Abstract
The ETn (Early Transposon) elements are among the most active murine mobile sequences, being responsible for a series of mutations by insertion in vivo. Yet they are noncoding, and it had long been suspected that ETn are mobilized in trans by coding-competent elements, most probably from the closely related MusD family of LTR-retrotransposons. A genome-wide in silico search for coding-competent MusD elements identified a total of nine such copies, which we cloned and marked to test their transpositional activity, using an ex vivo assay in heterologous cells. Three copies were found to be autonomous for transposition, with each gag, pro, and pol MusD gene absolutely required for mobility. These active MusD copies specifically trigger retrotransposition of marked ETn elements with high efficiency, by complementation in trans. Characterization of the structures of de novo transposed MusD and ETn marked elements, as well as of their integration sites, disclosed canonical retroviral-like retrotransposition, with 6-bp target site duplications common to both elements. These results highlight the parasitic molecular strategies that are used by the ETn elements for their mobility, and unambiguously identify their “master genes.”
LTR-retrotransposons have been found in all mammals in which they have been sought, and can represent several percent of the genome mass (Boeke and Stoye 1997; Lander et al. 2001; Waterston et al. 2002). The complete sequencing of the human genome has left only a remote possibility that some LTR-retrotransposons might still be active, with the exception of a few still fully coding elements from the HERV-K(HML2) family of endogenous retroviruses (Löwer et al. 1996; Mayer et al. 1999; Reus et al. 2001; Turner et al. 2001). In contrast, in the mouse genome, it has long been shown that several LTR-retrotransposons such as the IAP sequences and ETn (Early Transposon) elements are mobile, because they are responsible for most of the insertional mutagenesis events that have been revealed by phenotypic mutations in vivo (for review, see Kuff and Lueders 1988; Kuff 1990; Ostertag and Kazazian Jr. 2001; Baust et al. 2002). Recently, several IAP sequences with fully coding gag, pro, and pol genes and autonomous for transposition have been identified and characterized for their mobility in ex vivo assays (Dewannieux et al. 2004). Conversely, ETn elements have been involved in numerous germ-line and somatic mutations, indicating recent retrotransposition, although they do not harbor full-length retroviral genes (Sonigo et al. 1987). The ETn family comprises 5.7-kb elements that can be classified into two groups differing by the 3′-half of the LTR and the 5′-internal region (Shell et al. 1990): ETn I with 200 copies per haploid genome of C57BL/6J mice, and ETn II with 40 copies (Baust et al. 2003). Their expression is limited to embryos, peaking between embryonic days 3.5 (E3.5) and E7.5 (Brûlet et al. 1985), as well as to a few cell lines (Brûlet et al. 1983; Shell et al. 1987; Tanaka and Ishihara 2001). ETn elements are related to MusD elements, another murine LTR-retrotransposon family (Mager and Freeman 2000). The MusD family comprises a hundred copies of 7.5-kb-long elements containing gag, pro, and pol genes with similarities to the genes of betaretroviruses. They share with ETn elements almost identical LTRs as well as a similar 3′ internal region downstream from the pol gene, including a polypurine tract (Fig. 1A). MusD and ETn II have also a similar 5′ internal region upstream of the gag gene, including an intact primer binding site for tRNALys—also found in the ETn I elements (Kaghad et al. 1985; Baust et al. 2003)—and a possible retroviral packaging signal (described in Mager and Freeman 2000). It has been proposed that ETn elements derive from MusD copies by recombinatory replacement of the retroviral genes with sequences of unknown origin, and that they have colonized the mouse genome by retrotransposition via complementation in trans by active MusD elements (Mager and Freeman 2000). The present study was aimed at the search and characterization of “master” MusD copies, that is, of copies that encode all the enzymatic and structural proteins that are necessary for their own mobility, and also possibly responsible for the mobilization in trans of the ETn elements. A genome-wide in silico search for complete MusD copies with fully coding gag, pro, and pol genes resulted in the identification of nine elements, which we cloned and assayed for their functionality. Interestingly, three of the copies tested proved to be transposition-competent and behave as high-efficiency mobile elements in heterologous cells. We further demonstrate that mobilization of ETn elements is triggered by active MusD copies, thus strengthening the “parasitic” relationship between these two classes of elements.
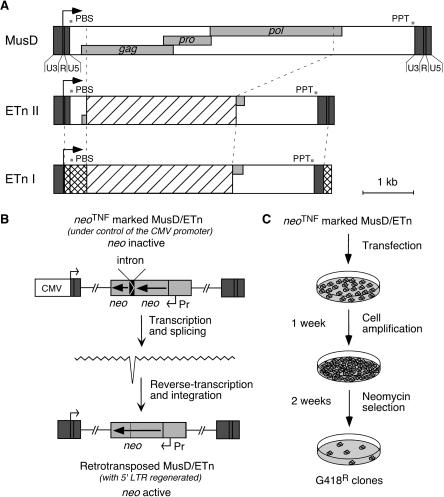
Figure 1.
Structure of the related MusD and ETn elements and rationale of the assay for retrotransposition. (A) Genomic organization of MusD, ETn type II and I elements. The LTRs (dark gray box) with a U3-R-U5 organization border three ORFs homologous to the retroviral gag, pro, and pol genes (light gray boxes). The transcription start site is marked with an arrow, and the signals necessary for retroviral reverse transcription are indicated: (PBS) primer binding site; (PPT) poly purine tract. ETn elements are closely related to MusD despite the replacement of the retroviral genes by a noncoding sequence (hatched box). ETn I elements differ from ETn II only in the LTRs and 5′-UTR region (squared boxes). (B) Schematic representation of a MusD/ETn element under the control of the CMV promoter and marked with the neoTNF reporter gene (light gray), in which the neo gene placed in backward orientation with its own promoter (Pr) is rendered inactive by the presence of a forward intron (top), which should be spliced out in the transposition RNA intermediate (middle), thus resulting in an active neo gene in the de novo MusD/ETn insertion (bottom). (C) Experimental procedure for detection of MusD/ETn retrotransposition. Cells were transfected either with a neoTNF-marked defective MusD/ETn together with an MusD expression vector, or with a full-length neoTNF-marked MusD alone. Cells were amplified for a week, and retrotransposition events were detected upon G418 selection.
Results
Search for MusD copies encoding proteins active for retrotransposition
The sequences of two previously described MusD elements (Mager and Freeman 2000) were used to screen the mouse genome to identify the different copies present in this organism. About a hundred copies were found in the Non-Redundant (nr) and High-Throughput Genomic Sequences (htgs) databases (December 2003 GenBank release), among which only nine (which we named MusD-1 to MusD-9) (see Table 1) were coding-competent for the three gag, pro, and pol genes and thus potentially autonomous for retrotransposition (no envelope gene has been identified for the MusD elements). The corresponding BAC clones were obtained from BACPAC Resources, to isolate the elements for further analysis.
Table 1.
Coding-competent MusD and selected ETn elements
| Name | Chromosome locationa | Accession no. | Position in sequence entryb | No. nucleotide differences between 5′ - and 3′-LTRs |
|---|---|---|---|---|
| MusD-1 | 16qC3 | AC034116 | 181,293-188,770 (+) | 0 |
| MusD-2 | 2qG3 | AC084696 | 132,348-139,824 (-) | 0 |
| MusD-3 | Xqa2 | AC102360 | 147,064-154,549 (+) | 2 |
| MusD-4 | 16qC1 | AC102635 | 84,839-92,292 (-) | 5 |
| MusD-5 | 3qE3 | AC122493 | 172,998-180,469 (+) | 8 |
| MusD-6 | 13qB3 | AC124426 | 9078-16,569 (+) | 0 |
| MusD-7 | 15qA2 | AC133203 | 105,627-113,076 (-) | 1 |
| MusD-8 | 18qA1 | AC134834 | 61,019-68,474 (-) | 4 |
| MusD-9 | XqA2 | AL844221 | 165,724-173,126 (+) | 1 |
| ETn IIα-1 | 11qA3 | AC102263 | 133,554-140,629 (-) | 0 |
| ETn IIβ-1 | 5qE2 | AC101839 | 68,218-73,760 (+) | 0 |
| ETn IIβ-2 | 1qC1 | AC107832 | 98,176-103,711 (+) | 0 |
| ETn IIβ-3 | 13qA1 | AC126548 | 122,156-127,693 (+) | 0 |
| ETn I-1 | XqA2 | AL590633 | 84,029-89,525 (+) | 0 |
Obtained by screening of the MGSC database released on December 2003 (http://www.ensembl.org/Mus_musculus/)
+ and -, orientation in the sequence entry
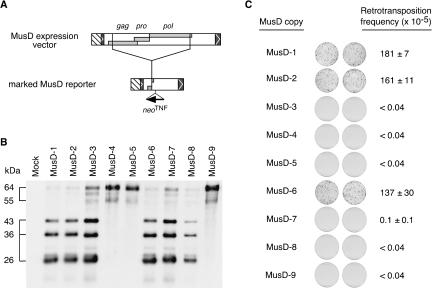
Because MusD elements are poorly transcribed (Baust et al. 2003; data not shown), we first replaced the U3 promoter domain within the 5′-LTR of these elements by the strong CMV promoter, in such a way as to preserve the natural retroviral start site and transcript structure (see Methods). As a first step, the resulting constructs were then assayed by transient transfection of heterologous human HeLa cells (not containing MusD sequences), to identify the copies with potentially functional gag and pro genes. Lysates from the transfected HeLa cells were analyzed by immunoblotting for the presence of Gag products, using an antiserum that we have raised against a recombinant full-length Gag polyprotein (see Methods). As illustrated in Figure 2B, two distinct patterns can be observed among the nine coding-competent MusD elements. For three of them (i.e., MusD-4, -5, -9), only high-molecular-mass fragments with a major band of 64 kDa most probably associated with the uncleaved Gag polyprotein precursor (63 kDa theoretical molecular mass) can be observed, whereas, for the six other copies, bands of lower molecular mass (of 43, 36, and 26 kDa) are generated, most probably associated with an active protease and thus strongly suggesting functional gag–pro genes. Further characterization of these copies was performed directly via a functional ex vivo assay for retrotransposition.
Figure 2.
Assay for MusD gene products and retrotransposition. (A) Structure of the MusD expression vector and neoTNF-marked reporter. (B) Western blot analysis of the Gag-related gene products from the nine coding-competent MusD copies (see Table 1 for copy and accession nos). Lysates of cells transfected with a control plasmid (pCMVβ; mock) or the expression vectors for the coding-competent MusD copies were electrophoresed in a denaturing polyacrylamide gel (SDS-PAGE), blotted, and hybridized with a rabbit anti-serum directed against the Gag polyprotein. The apparent molecular masses of the major bands are indicated. (C) Assay for retrotransposition of the defective neoTNF-marked MusD reporter upon transcomplementation by the coding-competent MusD copies. Retrotransposition was assayed in heterologous human cells upon cotransfection of the reporter with a control plasmid (pCMVβ) (data not shown) or expression vectors for the selected MusD elements. Results of the G418 selections are illustrated with images of the plates after the G418R foci have been fixed and stained. The retrotransposition frequencies, as derived from the number of foci per seeded cells, are given for each construct as the mean of two to four independent experiments, carried out with at least 3 × 106 cells, with standard errors indicated.
As schematized in Figure 2A, this assay makes use of a reporter MusD element marked with an indicator gene for retrotransposition, and of the vectors for the genomic MusD elements added in trans. The marked MusD reporter is derived from a genomic MusD copy (MusD-6; AC124426), which has been internally deleted to remain—tentatively—competent but not autonomous for transposition, and that we marked with the neoTNF indicator gene for retrotransposition (Esnault et al. 2002). In this reporter, as illustrated in Figure 1, B and C, a selectable neo gene becomes active only after a cycle of transcription and reverse transcription of the marked element, thus allowing detection and positive selection of the retrotransposition event. Assays for retrotransposition were therefore performed by cotransfection of human HeLa cells with this marked MusD reporter together with each of the expression vectors for the nine coding-competent MusD proviruses (Fig. 2A), followed by selection of the transfected cells for G418-resistant (G418R) clones. As illustrated in Figure 2C, this assay resulted in the generation of a high number of resistant clones, with a frequency in the 10–3 range, for three of the MusD copies, whereas no clones were obtained for either a control (CMVβ) (data not shown) or the other MusD copies. Interestingly, and consistent with the Western blot analysis in Figure 2B, the three copies active for retrotransposition in trans (i.e., MusD-1, -2, and -6) are among those displaying a cleaved Gag profile, whereas those with an unprocessed Gag (i.e., MusD-4, -5, and -9) are inactive. Yet the correlation is not perfect, as MusD-3, -7, and -8 elements disclose a cleaved Gag profile but no transpositional activity (see Discussion).
MusD copies autonomous for retrotransposition and characterization of de novo insertions
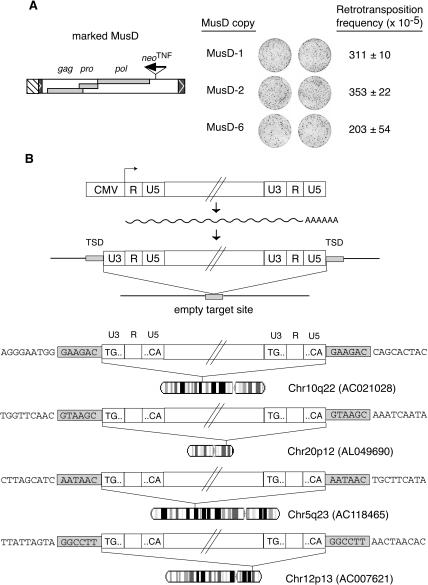
We then tried to determine whether the three “positive” MusD copies described above are able to promote their own mobilization, that is, whether these elements contain all the signals in cis, in addition to the proteins required in trans, for their efficient autonomous retrotransposition. The three MusD elements were therefore marked with the indicator gene, which was inserted without deletion just downstream from the pol gene to preserve all the element-coding sequences, as illustrated in Figure 3A. The marked MusD vectors were then introduced by tranfection into heterologous HeLa cells, and transfected cells were subjected to G418 selection. As illustrated in the figure, this resulted in the emergence of numerous resistant clones for all three MusD copies, at a frequency in the range of 2–3 × 10–3. To ascertain that the G418R clones obtained in this assay (as well as in the previous one) are, indeed, the consequence of retrotransposition of the marked elements, we checked by PCR analysis of the genomic DNA from isolated G418R clones that the intron present in the indicator gene of the transfected marked copy had been precisely spliced out in the transposed MusD copies: among a series of randomly selected G418R clones, all of them (i.e., 12/12) disclosed a PCR band of reduced size as expected forthe removal of the intron (and as further ascertained by nucleotide sequencing of the PCR fragments) (data not shown). The structure of the transposed elements and their insertion sites were analyzed further by carrying out inverted PCR on genomic DNA from some G418R clones, to confirm canonical retrotransposition. As illustrated in Figure 3B, the integration of four newly transposed MusD elements is characterized by generation of a target-site duplication of 6 bp (as observed for the naturally occurring genomic copies), and reconstitution of complete LTRs. The regenerated 5′-LTR does not contain any “residual” CMV promoter sequence but a complete U3 domain, as expected for a retroviral-like replicative cycle in which the ends of the proviral LTRs are reconstituted via strand transfers in the course of reverse transcription (for a review on retroviral replication, see Boeke and Stoye 1997). The integrated copies, as determined by PCR and partial sequencing, were found to be full-length structures.
Figure 3.
Assay for autonomous MusD retrotransposition and characterization of the de novo insertions. (A) Each of the three MusD elements encoding functional proteins, as determined in the assay in trans, was marked with the neoTNF indicator downstream from the pol ORF as schematized and assayed for its autonomous retrotransposition. Images of the plates and retrotransposition frequencies are given as in Figure 2 (two to three independent experiments, with standard errors indicated). (B) Structure and chromosomal localization of four transposed MusD copies. The complete characterization of transposed MusD copies and insertion sites was performed using G418R clones from MusD-1 and MusD-6 marked elements assayed for transposition in HeLa cells. The structures of the marked MusD expression vector, of the intermediate MusD transcript, and of the de novo insertion (with reconstituted LTRs and target site duplication, TSD) are schematized, together with the corresponding empty target site. The sequences of four insertions are given below, with the [TG... CA] LTR borders and flanking DNA sequences indicated; target-site duplications (grayed) are found in all four cases, associated with complete LTRs. The GenBank Accession no. of each insertion site is given together with its chromosomal localization (with the R bands in white and the G bands in dark gray).
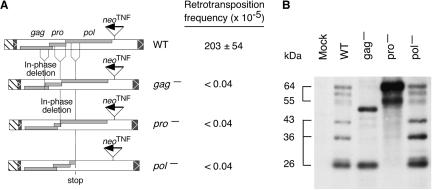
To characterize the role of the three MusD genes in retrotransposition, we next inactivated each of them independently (either by in-frame deletions for gag and pro so as to preserve translation of the downstream genes or by an out-of-frame deletion, thus introducing a prematurate stop codon for pol) (Fig. 4A). As illustrated in the figure, the frequency of G418R clones observed after transfection of each of the mutated copies was severely reduced (<10–6 compared with 10–3 for wild type), demonstrating the essential role of each gag, pro, and pol gene in MusD retrotransposition. We checked by Northern blot analysis that none of the introduced mutations had any effect on the level of transcription of the mutant proviruses (data not shown), and the level of expression of the Gag protein was controlled by Western blot analysis (see Fig. 4B). The latter analysis also clearly demonstrated that inactivation of the protease gene resulted in the disappearance of the low-molecular-mass (26, 36, and 43 kDa) Gag-related cleavage products of the wild-type element, and in the occurrence of high-molecular-mass bands as observed for some of the nonfunctional MusD copies in Figure 2.
Figure 4.
MusD gene products required for retrotransposition. (A) Assay for MusD retrotransposition was performed as in Figure 3, with either the wild-type neoTNF marked MusD-6 (WT) or the same element rendered defective for gag or pro (via an in-frame deletion in the corresponding gene so as not to alter translation of the downstream ORFs) or pol (via an out-of-frame deletion). The retrotransposition frequencies obtained for each construct are indicated. (B) Western blot analysis for cleavage of the MusD Gag polyprotein. Lysates of cells transfected with a control plasmid (mock) or the MusD constructs in A were electrophoresed (SDS-PAGE), blotted, and hybridized with a rabbit anti-serum directed against the Gag polyprotein. The apparent molecular mass for the major bands of the wild-type MusD is indicated.
Mobilization of ETn in trans by MusD elements
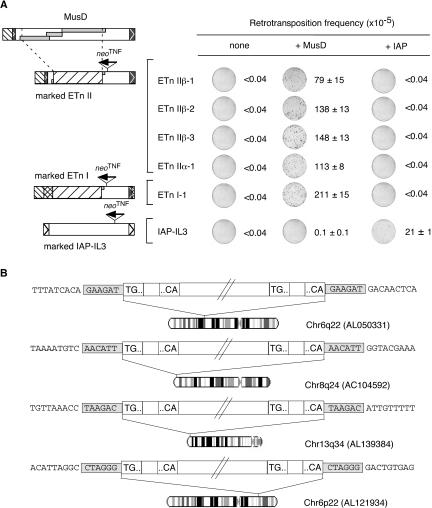
As previously demonstrated and illustrated in Figure 1A, ETn and MusD elements have several sequences in common, including almost identical 5′- and 3′-LTRs, a PolyPurine Tract (PPT), and a tRNALys Primer Binding Site (PBS) (see references in Baust et al. 2003). Yet, ETn elements are noncoding, and we therefore tested whether they could be mobilized in trans by one of the functional MusD elements that we have presently identified. To do so, we designed expression vectors for five ETn elements, which we selected from an in silico search in the mouse database on the basis of a 100% identity between their 5′- and 3′-LTRs (as a hint for “recent” integration). These selected elements included four type II ETns and one type I ETn for which BAC clones were obtained (see Table 1). These elements were placed under the control of the CMV promoter by replacement of the U3 domain of their 5′-LTR, as in the case of the MusD elements, and were marked with the neoTNF indicator gene as indicated (Fig. 5A). These marked elements were then introduced by transfection into HeLa cells to assay for their retrotransposition, either alone or together with an expression vector for one of the previously identified functional MusD element (MusD-6). As illustrated in Figure 5A, G418 selection of the transfected cells did not yield any G418R clone in the absence of MusD elements, but G418R clones were obtained at a high frequency upon cotransfection with the transposition-competent MusD vector. Retrotransposition was ascertained, as above for the MusD elements, by PCR analysis of randomly selected clones for the presence of copies with a spliced-out intron within the neoTNF indicator gene (data not shown but see below). Retrotransposition was observed for both the ETn II and the ETn I elements, with closely related rates. Transposition rates were also similar to those observed under identical experimental conditions upon complementation in trans of the marked defective MusD element (e.g., Fig. 2). To characterize further the trans-complementation process and determine whether ETn elements are specifically mobilized by MusD, we took advantage of the recent identification in our laboratory of active autonomous copies of another major family of murine LTR-retrotransposons, namely, the IAP sequences (Dewannieux et al. 2004), which are not related to MusD. A functional IAP was therefore assayed for complementation in trans of the ETn elements, as well of a control IAP marked copy. As illustrated in Figure 5A, cotransfection of the marked ETn with the IAP yielded no G418R clone, under conditions in which the IAP efficiently mobilized the marked IAP reporter. Conversely, it is noteworthy that MusD could not efficiently mobilize in trans the marked IAP, thus illustrating the genetic disjunction between these two families of LTR-retrotransposons.
Figure 5.
MusD-mediated retrotransposition of the ETn elements. (A) Assay for retrotransposition of neoTNF-marked ETn elements (schematized on the left with their copy number indicated) (see also Table 1) upon transcomplementation with the autonomous MusD-6 copy (+MusD) or a control plasmid (none). Images of the plates and retrotransposition frequencies are given as in Figure 2 (two to three independent experiments, with standard errors indicated). Additional experiments included assays with a neoTNF-marked defective IAP element (marked IAP-IL3) as well as complementation in trans with a functional IAP vector (RP23-92L23; +IAP) (Dewannieux et al. 2004). (B) Four ETn II insertions (originating from the ETn IIβ-1 element) are illustrated as in Figure 3, with the GenBank accession no. and chromosomal localization of each insertion site indicated.
Finally, the structure of newly transposed ETn copies and their insertion sites was determined as in Figure 3 for MusD copies, by inverted PCR on a series of four G418R clones. As illustrated in Figure 5B, partial sequencing of the transposed copies demonstrated bona fide retroviral-like retrotransposition, with reconstitution of complete ETn proviruses and LTRs (without any residual CMV promoter sequences) and generation of 6-bp duplications of the target sites, as observed for naturally occurring genomic ETn elements and for de novo retrotransposed MusD copies. This result is consistent with usage of the MusD retrotranspositon enzymatic machinery for the synthesis and integration of the ETn cDNA.
Discussion
Genome-wide screening of the mouse sequence databases for coding-competent MusD copies has revealed only nine such elements among the hundred copies of the MusD family. We show that only three of them are transposition-competent in an ex vivo assay in heterologous HeLa cells, and that all three gag, pro, and pol MusD genes are absolutely required for autonomous retrotransposition. We further demonstrate canonical retrotransposition of these elements, with splicing out of an intron contained in the indicator gene used to mark these elements, reconstitution of complete LTRs in the transposed copies, and precise integration of the copies into genomic DNA with generation of 6-bp duplications of the target site. Western blot analysis of the Gag-related products further demonstrates proteolytic cleavage of the Gag polyprotein precursor in the wild-type functional MusD copies, with inhibition of cleavage—associated with loss of retrotransposition competence—in the case of a protease mutant that we have generated. A similar Western blot analysis also demonstrates that three out of the six coding-competent but transposition-defective MusD copies generate uncleaved Gag products (MusD-4, -5, and -9). Interestingly, for two of these elements (MusD-4 and -9), mutations within the putative active site of the protease gene can be identified, consistent with the observed loss of transposition competence. For two other MusD copies that were found to be transposition-defective among the identified coding-competent elements, mutations could also be identified within critical regions of the integrase domain of the pol gene (within a conserved zinc-finger motif for MusD-3 and -7). Altogether, the present functional analysis of the genomic MusD copies discloses only a limited number of autonomous elements, clearly much lower than observed for the other major LTR-retrotransposons found in the mouse genome, that is, the IAP sequences, for which a similar analysis (Dewannieux et al. 2004) has led to an estimate of 300 functional copies (among 1000 elements). It is finally noteworthy that among the nine coding-competent MusD elements, the three autonomous MusD copies all disclose 100% identity between their 5′- and 3′-LTRs (Table 1), suggestive of recent transposition and consistent with their being functional elements. In agreement with this conclusion, it is of interest that two out of the three active MusD copies (i.e., MusD-1 and -2) were found to be present only in some, but not all, mouse strains at the corresponding orthologous loci (Baust et al. 2002).
The second major outcome of the present investigation is the unambigous demonstration of the capacity of the MusD elements to complement in trans the ETn elements for their efficient retrotransposition. Indeed, in agreement with the previously noticed remarkable sequence identities between noncoding domains of both MusD and ETn (Mager and Freeman 2000) and the absence of any coding gene within ETn, we show that active MusD copies can mobilize ETn elements (of both type I and II), with an efficiency similar to that of the noncoding MusD reporter element. The demonstrated transposition competence of the ETn type I element is noteworthy, because there exist some differences between ETn I and MusD in parts of their LTRs and 5′-UTRs, that is, within domains that include the tRNA Primer Binding Site (PBS) as well as, possibly, part of the packaging signal. This indicates that ETn type I elements should have conserved a homologous secondary structure for the latter function, despite nonidentity between the primary sequences of both elements. It is also consistent with the occurrence of a tRNALys PBS within the ETn type I elements, as already noted (Kaghad et al. 1985; Baust et al. 2003). This site is conserved among the ETn I and the ETn II/MusD elements despite marked differences in the surrounding sequences, and can, indeed, be found in the presently assayed ETn I copy (i.e., 5′-TGGCGAACCAACGTGGGAC-3′). The present demonstration of a high transposition rate for the ETn I element is also of interest because it has been reported that among 15 transposition events revealed by phenotypic mutations in mice and associated with MusD/ETn elements (Baust et al. 2002), none was associated with ETn type I elements (whereas seven could be unambiguously associated with type II ETn elements). A possible explanation could be that because ETn transposition requires MusD activation for trans-complementation (our present data), the probability that both a MusD element and an ETn element be simultaneously induced is higher in the case of the ETn II elements, which are more closely related to MusD than ETn I elements, especially within their LTRs. Yet ETn Is are present at a high copy number in mice—an even higher number than for ETn II, that is, 200 versus 40, respectively—consistent with our functional analysis and results.
Interestingly, we have also demonstrated that complementation in trans of the ETn elements by functional MusD sequences is, indeed, MusD-specific, with no effect of a transposition-competent IAP on ETn transposition, and conversely no effect of a transposition-competent MusD on IAP transposition. Finally, the close relationship between the ETn and “master” MusD elements is strengthened by the identity of the target-site signature (6-bp duplications) that can be observed both in the present ex vivo assay for newly transposed marked copies and in naturally occurring genomic copies. It is also noteworthy that, as observed for the IAP elements (Dewannieux et al. 2004) and in agreement with what has been described for the insertion sites of retroviruses (Bushman 2003), analysis of the eight characterized insertion sites for both the MusD and ETn marked elements did not reveal any sequence-specificity of the target, but in six cases out of eight disclosed insertions preferentially within actively transcribed domains, as inferred from an EST-based analysis (using the UCSC Genome Bioinformatic Site; http://genome.ucsc.edu) (data not shown).
In conclusion, the present results provide definite evidence for “parasitic” molecular strategies among endogenous elements, with the ETn family members “using” the MusD transpositional machinery for their mobility (see also Dewannieux et al. [2003] and Schmid [2003] for the LINE/Alu non-LTR retrotransposon pair). The identification of “active” MusD copies should further allow a refined structural characterization of these elements in relation with their retroviral origin before they became bona fide endogenous genomic elements.
Methods
Database mining
The sequences of two MusD elements (AF246632 and AF246633) (Mager and Freeman 2000) were used as query sequences to screen the Non-Redundant (nr) and High-Throughput Genomic Sequences (htgs) GenBank database at the NCBI site (http://www.ncbi.nlm.nih.gov; December 2003 release). All the identified copies were analyzed to isolate coding-competent elements (Table 1). ETn elements were similarly identified, using as query sequences Y17106 for the ETn II and M16478 for the ETn I subfamily, and only a few elements with 100% identical 5′- and 3′-LTRs (Table 1) were selected for the retrotransposition assay.
Oligonucleotides used for PCR amplification
All primer sequences are listed in Supplemental Table A.
Plasmids
The ETn and coding-competent MusD elements were derived from commercial BACs (BACPAC Resources). Accession number, position of the first-to-last nucleotides of the MusD and ETn proviruses, and strand orientation are indicated in Table 1.
Construction of MusD expression vectors
The R-U5 domain of MusD-6 (AC124426) was PCR-amplified from the corresponding BAC and cloned just 3′ to the transcription start site of the CMV promoter contained in a SacI–SalI fragment derived from pCMV-β (Clontech; pCMV/RU5 plasmid). The expression vectors for the nine coding-competent MusD elements were obtained either by inserting the corresponding NotI–NotI internal fragment excised from the BACs (U5-gag-pro-pol-U3-R-U5; 7.2 kb) into the pCMV/RU5 plasmid digested by NotI, or by replacing an internal fragment containing the three ORFs in an existing MusD expression vector when NotI sites were absent.
Construction of the neoTNF-marked MusD elements
The neoTNF-marked defective MusD was obtained upon deletion of a 4748-bp fragment encompassing the gag–pro–pol genes (nucleotides 843 to 5590) from the expression vector constructed with the MusD-6 copy, and insertion of a blunt-ended HindIII–XhoI fragment from pSVNeoTNF (Esnault et al. 2002) at the unique SwaI site (nucleotide 5839). Three full-length, coding-competent MusD elements were marked by insertion (without deletion) of the neoTNF reporter gene at the SwaI site (downstream from the pol gene) of the corresponding expression vectors. The gag–, pro–, and pol– MusD mutants were constructed using the marked MusD-6 copy by introducing in-frame deletions in the gag (492-nt deletion from nucleotides 1634 to 2125) and pro genes (630-nt deletion from nucleotides 2464 to 3093), and an out-of-frame 493-nt deletion in the pol gene (from nucleotides 3392 to 3884, thus introducing a 2-nt frameshift resulting in a premature stop codon).
Construction of the marked ETn elements
Five expression vectors for ETn elements were constructed using the same strategy as for the MusD expression vectors. Briefly, the R-U5 domain of each selected ETn was PCR-amplified from the corresponding BAC and cloned just 3′ to the transcription start site of the CMV promoter, and the internal domain of the corresponding elements was inserted as a NotI–NotI fragment (except for ETn I-1, which was PCR-amplified). These copies were then marked by insertion of the neoTNF reporter gene at a unique 3′-site (SwaI for ETn IIβ-1 and ETn I-1, BsmI for ETn IIα-1, ETn IIβ-2, and ETn IIβ-3).
Cells, transfection, and transposition assays
Human HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (GIBCO BRL), 100 μg/mL streptomycin and 100 U/mL penicillin. The day prior to transfection, cells were seeded at 5.5 × 105 cells per 60-mm dish. Transfection of HeLa cells was performed with 3 μg of DNA using Lipofectamine (GIBCO BRL), following the manufacturer's instructions. To assay for retrotransposition, transfected cells were expanded for 6 d, seeded at between 1 and 5 × 105 cells per 100-mm dish and allowed to settle for 24 h before adding G418 (560 μg/mL; GIBCO BRL). After a 10–15-d selection, G418R foci were fixed, stained, and counted, or individually picked and expanded.
Gag antiserum and Western blot analysis
A 1728-bp fragment (nucleotides 641 to 2368) from the MusD-1 copy (AC034116) and corresponding to the gag gene, was PCR-amplified and cloned into the pET19b vector (Novagen) opened by NdeI–BlpI, to allow prokaryotic expression as a histidine-tagged protein. The recombinant protein was then purified on a HiTrap column (Amersham) and used for rabbit immunization. For analysis of Gag products, HeLa cells transfected with the MusD expression vectors or a control plasmid were lysed in Laemmli buffer 48 h after transfection, and whole-cell lysates were separated by SDS-PAGE using gels containing 12% polyacrylamide (37.5:1 ratio of acrylamide to N,N-methylenebisacrylamide). Proteins were transferred to nitrocellulose membranes (Schleicher & Schuell) and incubated with the rabbit antiserum (1:1500 dilution). A goat antibody to rabbit IgG conjugated to horseradish peroxidase (Amersham) was used as the secondary antiserum.
DNAs and characterization of the transposed copies
Cells were lysed and incubated overnight in 10 mM Tris (pH 8), 10 mM EDTA (pH 8), 125 mM NaCl, 0.2% SDS, and 300 μg/mL proteinase K, and DNA was recovered by phenol-chloroform extraction and ethanol precipitation. For the characterization of the transposed copies by inverted PCR, DNA (5 μg) was digested using restriction enzymes cutting in neoTNF and in the 3′-flanking sequence at an unknown distance from the inserted MusD/ETn copies. Restriction fragments were blunt-ended (Klenow enzyme) and self-ligated at a high dilution (500 μL) using 3200 U of T4 Ligase (Biolabs). The circularized DNAs containing a spliced neoTNF and 3′-LTR were amplified by one round, or two, of PCR with sets of divergent primers. PCR products were screened for the presence of the NotI site (located in the MusD/ETn 3′-LTR), and positive fragments were sequenced to identify the 3′ cellular flanking sequence. Primers were then designed in the putative 5′-flanking region (according to the human genome sequence database at the NCBI site), and the 5′-joining region was finally PCR-amplified and sequenced.
Acknowledgments
We thank C. Lavialle for comments and critical reading of the manuscript. This work was supported by the CNRS and the Ligue Nationale Contre le Cancer (Equipe “labellisée”).
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2924904. Article published online before print in October 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Baust, C., Baillie, G.J., and Mager, D.L. 2002. Insertional polymorphisms of ETn retrotransposons include a disruption of the wiz gene in C57BL/6 mice. Mamm. Genome 13: 423-428. [DOI] [PubMed] [Google Scholar]
- Baust, C., Gagnier, L., Baillie, G.J., Harris, M.J., Juriloff, D.M., and Mager, D.L. 2003. Structure and expression of mobile ETnII retroelements and their coding-competent MusD relatives in the mouse. J. Virol. 77: 11448-11458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke, J.D. and Stoye, J.P. 1997. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In Retroviruses (eds. J.M. Coffin et al.), pp. 343-435. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- Brûlet, P., Kaghad, M., Xu, Y.-S., Croissant, O., and Jacob, F. 1983. Early differential tissue expression of transposon-like repetitive DNA sequences of the mouse. Proc. Natl. Acad. Sci. 80: 5641-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brûlet, P., Condamine, H., and Jacob, F. 1985. Spatial distribution of transcripts of the long repeated ETn sequence during early mouse embryogenesis. Proc. Natl. Acad. Sci. 82: 2054-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushman, F.D. 2003. Targeting survival. Integration site selection by retroviruses and LTR-retrotransposons. Cell 115: 135-138. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., Esnault, C., and Heidmann, T. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35: 41-48. [DOI] [PubMed] [Google Scholar]
- Dewannieux, M., Dupressoir, A., Harper, F., Pierron, G., and Heidmann, T. 2004. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 36: 534-539. Epub Apr. 25, 2004. [DOI] [PubMed] [Google Scholar]
- Esnault, C., Casella, J.F., and Heidmann, T. 2002. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 30: e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaghad, M., Maillet, L., and Brûlet, P. 1985. Retroviral characteristics of the long terminal repeat of murine ETn sequences. EMBO J. 4: 2911-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuff, E.L. 1990. Intracisternal A particles in mouse neoplasia. Cancer Cells 2: 398-400. [PubMed] [Google Scholar]
- Kuff, E.L. and Lueders, K.K. 1988. The intracisternal A-particle gene family: Structure and functional aspects. Adv. Cancer Res. 51: 183-276. [DOI] [PubMed] [Google Scholar]
- Lander, E.S., Linton, L.M., Birren, B., Nusbaum, C., Zody, M.C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860-921. [DOI] [PubMed] [Google Scholar]
- Löwer, R., Löwer, J., and Kurth, R. 1996. The viruses in all of us: Characteristics and biological significance of human endogenous retrovirus sequences. Proc. Natl. Acad. Sci. 93: 5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager, D.L. and Freeman, J.D. 2000. Novel mouse type D endogenous proviruses and ETn elements share long terminal repeat and internal sequences. J. Virol. 74: 7221-7229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, J., Sauter, M., Racz, A., Scherer, D., Mueller-Lantzsch, N., and Meese, E. 1999. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat. Genet. 21: 257-258. [DOI] [PubMed] [Google Scholar]
- Ostertag, E.M. and Kazazian Jr., H.H. 2001. Biology of mammalian L1 retrotransposons. Annu. Rev. Genet. 35: 501-538. [DOI] [PubMed] [Google Scholar]
- Reus, K., Mayer, J., Sauter, M., Scherer, D., Muller-Lantzsch, N., and Meese, E. 2001. Genomic organization of the human endogenous retrovirus HERV-K(HML-2.HOM) (ERVK6) on chromosome 7. Genomics 72: 314-320. [DOI] [PubMed] [Google Scholar]
- Schmid, C.W. 2003. Alu: A parasite's parasite? Nat. Genet. 35: 15-16. [DOI] [PubMed] [Google Scholar]
- Shell, B., Szurek, P., and Dunnick, W. 1987. Interruption of two immunoglobulin heavy-chain switch regions in murine plasmacytoma P3.26Bu4 by insertion of retroviruslike element ETn. Mol. Cell. Biol. 7: 1364-1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shell, B.E., Collins, J.T., Elenich, L.A., Szurek, P.F., Dunnick, W.A., Juriloff, D.M. and Mager, D.L. 1990. Two subfamilies of murine retrotransposon ETn sequences. Gene 86: 269-274. [DOI] [PubMed] [Google Scholar]
- Sonigo, P., Wain-Hobson, S., Bougueleret, L., Tiollais, P., Jacob, F., and Brûlet, P. 1987. Nucleotide sequence and evolution of ETn elements. Proc. Natl. Acad. Sci. 84: 3768-3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, I. and Ishihara, H. 2001. Enhanced expression of the early retrotransposon in C3H mouse-derived myeloid leukemia cells. Virology 280: 107-114. [DOI] [PubMed] [Google Scholar]
- Turner, G., Barbulescu, M., Su, M., Jensen-Seaman, M.I., Kidd, K.K., and Lenz, J. 2001. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr. Biol. 11: 1531-1535. [DOI] [PubMed] [Google Scholar]
- Waterston, R.H., Lindblad-Toh, K., Birney, E., Rogers, J., Abril, J.F., Agarwal, P., Agarwala, R., Ainscough, R., Alexandersson, M., An, P., et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520-562. [DOI] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://genome.ucsc.edu; UCSC Genome Bioinformatic Site.
- http://www.ensembl.org/Mus_musculus; Ensembl Mouse Genome Server.
- http://www.ncbi.nlm.nih.gov; National Center for Biotechnology Information.