Abstract
Because bacterial chromosomes are tightly packed with genes and were traditionally viewed as being optimized for size and replication speed, it was not surprising that the early annotations of sequenced bacterial genomes reported few, if any, pseudogenes. But because pseudogenes are generally recognized by comparisons with their functional counterparts, as more genome sequences accumulated, many bacterial pathogens were found to harbor large numbers of truncated, inactivated, and degraded genes. Because the mutational events that inactivate genes occur continuously in all genomes, we investigated whether the rarity of pseudogenes in some bacteria was attributable to properties inherent to the organism or to the failure to recognize pseudogenes. By developing a program suite (called Ψ-Φ, for Ψ-gene Finder) that applies a comparative method to identify pseudogenes (attributable both to misannotation and to nonrecognition), we analyzed the pseudogene inventories in the sequenced members of the Escherichia coli/Shigella clade. This approach recovered hundreds of previously unrecognized pseudogenes and showed that pseudogenes are a regular feature of bacterial genomes, even in those whose original annotations registered no truncated or otherwise inactivated genes. In Shigella flexneri 2a, large proportions of pseudogenes are generated by nonsense mutations and IS element insertions, events that seldom produce the pseudogenes present in the other genomes examined. Almost all (>95%) pseudogenes are restricted to only one of the genomes and are of relatively recent origin, suggesting that these bacteria possess active mechanisms to eliminate nonfunctional genes.
The recognition of pseudogenes (Ψ-genes) within eukaryotic genomes was based originally on comparisons of the sequences of the members of multigene families (Jacq et al. 1977; Vanin et al. 1980). But because bacterial genomes contain few duplicated genes and generally low amounts of nonfunctional DNA (Mira et al. 2001; Rogozin et al. 2002), the presence of pseudogenes in their genomes was originally assumed to be rare (Lawrence et al. 2001). In light of the fact that bacterial genes can be quite short, that the functional analysis of all hypothetical Coding Sequences (CDSs) is not feasible, and that relatively few sequences were then available for comparison, it is not surprising that the initial annotations of fully sequenced bacterial genomes reported virtually no pseudogenes.
Several bacterial pseudogenes were discovered by experimental or comparative analysis (e.g., Delorme et al. 1993; Lai et al. 1996; Simonet et al. 1996; Zhu et al. 1999); however, among the unprecedented discoveries from complete genome sequencing was that certain bacteria, particularly pathogens, contain substantial numbers of pseudogenes. For example, the genome of typhus bacterium Rickettsia prowazekii contains a large noncoding fraction, formed, in part, by recognizable and degraded pseudogenes (Andersson et al. 1998); and in Mycobacterium leprae, the causative agent of leprosy, pseudogenes constitute 27% of the 3.3-Mb genome (Cole et al. 2001). Additionally, the enteric pathogens Shigella flexneri (Jin et al. 2002; Wei et al. 2003), Salmonella enterica (McClelland et al. 2001; Parkhill et al. 2001a), and Yersinia pestis (Parkhill et al. 2001b; Deng et al. 2002) each harbors a large number, often hundreds, of pseudogenes.
As in eukaryotes, bacterial pseudogenes can originate by diverse mechanisms, including the disruption of a reading frame or promoter regions by point mutations, frameshifts, or the integration of transposable elements. When such mutations occur in genes that are no longer required, they can be maintained in the genome for some time but are gradually degraded and eliminated by deletions (Andersson and Andersson 1999a,b; Mira et al. 2001). Changes in bacterial lifestyle often lead to modifications in the utility of some genes, and the high proportion of pseudogenes in pathogens is related both to the dispensability of previously useful genes in the host environment (Andersson and Andersson 1999b; Jin et al. 2002) and to a population structure that promotes the maintenance of deleterious mutations (Andersson and Hughes 1996; Cooper and Lenski 2000; Mira et al. 2001). Because mutational events that inactivate genes arise continuously in all genomes, the occurrence of pseudogenes might be exaggerated in, but should not be limited to, pathogens. Thus, there is little reason to expect that all CDSs within a bacterial genome will encode functional proteins.
If all organisms generate pseudogenes, why were such regions so rarely detected in the bacterial genomes that were among the first to be completely sequenced? There are three possible explanations: First, it is possible that every gene in these genomes is under strong selective constraints, such that any inactivated gene is associated with a fitness decrement causing the organism to be eliminated from the population. Second, pseudogenes could be produced but then degraded and removed by the deletional processes known to be present in bacterial genomes (Andersson and Andersson 1999a,b; Mira et al. 2001). Finally, it could be that these genomes harbor large numbers of pseudogenes but that they have yet to be recognized. Therefore, the functional status of annotated ORFs in each genome needs to be evaluated.
Many annotation tools, particularly those relying on some minimal length threshold for the recognition of ORFs (e.g., GLIMMER; Salzberg et al. 1998; Delcher et al. 1999), might be inaccurate in their assignments of functional genes. However, the increased availability of sequenced genomes now allows discrimination between functional and disrupted or degraded regions through comparisons of homologs in related strains and species. For example, such an approach based on the already annotated CDSs from two genomes detected 59 pseudogenes in Escherichia coli MG1655, a nonpathogenic strain originally thought to contain only a single pseudogene (Homma et al. 2002). And through comparisons of additional genomes, as well as considering the unannotated portions of genomes, it is likely that numbers of previously unidentified pseudogenes will vastly increase.
To determine the extent to which the lack of pseudogenes in many bacterial genomes is a function of biological processes or of erroneous CDS assignments, we developed Ψ-Φ (short for Ψ-gene Finder), a suite of programs designed to identify pseudogenes (attributable both to misannotation and to nonrecognition) through comparative analyses of related genomes. Applying Ψ-Φ to sequenced members of the E. coli/Shigella clade, in which current annotations report from one pseudogene in E. coli MG1655 to 254 in S. flexneri 2a, we resolved at least 100 additional disrupted CDSs in each genome. This method greatly enhances gene recognition and annotation by identifying incorrectly annotated genes, by detecting new pseudogenes in previously unannotated regions, and even by discovering new potentially functional ORFs.
Results
Genome inventories of pseudogenes
The Ψ-Φ program suite, developed to compare the coding capacity of complete genomes, detected 98, 142, 98, and 168 new pseudogenes in the genomes of E. coli MG1655, E. coli O157:H7, and E. coli CFT073 and S. flexneri 2a, respectively (see Supplemental material for the list of the new pseudogenes in each genome). Given that the numbers of pseudogenes reported in the original annotations of these genomes ranged from only one in E. coli MG1655 (Blattner et al. 1997) to 254 in S. flexneri 2a (Jin et al. 2002), such systematic comparisons will identify numerous truncated and inactivated CDSs both in genomes thought to be lacking in as well as those replete with pseudogenes.
Considering all recognized pseudogenes, including those previously annotated and those newly identified by Ψ-Φ, these enteric genomes display very different spectra of mutations that led to the formation of pseudogenes. Events that inactivate a gene fall into five general classes: nonsense mutations, frameshifts, IS insertions, other insertions or deletions, and truncations (Fig. 1A). Those classified as “frameshifts” are generally very small insertions or deletions (typically only 1 or 2 nt in length), whereas those labeled as “other insertions” are >10 bp in length, and “truncations” include large deletions at either or both the beginning and end of a CDS.
Figure 1.
(A) Numbers and types of mutations forming pseudogenes. Bars include those pseudogenes reported in the original annotation (white) and those recognized subsequently by comparative analyses (black). (The specific mutations labeled as “deletions” in S. flexneri 2a were not provided in the original annotation.) Asterisks denote genomes for which some of the newly recognized pseudogenes include many of those reported in Homma et al. (2002). (B) Locations of newly recognized pseudogenes in relation to annotated CDSs and nonannotated regions.
S. flexneri 2a, which harbors the highest number of insertion sequences (n = 314) (Jin et al. 2002), has the highest frequency of CDSs interrupted by IS element insertions (79/422 vs. three to six in the other species); however, it should be noted that nonsense mutations constitute the largest proportion (155/422) of gene-inactivating events in this genome. In contrast, in both E. coli MG1655 and E. coli CFT073, most pseudogenes originate from frameshifts and truncations, and only low proportions are caused by either nonsense mutations or IS insertions (Fig. 1A).
Newly discovered pseudogenes
Despite their close relationships and broadly overlapping gene inventories, the source of the newly identified pseudogenes varies among these four genomes (Fig. 1B). In each of these genomes, application of Ψ-Φ led to the discovery of substantial numbers of previously unrecognized truncations, that is, pseudogenes formed by deletions of large portions of the original CDS, and these truncations occur with approximately equal frequencies in annotated coding regions and intergenic regions. The majority of new pseudogenes in E. coli CFT073 represents incorrect annotations of CDSs, that is, they were considered functional in the original genome annotation, whereas in S. flexneri 2a, most of the newly detected pseudogenes reside in intergenic regions. Figure 2 depicts examples of three of the newly recognized pseudogenes in Shigella that were generated by different mechanisms.
Figure 2.
Examples of newly discovered pseudogenes in the S. flexneri 2a genome and their counterparts in E. coli MG1655. (A) Pseudogene formed by integration of an IS element. (B) Nonsense mutation (star) in the S. flexneri homolog of E. coli b2107 forming a truncated CDS (annotated as yohN). (C) Frameshift mutation in the S. flexneri homolog of the E. coli hyfG gene, which resulted in the incorrect annotation of two CDSs in Shigella.
Gene function and pseudogene formation
The majority of pseudogenes (∼80%) is formed from intact homologs whose functional status in other genomes is listed as hypothetical, putative, or unknown. In only very few cases have informational genes, that is, those known to be involved in transcription, translation, and related processes (Jain et al. 1999), become pseudogenes. The dnaA gene, whose product controls the initiation of chromosome replication, has been inactivated by independent events in S. flexneri 2a and E. coli CFT073. Despite its role in DNA replication, dnaA is not essential (Kogoma and von Meyenburg 1983), and it has also been eliminated from the reduced genomes of the insect endosymbionts Blochmannia floridanus (Gil et al. 2003) and Wigglesworthia glossinidia (Akman et al. 2002). In addition to dnaA, genes involved in DNA metabolism (lhr) (Reuven et al. 1995) and DNA repair (ung) (Duncan and Miller 1980) are among the Shigella pseudogenes, and in E. coli O157:H7, the DNA recombination exonuclease gene recE (Porter 1983) has been truncated.
Evolution of pseudogenes
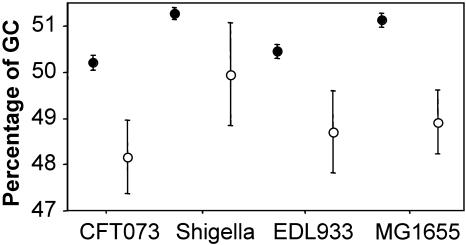
On average, the CDSs in each genome that have pseudogene counterparts in other genomes have a lower G+C content than the rest of the genes in the genome (Fig. 3). The lower G+C content reflects the fact that these genes display less adaptive codon usage bias (CAIgenes with pseudogene homologs = 0.32 vs. CAIgenes without pseudogene homologs = 0.37 in E. coli MG1655), and these reduced selective constraints are also evident in their evolutionary rates. Using comparisons between E. coli and Salmonella enterica homologs to obtain estimates of nonsynonymous (Ka) and synonymous (Ks) substitution rates, both the Ka and Ks values are significantly higher for genes that became pseudogenes in one or more genomes (Mann-Whitney U-test, p < 0.001), which usually results from reduced functional constraints. Note, however, that Ka/Ks ratios are still much less than 1, indicating that these genes were functional prior to becoming pseudogenes.
Figure 3.
Average G+C content of genes that do (open circles) or do not (shaded circles) have pseudogene counterparts in the other three genomes examined. Note that in all genomes, genes that become pseudogenes are of significantly lower %G+C.
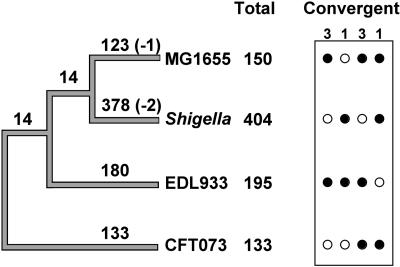
We reconstructed the phylogenetic relationships of these strains using the consensus of Neighbor-Joining trees of all orthologs in the four species, using S. enterica as the outgroup. This tree allows us to trace the origin of each pseudogene during the evolutionary diversification of lineages (Fig. 4). About 95% of the pseudogenes are young and are represented in only one of the genomes. However, there are, in total, 14 pseudogenes that are shared by E. coli MG1655 and S. flexneri 2a (which are sister taxa), and another 14 are ancestral to E. coli MG1655, Shigella, and E. coli O157:H7 (with one subsequently lost by E. coli MG1655 and two lost by S. flexneri). For eight pseudogenes, the pattern of distribution of functional and nonfunctional homologs required either a recombination event between non-sister taxa or the independent occurrence of the same inactivating mutation in two lineages.
Figure 4.
Phylogeny of the genomes examined showing numbers of contemporary pseudogenes that arose on each branch. In a few cases, the distribution of a pseudogene allowed inference of its loss by a particular lineage (negative numbers). Total numbers of pseudogenes (that can be mapped onto the tree) are shown after strain designations. Convergent pseudogenes correspond to those that occur in the same gene with the same mutations but that arose independently based on their phylogenetic distribution. The black circles represent the presence of the convergent pseudogenes in the given species, and the white circles correspond to their absence. The numbers over the box indicate the number of pseudogenes with the given pattern.
Discussion
Because bacterial genomes were traditionally viewed as being optimized for size and replication speed, there was little expectation that they would harbor substantial amounts of noncoding DNA or nonfunctional genes. And when combined with the fact that most known pseudogenes were originally identified by comparative analyses against functional counterparts, it is not surprising that pseudogenes went unrecognized in genomes for which no broadly overlapping genomic sequences were available. However, based on comparisons of full genome sequences, we find that pseudogenes are a regular feature of bacterial genomes, even in those previously thought to have few, if any, truncated and otherwise inactivated genes. By analyzing the genomes of four sequenced representatives of the E. coli/Shigella clade, we detected hundreds of previously unrecognized pseudogenes, some of which (∼20%) correspond to genes of known or assigned function in one of the other three strains.
This comparative approach is made feasible only by the recent availability of the complete sequences of closely related bacterial species, and we designed the program suite Ψ-Φ to perform such systematic searches for pseudogenes among any pair of genome sequences. Extending our analyses to other groups for which there are several sequenced genomes, we have detected numerous new pseudogenes in Yersinia pestis, Streptococcus pyogenes, and Staphylococcus aureus (E. Lerat and H. Ochman, in prep.). Naturally, the ability to discover previously unrecognized pseudogenes relies on several factors, including the manner in which the genome was originally annotated, the level of sequencing artifacts, and the overall similarity to the genomes available for comparison.
Only a subset of the pseudogenes, that is, truncated or otherwise disrupted CDSs, are recognized by this approach, whereas pseudogenes produced by missense mutations that abolish protein function (but maintain protein length) and by regulatory mutations that prevent transcription will go undetected. Collectively, the genes inactivated by the types of mutations identified using Ψ-Φ—that is, those containing premature stop codons, small indels that alter the reading frame, and large insertions or deletions that fragment the protein—are likely to constitute the more prevalent classes of pseudogenes in the genome. The frequency of mutations in noncoding regions or of amino acid replacements that sequester gene function is relatively low because the critical sites within proteins and regulatory regions span only a small proportion of the total length of the gene. Comprehensive studies of the lac repressor gene by site-directed mutagenesis showed that about half of the missense substitutions will completely destroy protein function, and that many of these involve radical changes from the original amino acid (Gordon et al. 1988; Kleina and Miller 1990). In any event, only the very recent pseudogenes produced by such events will be missed, because once inactivated, the pseudogenes will subsequently incur nonsense, frameshift, or other mutations that can be detected by Ψ-Φ.
Aside from the classes of pseudogenes that might go unrecognized, there is also the possibility that some of the putative pseudogenes defined by Ψ-Φ are still operative despite considerable alteration from their functional counterparts. Among those pseudogenes identified by Ψ-Φ, those involving IS insertions or large truncations unambiguously produce nonfunctional genes, whereas some nonsense mutations or frameshifts, depending on their specific location, might not disrupt gene function. In this regard, we consider as pseudogenes only those cases in which an internal stop codon results in a protein that is <80% of the length of its functional counterpart or in which a frameshift has altered >20% of the amino acid sequence. (It is possible that some small percentage of sequences meeting these criteria are not actual pseudogenes, as would be the case if the reference gene is annotated as being >20% of its actual length.)
All strains of E. coli investigated, regardless of their genome sizes or virulence potential, contain hundreds of pseudogenes corresponding to 3.7% in E. coli MG1655, 3.8% in E. coli O157:H7, and 2.8% in E. coli CFT073 of the coding capacity. The genome sizes of two pathogenic strains (EDL933 and CFT073) are larger than that of MG1655, owing, in part, to the independent integration of numerous DNA segments, some of which encode virulence determinants. In that most of these sequences are acquired and under no (or very weak) functional constraints, they would serve as a good source of pseudogenes. However, the functions of almost all these strain-specific sequences are unknown, and their lack of homologs renders the identification of pseudogenes by comparative analysis unfeasible; therefore, each of these pathogenic strains might harbor numerous additional pseudogenes that will never be recognized. This might explain why we detected as many pseudogenes in the nonpathogenic strain E. coli MG1655 as in the pathogenic strains.
The majority of newly discovered pseudogenes in Shigella were detected in intergenic regions. These were not found during the original annotation presumably because the large numbers of IS elements prevented recognition of short, truncated genes that did not reach some threshold length. In contrast, in E. coli CFT073, whose annotation includes a higher quantity of small (<200 bp) CDSs than those of the other sequenced enteric genomes, most new pseudogenes correspond to misidentified genes. Because most genome annotation involves a search for ORFs of some prescribed (and sometimes arbitrary) minimal length cutoff, we suspect that the numbers of pseudogenes are inaccurate for those genomes in which systematic comparisons with closely related genomes either were not performed or were impossible. E. coli MG1655 is a prime example: the original annotation reported a single putative pseudogene, whereas the conservative application of Ψ-Φ detected >150, and these numbers are likely to increase as more genomes become available.
Among the enteric genomes that we queried, the largest numbers of pseudogenes (for both newly discovered and those already annotated) were detected in S. flexneri, in which at least 10% of the CDSs are inactivated. The accumulation of pseudogenes in recent and/or intracellular pathogens is well established and reflects a process by which the effectiveness of selection is decreased by reductions in effective population sizes, coupled with the redundancy of many previously useful genes, in the host environment. Moreover, the inefficiency of selection operating in Shigella also accounts for the increases in the quantities of insertion sequences (with a concomitant production of pseudogenes created by IS insertions) relative to those observed in related genomes. These increases in the numbers of transposable elements and of pseudogenes denote an intermediate phase in the evolution of host-associated bacteria (Moran 2002; Dale et al. 2003) because the derived genomes of intracellular obligate symbionts, such as Buchnera, Wigglesworthia, and Blochmania, are very highly reduced (with genomes' sizes in the range of 500 kb) and contain very few pseudogenes or IS elements (Shigenobu et al. 2000; Akman et al. 2002; Gil et al. 2003). Because the mutational process in bacteria is biased toward deletions, pseudogenes ultimately diverge and erode beyond recognition, and are eventually eliminated (Andersson and Andersson 1999b; Lawrence et al. 2001; Mira et al. 2001). This situation is very pronounced in M. leprae, whose reduced genome contains >1000 pseudogenes (Cole et al. 2001) and whose annotated spacers, which likely correspond to very highly degraded genes, are nearly five times longer than those present in all other bacterial genomes (Mira et al. 2001).
Despite the close relationships of the strains that we analyzed (averaging only 1% sequence divergence), there is almost no overlap in the sets of pseudogenes harbored by each, indicating that these genes became truncated or inactivated relatively recently (Fig. 4). Given that mutations (and therefore, pseudogenes) are continually being formed over the entire history of a lineage, the paucity of pseudogenes that are ancestral to sister taxa is puzzling. It could be that older pseudogenes are extremely degraded, thereby preventing recognition by our comparative analyses; but the close relationships of the strains examined suggest that there was not sufficient time for such extensive sequence divergence. Rather, it appears that the older pseudogenes were eliminated from these genomes as a result of a genome-wide mechanism that promotes removal of nonfunctional sequences (Lawrence et al. 2001) or because the loss-of-gene functions was deleterious. Because pseudogenes represent an initial phase in the process of genome degradation occurring in bacterial pathogens and symbionts (Moran 2002), the preponderance of strain-specific pseudogenes could reflect recent changes in the lifestyles of each of these enterics. If this view is correct, we expect, in addition to the ongoing disruption and inactivation of genes, there to be waves of pseudogene formation over the history of a lineage, defined by the major, long-term shifts in selective pressures and/or population sizes. In this regard, the inventories and age distributions of pseudogenes in other groups of bacteria, particularly those in more long-standing pathogenic lineages, should be distinct from those observed in E. coli and should reflect aspects of their particular associations with hosts.
Methods
Because pseudogenes can be produced by point mutations that introduce stop codons, indels, and frameshifts, and insertions of transposable elements—all of which prevent the production of a full-length protein—computational procedures must incorporate a means for detecting products of each of these types of events. (Although pseudogenes can also result from missense mutations and from the alteration of regulatory sequences, the present analysis is limited to the detection and recovery of truncated CDSs.)
Genome comparisons
We retrieved conceptually translated proteins from the annotated genome sequences of E. coli MG1655 (GenBank accession no. U00096) (Blattner et al. 1997), E. coli CFT073 (AE014075) (Welch et al. 2002), E. coli EDL933 (AE005174) (Perna et al. 2001), and Shigella flexneri 2a (AE005674) (Jin et al. 2002). After eliminating all sequences annotated as phage or insertion sequences, the set of proteins encoded by each genome was used to query the nucleotide sequences of the three other genomes via TBLASTN (Altschul et al. 1997). These cross-comparisons recover all regions in the subject genomes matching the annotated CDSs in each species, and the orthology of matches is established though validations provided in the original annotations or by correspondence of neighboring genes.
Finding pseudogenes with Ψ-Φ
We developed Ψ-Φ (available upon request from the authors), a program suite, implemented in two modules and written in perl, to search for pseudogenes in bacterial genomes. The first module is designed both to analyze TBLASTN output to retrieve matches according to their identity to query proteins and to obtain the lengths of query CDSs by searching in the GenBank file of the query species. From this analysis, we retain matches with 80% to 100% protein sequence identity to the query protein. When a query sequence has two matches in close proximity in the genome (as might result from frameshifts or insertion), the matches are merged. The distance for merging such matches can be established empirically, and for the present analysis, we merged matches if they were <300 nt apart.
The second module of the program compares the output of the first module (i.e., all selected sequence matches to the CDSs in a query genome) to the GenBank file of the genome in which the search is being performed (i.e., the target genome) to identify and retain sequences whose characteristics denote assignment as a potential pseudogene. To recognize probable pseudogenes, we first determine if a sequence match overlaps an already annotated protein-coding gene in the target genome, or whether matches reside in or embrace intergenic regions (and therefore are indicative of misannotation). In cases in which a match corresponds to an intergenic region, we consider the length of the match relative to that of the query CDS, the probability of the match (based on BLAST E-values), and the occurrence of a premature stop codon to differentiate classes of probable pseudogenes for further curation. This procedure differentiates among intergenic matches with internal stop codons, intergenic matches of lengths <80% of the query CDS, and intergenic matches that are very short (<30% of the length of the query). In this analysis, we applied an E-value cutoff of <10e–15. (Note that any matches to intergenic sequences that do not display these characteristics are likely to correspond to functional, but nonannotated, genes.) In each case, the orthology is inferred from gene context, that is, when the neighboring genes are the same in the two compared genomes.
When a sequence match encompasses a previously annotated gene in the subject genome, we first asked if it has been designated as a pseudogene in the genome annotation. Those not already identified as such in current databases are further classified by the procedures described above, which consider E-values, length disparities, and the presence of premature stop codons. First, we cull all matches that contain an internal stop codon and determine the type of inactivating mutation (point mutation or frameshift). (In a few cases in which the target CDSs were of the same length [±3 bp] as the query but contained an internal stop codon, the genes were already labeled as containing a natural frameshift or a selenocysteine codon rather than as pseudogenes in the available annotations.) When the target is <80% of the length of the protein query and has an E-value <10e–15, but has no internal stop codon, we evaluate the BLAST match lengths for chance similarities, and we retrieve those whose lengths are 30% to 80% of that of the query for later identification of the event that truncated the CDSs. Even those matching over smaller regions (<30% of the query) could result from CDSs that are split by a large insertion, thus we identify such events by searching for query CDSs that have two matches in proximity (within a few kilobases). Usually such cases included one very short match and another match that covered >80% of the length of the query CDS.
The resulting list of matches, now annotated with respect to their lengths and the occurrence of stop codons relative to the query CDSs, are then curated to identify likely pseudogenes based on various criteria, including the position of the stop codon, the length of the truncated region, a shift in reading frame, or the presence of a large insertion or deletion. Such identifications of the mutations leading to the inactivation of the pseudogenes are obtained by aligning the nucleotide sequences of putative pseudogenes with their query CDS in CLUSTALW 1.8 (Thompson et al. 1994).
To evaluate the total spectrum of events that generate pseudogenes in each of the subject genomes, the sets of pseudogenes derived from Ψ-Φ were compared and combined with those reported in the original annotations—E. coli MG1655 (Blattner et al. 1997); E. coli EDL933 (Perna et al. 2001); S. flexneri 2a (Jin et al. 2002)—and in subsequent publications (e.g., Homma et al. 2002). (Because Welch et al. [2002] provide no information about the mutations that led to the formation of the E. coli CFT07 pseudogenes, we reconstructed these events by comparing the annotated pseudogenes with their functional homologs in E. coli MG1655. Of the 55 pseudogenes originally reported, it was possible to distinguish the disruptive feature for 46 in this manner; but in nine cases [dubbed “possible pseudogenes”], there were no homologs available for comparison.)
Acknowledgments
The authors thank V. Daubin for comments on earlier versions of this manuscript. This research is supported by grant GM56120 from the National Institutes of Health to H.O.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2925604. Article published online before print in October 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M., and Aksoy, S. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32: 402-407. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25: 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, J.O. and Andersson, S.G. 1999a. Genome degradation is an ongoing process in Rickettsia. Mol. Biol. Evol. 16: 1178-1191. [DOI] [PubMed] [Google Scholar]
- ———. 1999b. Insights into the evolutionary process of genome degradation. Curr. Opin. Genet. Dev. 9: 664-671. [DOI] [PubMed] [Google Scholar]
- Andersson, D.I. and Hughes, D. 1996. Muller's ratchet decreases fitness of a DNA-based microbe. Proc. Natl. Acad. Sci. 93: 906-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, S.G., Zomorodipour, A., Andersson, J.O., Sicheritz-Pontén, T., Alsmark, U.C.M., Podowski, R.M., Näslund, A.K., Eriksson, A.-S., Winkler, H.H., and Kurland, C.G. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396: 133-143. [DOI] [PubMed] [Google Scholar]
- Blattner, F.R., Plunkett III, G., Bloch, C.A., Perna, N.T., Burland, V., Riley, M., Collado-Vides, J., Glasner, J.D., Rode, C.K., Mayhew, G.F., et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453-1462. [DOI] [PubMed] [Google Scholar]
- Cole, S.T., Eiglmeier, K., Parkhill, J., James, K.D., Thomson, N.R., Wheeler, P.R., Honoré, N., Garnier, T., Churcher, C., Harris, D., et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409: 1007-1011. [DOI] [PubMed] [Google Scholar]
- Cooper, V.S. and Lenski, R.E. 2000. The population genetics of ecological specialization in evolving Escherichia coli populations. Nature 407: 736-739. [DOI] [PubMed] [Google Scholar]
- Dale, C., Wang, B., Moran, N.A., and Ochman, H. 2003. Loss of DNA recombinational repair enzymes in the initial stages of genome degeneration. Mol. Biol. Evol. 20: 1188-1194. [DOI] [PubMed] [Google Scholar]
- Delcher, A.L., Harmon, D., Kasif, S., White, O., and Salzberg, S.L. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 27: 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delorme, C., Godon, J.J., Ehrlich, S.D., and Renault, P. 1993. Gene inactivation in Lactococcus lactis: Histidine biosynthesis. J. Bacteriol. 175: 4391-4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, W., Burland, V., Plunkett III, G., Boutin, A., Mayhew, G.F., Liss, P., Perna, N.T., Rose, D.J., Mau, B., Zhou, S., et al. 2002. Genome sequence of Yersinia pestis KIM. J. Bacteriol. 184: 4601-4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan, B.K. and Miller, J.H. 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287: 560-561. [DOI] [PubMed] [Google Scholar]
- Gil, R., Silva, F.J., Zientz, E., Delmotte, F., Gonzalez-Candelas, F., Latorre, A., Rausell, C., Kamerbeek, J., Gadau, J., Holldobler, B., et al. 2003. The genome sequence of Blochmannia floridanus: Comparative analysis of reduced genomes. Proc. Natl. Acad. Sci. 100: 9388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, A.J., Burns, P.A., Fix, D.F., Yatagai, F., Allen, F.L., Horsfall, M.J., Halliday, J.A., Gray, J., Bernelot-Moens, C., and Glickman, B.W. 1988. Missense mutation in the lacI gene of Escherichia coli. Inferences on the structure of the repressor protein. J. Mol. Biol. 200: 239-251. [DOI] [PubMed] [Google Scholar]
- Homma, K., Fukuchi, S., Kawabata, T., Ota, M., and Nishikawa, K. 2002. A systematic investigation identifies a significant number of probable pseudogenes in the Escherichia coli genome. Gene 294: 25-33. [DOI] [PubMed] [Google Scholar]
- Jacq, C., Miller, J.R., and Brownlee, G.G. 1977. A pseudogene structure in 5S DNA of Xenopus laevis. Cell 12: 109-120. [DOI] [PubMed] [Google Scholar]
- Jain, R., Rivera, M.C., and Lake, J.A. 1999. Horizontal gene transfer among genomes: The complexity hypothesis. Proc. Natl. Acad. Sci. 96: 3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, Q., Yuan, Z., Xu, J., Wand, Y., Shen, Y., Lu, W., Wang, J., Liu, H., Yang, F., Zhang, X., et al. 2002. Genome sequence of Shigella flexneri 2a: Insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30: 4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleina, L.G. and Miller, J.H. 1990. Genetic studies of the lac repressor. XIII. Extensive amino acid replacements generated by the use of natural and synthetic nonsense suppressors. J. Mol. Biol. 212: 295-318. [DOI] [PubMed] [Google Scholar]
- Kogoma, T. and von Meyenburg, K. 1983. The origin of replication, oriC, and the dnaA protein are dispensable in stable DNA replication (sdrA) mutants of Escherichia coli K-12. EMBO J. 2: 463-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C.Y., Baumann, P., and Moran, N. 1996. The endosymbiont (Buchnera sp.) of the aphid Diuraphis noxia contains plasmids consisting of trpEG and tandem repeats of trpEG pseudogenes. Appl. Environ. Microbiol. 62: 332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence, J.G., Hendrix, R.W., and Casjens, S. 2001. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 9: 535-540. [DOI] [PubMed] [Google Scholar]
- McClelland, M., Sanderson, K.E., Spieth, J., Clifton, S.W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., et al. 2001. Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature 413: 852-856. [DOI] [PubMed] [Google Scholar]
- Mira, A., Ochman, H., and Moran, N.A. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17: 589-596. [DOI] [PubMed] [Google Scholar]
- Moran, N.A. 2002. Microbial minimalism: Genome reduction in bacterial pathogens. Cell 108: 583-586. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Dougan, G., James, K.D., Thomson, N.R., Pickard, D., Wain, J., Churcher, C., Mungall, K.L., Bentley, S.D., Holden, M.T., et al. 2001a. Complete genome sequence of a multiple drug resistant Salmonella enterica serovar Typhi CT18. Nature 25: 848-852. [DOI] [PubMed] [Google Scholar]
- Parkhill, J., Wren, B.W., Thomson, N.R., Titball, R.W., Holden, M.T., Prentice, M.B., Sebaihia, M., James, K.D., Churcher, C., Mungall, K.L., et al. 2001b. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413: 523-527. [DOI] [PubMed] [Google Scholar]
- Perna, N.T., Plunkett III, G., Burland, V., Mau, B., Glasner, J.D., Rose, D.J., Mayhew, G.F., Evans, P.S., Gregor, J., et al. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409: 529-533. [DOI] [PubMed] [Google Scholar]
- Porter, R.D. 1983. Specialized transduction with λ plac5: Involvement of the RecE and RecF recombination pathways. Genetics 105: 247-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuven, N.B., Koonin, E.V., Rudd, K.E., and Deutscher, M.P. 1995. The gene for the longest known Escherichia coli protein is a member of helicase superfamily II. J. Bacteriol. 177: 5393-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogozin, I.B., Makarova, K.S., Natale, D.A., Spiridonov, A.N., Tatusov, R.L., Wolf, Y.I., Yin, J., and Koonin, E.V. 2002. Congruent evolution of different classes of non-coding DNA in prokaryotic genomes. Nucleic Acids Res. 30: 4264-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzberg, S.L., Delcher, A.L., Kasif, S., and White, O. 1998. Microbial gene identification using interpolated Markov models. Nucleic Acids Res. 26: 544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y., and Ishikawa, H. 2000. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407: 81-86. [DOI] [PubMed] [Google Scholar]
- Simonet, M., Riot, B., Fortineau, N., and Berche, P. 1996. Invasin production by Yersinia pestis is abolished by insertion of an IS200-like element within the inv gene. Infect. Immun. 64: 375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.L. 1994. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22: 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanin, E.F., Goldberg, G.I., Tucker, P.W., and Smithies, O. 1980. A mouse α-globin-related pseudogene lacking intervening sequences. Nature 286: 222-226. [DOI] [PubMed] [Google Scholar]
- Wei, J., Goldberg, M.B., Burland, V., Venkatesan, M.M., Deng, W., Fournier, G., Mayhew, G.F., Plunkett III, G., Rose, D.J., Darling, A., et al. 2003. Complete genome sequence and comparative genomics of Shigella flexneri serotype 2a strain 2457T. Infect. Immun. 71: 2775-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch, R.A., Burland, V., Plunkett III, G., Redfor, P., Roesch, P., Rasko, D., Buckles, E.L., Liou, S.-R., Boutin, A., Hackett, J., et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. 99: 17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, P., Morelli, G., and Achtman, M. 1999. The opcA and (psi)opcB regions in Neisseria: Genes, pseudogenes, deletions, insertion elements and DNA islands. Mol. Microbiol. 33: 635-650. [DOI] [PubMed] [Google Scholar]