more carefully to confirm that the perceived error was not due to an alignment inaccuracy; all such cases were found to reflect true errors. Detected gaps and errors were catalogued relative to annotated repetitive sequence and exons (Thomas et al. 2003). Note that any region annotated as a CDS or exon, excluding Genscan (Burge and Karlin 1997) predictions, was considered exonic sequence. “Simple repeats” reflect sequences within the RepeatMasker “simple” library (see www.repeatmasker.org), whereas “total repeats” reflect the total repetitive sequences (of all types) detected by RepeatMasker.
We investigated the statistical significance of finding gaps and errors (in comparative-grade finished sequence) within repetitive sequences and exons. Specifically, 4000 data sets were computationally generated whereby the same number and size of gaps or the same number of errors (as observed in the comparative-grade finished sequence) were randomly distributed across the human-grade finished sequence. The Perl script used for these simulations is available at www.nisc.nih.gov/data. For each data set, the number of randomly placed gaps or errors falling within annotated repetitive sequences or exons was calculated, with the resulting data for all 4000 simulated data sets then plotted as a histogram (Fig. 2). These analyses helped to assess if the observed results were likely to have occurred by chance.
Several of the Perl scripts used to perform these analyses called methods from the Bioperl toolkit (Stajich et al. 2002).
Analysis of whole-genome shotgun assemblies
Four available assemblies generated by whole-genome shotgun sequencing were examined: (1) mouse (WGSAv3, providing ∼6.5-fold coverage) (Mouse Genome Sequencing Consortium 2002); (2) rat (RGSC 3.1, providing ∼6.9-fold coverage) (Rat Genome Sequencing Project Consortium 2004); (3) chicken (GenBank no. AADN01000000, providing ∼6.6-fold coverage); and (4) chimpanzee (GenBank no. AADA01000000, providing approximately sixfold coverage). For each species, SSAHA (Ning et al. 2001) was used to locate the genomic segment from the whole-genome assembly that corresponded to the BAC-derived sequence generated for the greater CFTR region (Thomas et al. 2003); the corresponding assembled sequence, the primary scaffold, and the consensus quality scores were then extracted from the whole-genome sequence assembly, and analyses were performed using methods analogous to those used for the BAC-based sequences.
Acknowledgments
We thank numerous people associated with the NISC Comparative Sequencing Program, in particular the dedicated technicians and other staff involved in BAC isolation, mapping, and sequencing. We also thank Drs. Richard Gibbs and Jeff Touchman for critical reading of the manuscript. Finally, we thank the producers of the unpublished chimpanzee and chicken whole-genome shotgun sequences, which we used for some analyses; specifically, the Broad Institute of MIT and Harvard University (for the chimpanzee sequence) and the Genome Sequencing Center of Washington University School of Medicine (for the chimpanzee and chicken sequences).
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2648404. Article published online before print in October 2004.
References
- Adams, M.D., Celniker, S.E., Holt, R.A., Evans, C.A., Gocayne, J.D., Amanatides, P.G., Scherer, S.E., Li, P.W., Hoskins, R.A., Galle, R.F., et al. 2000. The genome sequence of Drosophila melanogaster. Science 287: 2185-2195. [DOI] [PubMed] [Google Scholar]
- Aparicio, S., Chapman, J., Stupka, E., Putnam, N., Chia, J., Dehal, P., Christoffels, A., Rash, S., Hoon, S., Smit, A., et al. 2002. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science 297: 1301-1310. [DOI] [PubMed] [Google Scholar]
- Ashurst, J.L. and Collins, J.E. 2003. Gene annotation: Prediction and testing. Annu. Rev. Genomics Hum. Genet. 4: 69-88. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Gu, Z., Clark, R.A., Reinert, K., Samonte, R.V., Schwartz, S., Adams, M.D., Myers, E.W., Li, P.W., and Eichler, E.E. 2002. Recent segmental duplications in the human genome. Science 297: 1003-1007. [DOI] [PubMed] [Google Scholar]
- Bailey, J.A., Church, D.M., Ventura, M., Rocchi, M., and Eichler, E.E. 2004. Analysis of segmental duplications and genome assembly in the mouse. Genome Res. 14: 789-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birren, B., Mancino, V., and Shizuya, H. 1998. Bacterial artificial chromosomes. In Genome analysis: a laboratory manual: Cloning systems, vol. 3 (eds. B. Birren et al.), pp. 241-295. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Bouck, J., Miller, W., Gorrell, J.H., Muzny, D., and Gibbs, R.A. 1998. Analysis of the quality and utility of random shotgun sequencing at low redundancies. Genome Res. 8: 1074-1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. 1998. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science 282: 2012-2018. [DOI] [PubMed] [Google Scholar]
- Chissoe, S.L., Marra, M.A., Hillier, L., Brinkman, R., Wilson, R.K., and Waterston, R.H. 1997. Representation of cloned genomic sequences in two sequencing vectors: Correlation of DNA sequence and subclone distribution. Nucleic Acids Res. 25: 2960-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper, G.M. and Sidow, A. 2003. Genomic regulatory regions: Insights from comparative sequence analysis. Curr. Opin. Genet. Dev. 13: 604-610. [DOI] [PubMed] [Google Scholar]
- DeSilva, U., Elnitski, L., Idol, J.R., Doyle, J.L., Gan, W., Thomas, J.W., Schwartz, S., Dietrich, N.L., Beckstrom-Sternberg, S.M., McDowell, J.C., et al. 2002. Generation and comparative analysis of ∼3.3 Mb of mouse genomic sequence orthologous to the region of human chromosome 7q11.23 implicated in Williams syndrome. Genome Res. 12: 3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth, R.E., Jamison, D.C., Touchman, J.W., Chissoe, S.L., Braden Maduro, V.V., Bouffard, G.G., Dietrich, N.L., Beckstrom-Sternberg, S.M., Iyer, L.M., Weintraub, L.A., et al. 2000. Comparative genomic sequence analysis of the human and mouse cystic fibrosis transmembrane conductance regulator genes. Proc. Natl. Acad. Sci. 97: 1172-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B. and Green, P. 1998. Base-calling of automated sequencer traces using Phred. II. Error probabilities. Genome Res. 8: 186-194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. 1998. Base-calling of automated sequencer traces using Phred. I: Accuracy assessment. Genome Res. 8: 175-185. [DOI] [PubMed] [Google Scholar]
- Felsenfeld, A., Peterson, J., Schloss, J., and Guyer, M. 1999. Assessing the quality of the DNA sequence from the Human Genome Project. Genome Res. 9: 1-4. [PubMed] [Google Scholar]
- Frazer, K.A., Elnitski, L., Church, D.M., Dubchak, I., and Hardison, R.C. 2003. Cross-species sequence comparisons: A review of methods and available resources. Genome Res. 13: 1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, D., Abajian, C., and Green, P. 1998. Consed: A graphical tool for sequence finishing. Genome Res. 8: 195-202. [DOI] [PubMed] [Google Scholar]
- Gordon, D., Desmarais, C., and Green, P. 2001. Automated finishing with Autofinish. Genome Res. 11: 614-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, E.D. 2001. Strategies for the systematic sequencing of complex genomes. Nat. Rev. Genet. 2: 573-583. [DOI] [PubMed] [Google Scholar]
- Hite, J.M., Eckert, K.A., and Cheng, K.C. 1996. Factors affecting fidelity of DNA synthesis during PCR amplification of d(C-A)n.d(G-T)n microsatellite repeats. Nucleic Acids Res. 24: 2429-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- ———. 2004. Finishing the euchromatic sequence of the human genome. Nature (in press). [DOI] [PubMed]
- Ishiura, M., Hazumi, N., Koide, T., Uchida, T., and Okada, Y. 1989. A recB recC sbcB recJ host prevents recA-independent deletions in recombinant cosmid DNA propagated in Escherichia coli. J. Bacteriol. 171: 1068-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Istrail, S., Sutton, G.G., Florea, L., Halpern, A.L., Mobarry, C.M., Lippert, R., Walenz, B., Shatkay, H., Dew, I., Miller, J.R., et al. 2004. Whole-genome shotgun assembly and comparison of human genome assemblies. Proc. Natl. Acad. Sci. 101: 1916-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith, J.M., Cochran, D.A.E., Lala, G.H., Adams, P., Bryant, D., and Mitchelson, K.R. 2004. Unlocking hidden genomic sequence. Nucleic Acids Res. 32: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan, J.E., Rowbottom, L., Liloglou, T., Field, J.K., and Risk, J.M. 2002. Sequencing of difficult templates containing poly(A/T) tracts: Closure of sequence gaps. BioTechniques 33: 276-280. [DOI] [PubMed] [Google Scholar]
- Margulies, E.H., NISC Comparative Sequencing Program, and Green, E.D. 2003a. Detecting highly conserved regions of the human genome by multispecies sequence comparisons. Cold Spring Harbor Symp. Quant. Biol. LXVIII: 255-263. [DOI] [PubMed] [Google Scholar]
- Margulies, E.H., Blanchette, M., NISC Comparative Sequencing Program, Haussler, D., and Green, E.D. 2003b. Identification and characterization of multi-species conserved sequences. Genome Res. 13: 2507-2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray, A.A., Sulston, J.E., and Quail, M.A. 1998. Short-insert libraries as a method of problem solving in genome sequencing. Genome Res. 8: 562-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, W., Makova, K.D., Nekrutenko, A., and Hardison, R.C. 2004. Comparative Genomics. Annu. Rev. Genomics Hum. Genet. 5: 15-56. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
- Mytelka, D.S. and Chamberlin, M.J. 1996. Analysis and suppression of DNA polymerase pauses associated with a trinucleotide consensus. Nucleic Acids Res. 24: 2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning, Z., Cox, A.J., and Mullikin, J.C. 2001. SSAHA: A fast search method for large DNA databases. Genome Res. 11: 1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobrega, M.A. and Pennacchio, L.A. 2004. Comparative genomic analysis as a tool for biological discovery. J. Physiol. 554: 31-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer, L.E. and McCombie, W.R. 2002. On the importance of being finished. Genome Biol. 3: 2010.1-2010.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio, L.A. and Rubin, E.M. 2003. Comparative genomic tools and databases: Providing insights into the human genome. J. Clin. Invest. 111: 1099-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428: 493-521. [DOI] [PubMed] [Google Scholar]
- Razin, S.V., Ioudinkova, E.S., Trifonov, E.N., and Scherrer, K. 2001. Non-clonability correlates with genomic instability: A case study of a unique DNA region. J. Mol. Biol. 307: 481-486. [DOI] [PubMed] [Google Scholar]
- Sanger, F., Coulson, A.R., Hong, G.F., Hill, D.F., and Petersen, G.B. 1982. Nucleotide sequence of the bacteriophage λ DNA. J. Mol. Biol. 162: 729-773. [DOI] [PubMed] [Google Scholar]
- Schwartz, S., Zhang, Z., Frazer, K.A., Smit, A., Riemer, C., Bouck, J., Gibbs, R., Hardison, R., and Miller, W. 2000. PipMaker: A web server for aligning two genomic DNA sequences. Genome Res. 10: 577-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, S., Elnitski, L., Li, M., Weirauch, M., Riemer, C., Smit, A., NISC Comparative Sequencing Program, Green, E.D., Hardison, R.C., and Miller, W. 2003. MultiPipMaker and supporting tools: Alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 31: 3518-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shizuya, H., Birren, B., Kim, U.-J., Mancino, V., Slepak, T., Tachiiri, Y., and Simon, M. 1992. Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor–based vector. Proc. Natl. Acad. Sci. 89: 8794-8797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich, J.E., Block, D., Boulez, K., Brenner, S.E., Chervitz, S.A., Dagdigian, C., Fuellen, G., Gilbert, J.G., Korf, I., Lapp, H., et al. 2002. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 12: 1611-1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein, L. 2001. Genome annotation: From sequence to biology. Nat. Rev. Genet. 2: 493-503. [DOI] [PubMed] [Google Scholar]
- Stein, L.D., Bao, Z., Blasiar, D., Blumenthal, T., Brent, M.R., Chen, N., Chinwalla, A., Clarke, L., Clee, C., Coghlan, A., et al. 2003. The genome sequence of Caenorhabditis briggsae: A platform for comparative genomics. PLoS. Biol. 1: 166-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, J.W., Touchman, J.W., Blakesley, R.W., Bouffard, G.G., Beckstrom-Sternberg, S.M., Margulies, E.H., Blanchette, M., Siepel, A.C., Thomas, P.J., McDowell, J.C., et al. 2003. Comparative analyses of multi-species sequences from targeted genomic regions. Nature 424: 788-793. [DOI] [PubMed] [Google Scholar]
- Venter, J.C., Adams, M.D., Myers, E.W., Li, P.W., Mural, R.J., Sutton, G.G., Smith, H.O., Yandell, M., Evans, C.A., Holt, R.A., et al. 2001. The sequence of the human genome. Science 291: 1304-1351. [DOI] [PubMed] [Google Scholar]
- Wilson, R.K. and Mardis, E.R. 1997a. Fluorescence-based DNA sequencing. In Genome analysis: A laboratory manual: Analyzing DNA, vol. 1 (eds. B. Birren et al.), pp. 301-395. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- ———. 1997b. Shotgun sequencing. In Genome analysis: A laboratory manual: Analyzing DNA, vol. 1 (eds. B. Birren et al.), pp. 397-454. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
WEB SITE REFERENCES
- www.nisc.nih.gov/data; source of Supplemental data and information.
- www.nisc.nih.gov; NIH Intramural Sequencing Center (NISC) home page and source of information for the NISC Comparative Sequencing Program.
- www.genome.wustl.edu/Overview/finrulesname.php?G16=1; quality specifications for the finished human genome sequence.
- www.ncbi.nlm.nih.gov/HTGS; definitions of different phases of genomic sequence.
- www.phrap.org; source of Phred, Phrap, Consed, and Cross_Match software.
- www-shgc.stanford.edu/informatics/orchid.html; source of Orchid software.
- bio.cse.psu.edu/pipmaker; source of PipMaker software.
- genome.wustl.edu/Overview/computerguid.php?commands=1; source of Pal software.
- www.repeatmasker.org; source of RepeatMasker software.