Abstract
Medulloblastoma is the most common malignant brain tumor in childhood, and development of targeted therapies is highly desired. Although the molecular mechanisms of malignant transformation are not fully understood, it is known that medulloblastomas may arise from cerebellar granule neuron precursors. The homeodomain transcription factor Barhl1 is known to regulate migration and survival of granule cell precursors, but its functional role in medulloblastoma is unknown. We show here that the expression of BARHL1 is significantly upregulated during human cerebellar development and in human medulloblastoma samples as compared with the normal adult cerebellum. We also detected high levels of Barhl1 expression in medulloblastomas of Math1-cre:SmoM2 mice, a mouse model for Sonic hedgehog-associated medulloblastomas that we developed previously. To investigate Barhl1 function in vivo during tumor development, we generated Barhl1−/−Math1-cre:SmoM2 mice. Interestingly, tumors that developed in these mice displayed increased mitotic activity and decreased neuronal differentiation. Moreover, survival of these mice was significantly decreased. Similarly, low expression of BARHL1 in human medulloblastoma cases was associated with a less favorable prognosis for patients. These results suggest that the expression of Barhl1 decelerates tumor growth both in human and in murine medulloblastomas and should be further investigated with respect to potential implications for individualized therapeutic strategies.
Keywords: cerebellum, medulloblastoma, Barhl1, survival, Sonic hedgehog, tumor
Introduction
Medulloblastoma is the most frequent malignant brain tumor in childhood. Although centralized and evidence-based treatment protocols have significantly improved the outcome of patients, still up to 30% of children succumb to their disease within 5 years (Gilbertson, 2004). Major issues that challenge patients and physicians are the strong tendency of this cancer to metastasize along the cerebrospinal axis together with the fact that specific treatment options based on a tumor’s molecular background are still lacking. In this context, it was essential to find that many familial and sporadic medulloblastomas are caused by a constitutive activation of either the Sonic hedgehog (Shh) or the Wnt/β-catenin signaling pathway, and researchers are now trying to establish well-tolerated drugs that may inhibit these pathways (Crawford et al., 2007; Scales and de Sauvage, 2009). Apart from these two well-defined groups of medulloblastomas, more recent studies identified additional groups that are distinguishable by array-based expression analysis (Kool et al., 2008; Northcott et al., 2011). Nevertheless, the diagnosis of medulloblastomas is still established according to the WHO (World Health Organization) criteria for the classification of central nervous system tumors, and these criteria are solely based on standard histology and immunohistochemistry (Louis, 2007). Interestingly, the desmoplastic and the rare extensively nodular variant of medulloblastomas, which typically show tumor nodules surrounded by very cell-dense areas containing reticulin fibers, are largely overlapping with tumors that are molecularly characterized by constitutive Shh signaling. All other molecular subgroups of medulloblastomas are mainly covered by histologically classic medulloblastomas (Kool et al., 2008; Northcott et al., 2011). Medulloblastomas of all groups display a neuronal protein expression signature and they are believed to arise from neuronal precursor cells. It is still unclear whether different subtypes of medulloblastomas may arise from different precursor populations, and indeed, recent advances have now proven in vivo that brainstem precursors may give rise to Wnt-associated medulloblastomas (Gibson et al., 2010), whereas granule neuron precursors are susceptible to develop Shh-associated medulloblastomas (Schüller et al., 2008). The mechanisms that control proliferation, differentiation and migration of these precursors have therefore been of particular interest to cancer researches, and the understanding of these mechanisms is considered crucial for the development of novel treatment strategies.
One of the transcription factors that regulate both the survival and the migration of cerebellar granule neuron precursors is the homeobox gene Barhl1 (Li et al., 2004). Barhl1 is a mammalian homolog of the Drosophila BarH1 and BarH2 genes, which encode homeodomain proteins that are primarily expressed in migrating neurons settling in specific domains within the diencephalon, rhombencephalon and spinal cord (Bulfone et al., 2000). Loss of Barhl1 function causes attenuated cerebellar foliation, defective radial migration and impaired survival of cerebellar granule neurons (Li et al., 2004). Interestingly, BARHL1 was also identified to be expressed in medulloblastomas (Yokota et al., 2004), but its functional significance for tumor development and progression has not yet been investigated.
We show here that BARHL1 is heterogeneously expressed in human medulloblastomas. High levels of BARHL1 expression are significantly correlated with longer survival in a series of 44 medulloblastoma patients. Similarly, deletion of Barhl1 in a mouse model of medulloblastoma results in decreased differentiation of tumor cells and in a significantly shorter survival of the mice. Therefore, our data suggest that Barhl1 may have tumor-suppressive roles in medulloblastoma.
Results
BARHL1 is heterogeneously expressed in human medulloblastomas
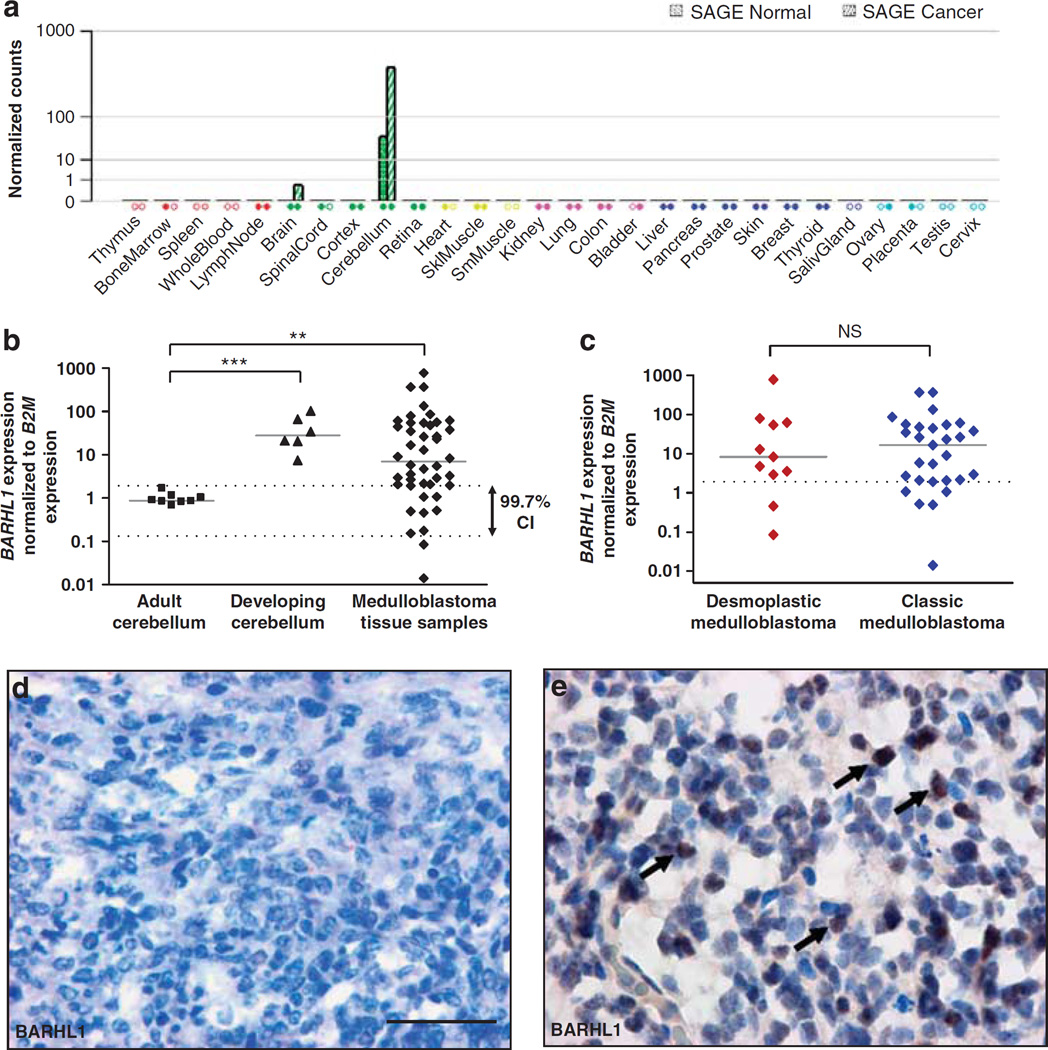
Barhl1 was found to have essential roles for cerebellar granule neuron precursors and for the overall development of the cerebellum (Li et al., 2004) Medulloblastomas derive, at least in part, from these precursor cells, and we therefore decided to investigate whether human medulloblastomas express BARHL1 and how this would functionally be involved in tumorigenesis. In a first step, we looked at BARHL1 expression in a series of 19 normal tissues and 17 different types of cancer. Serial analysis of gene expression (SAGE) analysis revealed that BARHL1 was highly expressed in the cerebellum and even stronger in the medulloblastoma, but only weakly in other types of brain cancer. Several types of normal tissues and tumors outside the central nervous system did not show any significant expression of BARHL1 (Figure 1a). Next, we used real-time reverse transcriptase (RT)–PCR to confirm and extend the expression pattern of BARHL1 in the human cerebellum and in the medulloblastoma. As demonstrated in Figure 1b, the expression of BARHL1 was ~30-fold higher in the developing cerebellum (n = 6) as compared with samples of the adult cerebellum (n = 8, P<0.001). Surgically removed medulloblastoma tissue samples (n = 44) were characterized by a more heterogeneous expression pattern with many cases showing strong BARHL1 expression (P = 0.003, Figure 1b). The median was 7.05 (range, 0.01–778.79) as compared with the median of adult cerebellum samples which was set at 1 (Figure 1b, Table 1). Patient’s age and levels of BARHL1 expression did not show any significant correlation (r = 0.127, P = 0.411, Supplementary Figure S1). Our series included 29 classic medulloblastomas and 11 desmoplastic medulloblastomas, but we did not detect any significant differences with regard to the expression of BARHL1 in these two histological subtypes (P = 0.952, Figure 1c). To confirm our RNA expression data, we performed immunohistochemistry on snap-frozen tissues expressing high or low levels of BARHL1. Medulloblastoma samples that were shown to highly express BARHL1 by real-time RT–PCR and that were attributed to the high BARHL1 group (see Figure 5) exhibited a strong nuclear staining for the transcription factor BARHL1, whereas medulloblastoma samples belonging to the low BARHL1 group (see Figure 5) did not or barely stained for BARHL1 (Figures 1d and e). Finally, quantitative real-time RT–PCR on genomic DNA from medulloblastoma samples (n = 32) was used to exclude the fact that genomic amplifications were the reason for BARHL1 overexpression on the mRNA level (Supplementary Figure S2).
Figure 1.
BARHL1 expression in normal human tissues and tumor samples. (a) Expression of BARHL1 in different tissues indicating specificity of high expression in cerebellar tissue and medulloblastoma. Serial analysis of gene expression data for BARHL1 (SAGE tag: AGCCCGTGAC) is based on data extracted from the Cancer Genome Anatomy project (CGAP) and from the Genomics institute of the Novartis Research Foundation (GNF) SymAtlas and was obtained from http://www.genecards.org/index.shtml. Filled diamonds = insignificant expression after thresholding and normalization. Empty diamonds = no data. (b) Real-time RT–PCR of the adult cerebella (n = 8), developing cerebella (n = 6) and human medulloblastoma tissue samples (n = 44). BARHL1 expression was normalized to Beta-2-microglobulin (B2M) expression. BARHL1 expression was significantly upregulated in the developing cerebella and in medulloblastomas as compared with the adult cerebella (P<0.001 and P = 0.003, respectively). Mean of expression in the adult cerebella was set at 1; dotted lines show mean ± 3*s.d. of expression in the adult cerebella; solid lines represent median of each group. Two asterisks: P≤0.01, three asterisks: P≤0.001. (c) Real-time RT–PCR analysis of BARHL1 expression in desmoplastic (n = 11) and classic (n = 29) medulloblastomas. BARHL1 overexpression (defined as >mean expression of normal adult cerebellum + 3*s.d., see dotted line) was found in both histological subtypes without statistically significant differences (P = 0.952). Solid lines: median in each group. CI: confidence interval, NS: not significant (d, e) Immunohistochemistry for BARHL1. (Panel d) Absence of staining for BARHL1 in medulloblastoma samples exhibiting low levels of BARHL1 mRNA. (Panel e) Arrows point at strong nuclear staining of medulloblastoma cells in medulloblastoma samples that were shown to highly express BARHL1.
Table 1.
Summary of tumor pathology and clinical data in examined medulloblastoma cases
| Sample number |
MB subtype | Sex | Age (years) |
Follow- up (months) |
State | Relative BARHL1 mRNA expression |
|---|---|---|---|---|---|---|
| 1 | Classic | F | 0.6 | 12.7 | DOD | 37.87 |
| 2 | Classic | M | 2.4 | 21.0 | DOD | 23.13 |
| 3 | Classic | F | 3.6 | 21.1 | DOD | 5.48 |
| 4 | Classic | F | 3.7 | 5.4 | NED | 133.63 |
| 5 | Classic | F | 3.8 | 47.1 | NED | 9.02 |
| 6 | Classic | M | 4.1 | 35.4 | NED | 0.01 |
| 7 | Classic | M | 4.9 | 75.7 | DOD | 16.69 |
| 8 | Classic | M | 5.4 | 135.6 | NED | 47.93 |
| 9 | Classic | F | 5.5 | 28.3 | DOD | 26.14 |
| 10 | Classic | M | 5.5 | 137.5 | NED | 60.78 |
| 11 | Classic | M | 5.9 | 1.1 | NED | 44.54 |
| 12 | Classic | F | 6.1 | 145.2 | NED | 1.92 |
| 13 | Classic | M | 6.2 | 22.2 | DOD | 2.96 |
| 14 | Classic | M | 6.7 | 33.4 | NED | 366.14 |
| 15 | Classic | M | 7.0 | 113.2 | NED | 2.07 |
| 16 | Classic | M | 7.0 | 142.8 | NED | 2.08 |
| 17 | Classic | M | 7.9 | 24.1 | DOD | 0.52 |
| 18 | Classic | M | 8.5 | 36.5 | DOD | 26.54 |
| 19 | Classic | M | 8.8 | 8.8 | NED | 367.04 |
| 20 | Classic | M | 9.3 | 50.3 | NED | 56.40 |
| 21 | Classic | M | 9.9 | 79 | NED | 0.49 |
| 22 | Classic | M | 10.6 | 23.7 | NED | 1.07 |
| 23 | Classic | M | 16.5 | 18.3 | NED | 1.07 |
| 24 | Classic | M | 22.3 | 35.3 | NED | 2.68 |
| 25 | Classic | F | 23.6 | 80.8 | NED | 54.73 |
| 26 | Classic | F | 25.5 | 133.7 | NED | 86.59 |
| 27 | Classic | F | 29.3 | 19.5 | NED | 5.84 |
| 28 | Classic | M | 37.5 | 4.8 | DOD | 35.06 |
| 29 | Classic | M | 39.7 | 24.4 | DOD | 2.15 |
| 30 | Desmoplastic | F | 1.6 | 66.9 | NED | 2.92 |
| 31 | Desmoplastic | F | 2.2 | 128.0 | NED | 62.27 |
| 32 | Desmoplastic | M | 2.5 | 7.0 | DOD | 0.08 |
| 33 | Desmoplastic | M | 7.5 | 14.2 | DOD | 3.56 |
| 34 | Desmoplastic | M | 10.0 | 101.4 | NED | 78.99 |
| 35 | Desmoplastic | F | 14.3 | 62.7 | DOD | 778.79 |
| 36 | Desmoplastic | M | 22.8 | 47.3 | NED | 8.26 |
| 37 | Desmoplastic | F | 25.5 | 75.5 | NED | 0.46 |
| 38 | Desmoplastic | F | 26.5 | 46.0 | NED | 54.33 |
| 39 | Desmoplastic | F | 30.5 | 46.4 | NED | 4.72 |
| 40 | Desmoplastic | F | 30.7 | 17.7 | NED | 12.91 |
| 41 | Extensively nodular |
F | 0.7 | 107.3 | NED | 0.15 |
| 42 | Extensively nodular |
F | 2.2 | 175.6 | NED | 3.27 |
| 43 | Extensively nodular |
M | 2.7 | 5.6 | NED | 1.97 |
| 44 | Extensively nodular |
F | 2.9 | 78.4 | NED | 0.18 |
Abbreviations: DOD, death of disease; F, female; M, male; NED, no evidence of disease.
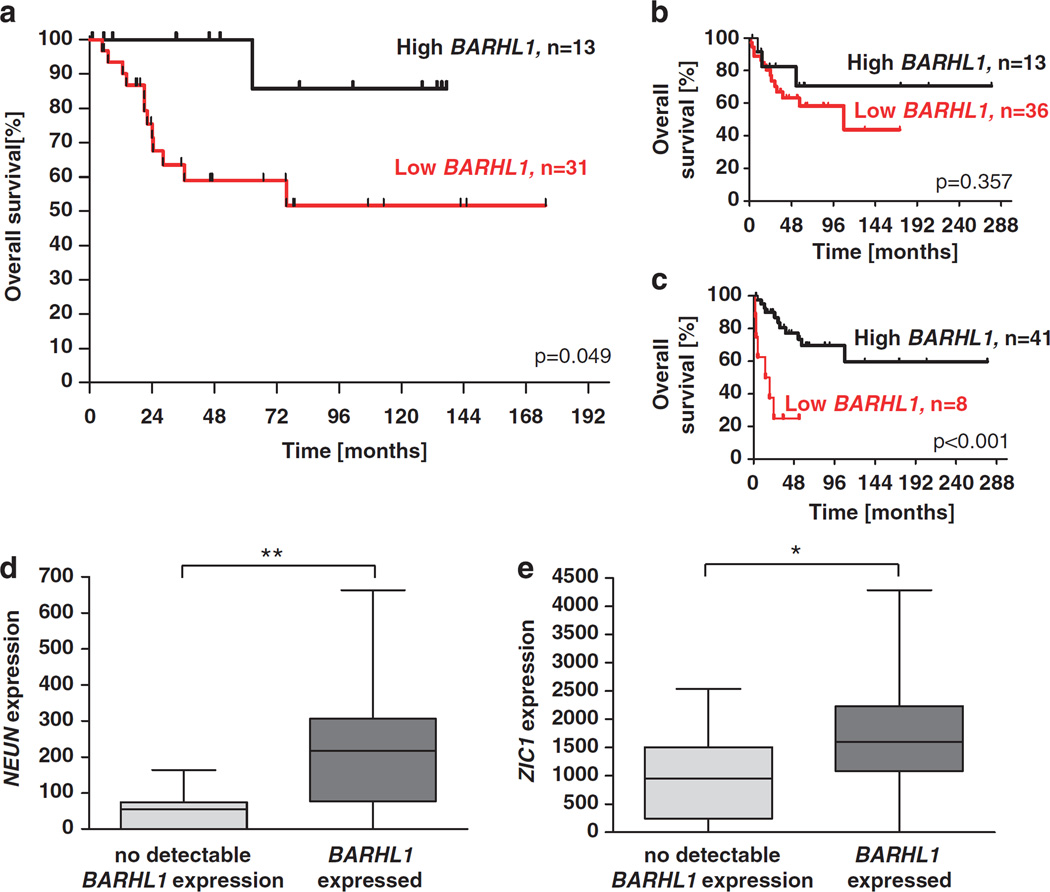
Figure 5.
Characterization of BARHL1 expression in human medulloblastoma. (a) Kaplan–Meier analysis of patients with medulloblastoma that express high or low levels of BARHL1. The 13 patients exhibiting the highest relative BARHL1 expression values (high BARHL1) were compared with the rest of the group (low BARHL1, n = 31). High levels of BARHL1 were significantly correlated with a better prognosis (P = 0.049). (b, c) BARHL1 expression and survival in a different set of medulloblastoma cases (Kool et al., 2008). Applying a similar threshold than in our patient group (expression cutoff at third quartile) shows a trend for favorable survival in the high BARHL1 group (n = 13) when compared with the rest of the group (low BARHL1, n = 36, P = 0.357, panel b). A significantly prolonged overall survival was found in the 41 patients showing high BARHL1 expression levels when compared with the remaining patients (low BARHL1, P<0.001, panel c). (d, e) Expression of differentiation markers with respect to BARHL1 expression in the set of medulloblastoma cases from Kool et al. (2008). Expression levels of NEUN and ZIC1 were significantly increased in the 50 medulloblastoma cases expressing BARHL1 when compared with the 12 medulloblastoma samples with no detectable BARHL1 expression (P = 0.001 and P = 0.021, respectively). One asterisk: P≤0.05, two asterisks: P≤0.01.
Shh-induced mouse medulloblastoma and its cerebellar precursor cells express Barhl1
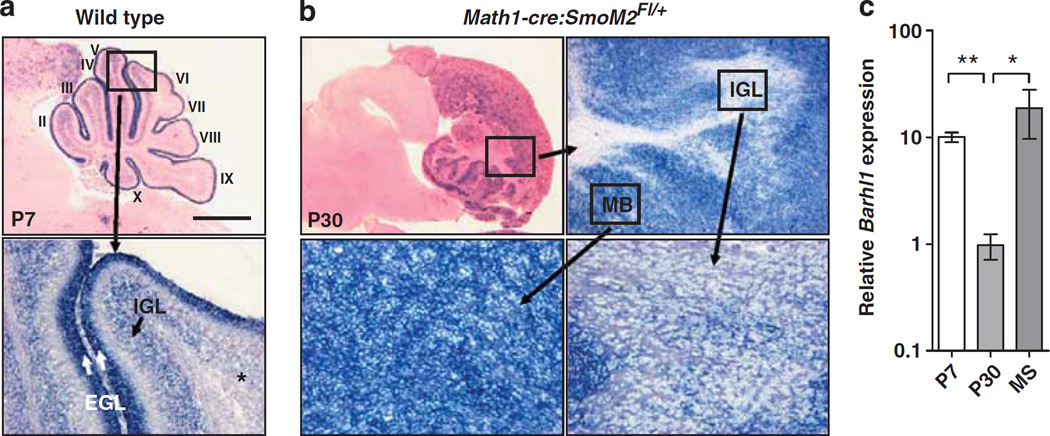
To further investigate the functional role of Barhl1 in tumor development, we used Math1-cre:SmoM2Fl/+ mice as a mouse model for Shh-associated medulloblastomas (Schüller et al., 2008). To characterize Barhl1 expression during development and in Shh-driven mouse medulloblastomas, we performed in situ hybridizations for Barhl1 on WT and Math1-cre:SmoM2Fl/+ mice. At postnatal day 7 (P7), Barhl1 was strongly expressed in cerebellar granule neurons (Figure 2a). Granule neuron precursors were labeled intensely throughout the external granule cell layer of the entire cerebellum, but the Barhl1 expression was discontinuous in mature granule neurons of the internal granule cell layer. Barhl1 expression in the internal granule cell layer was seen in cerebellar lobes II–V and X and to some extent in lobe VI, whereas it was absent in lobes VII, VIII and IX (Figure 2a). In medulloblastomas from Math1-cre:SmoM2Fl/+ mice, Barhl1 expression was strongly present in tumor tissues, and a weaker expression was seen in granule neurons of the internal granule cell layer (Figure 2b). To confirm these findings, we used real-time RT–PCR to quantify Barhl1 expression (Figure 2c). In agreement with the in situ experiments, Barhl1 expression levels were ~10 times higher during cerebellar development (P7) in comparison with the adult cerebellum (P30), and high expression was found in medulloblastomas from Math1-cre:SmoM2Fl/+ mice.
Figure 2.
Barhl1 is strongly expressed in the granule cell lineage and in tumors from Math1-cre:SmoM2Fl/+ mice. (a) Sagittal sections from P7 wild-type mice display strong expression of Barhl1 in cerebellar granule neurons. Although granule neuron precursors are labeled throughout the EGL of the entire cerebellum, Barhl1 expression in mature granule neurons of the IGL is restricted to lobes II–V and X (see high-power image with asterisk indicating Barhl1-negative granule neurons). (b) Sagittal cerebellar sections from Math1-cre:SmoM2Fl/+ mice. Top left: H&E staining of a characteristic medulloblastoma in a Math1-cre:SmoM2Fl/+ mouse. Top right: In situ hybridization for Barhl1 in the cerebellum of a Math1Cre:SmoM2Fl/+ mouse with strong expression in the tumor tissue (lower left) and weak expression in the IGL tissue (lower right, see asterisks). (c) Relative quantification of Barhl1 expression using real-time RT–PCR with results normalized to B2m. Barhl1 expression levels were significantly higher in the cerebella on P7 and in medulloblastomas from Math1-cre:SmoM2Fl/+ (MS) mice as compared with the adult cerebella (P30, P = 0.002 and P = 0.05, respectively). EGL, external granule cell layer; IGL, internal granule cell layer; MB, medulloblastoma. One asterisk: P≤0.05, two asterisks: P≤0.01. Scale bar is 1 mm and 100 µm for panel a, 1.5 mm for the H&E stain, 200 µm for the Barhl1 low power and 50 µm for the Barhl1 high-power images in panel b.
Deletion of Barhl1 in mouse medulloblastoma results in decreased differentiation of tumor cells and shortened survival
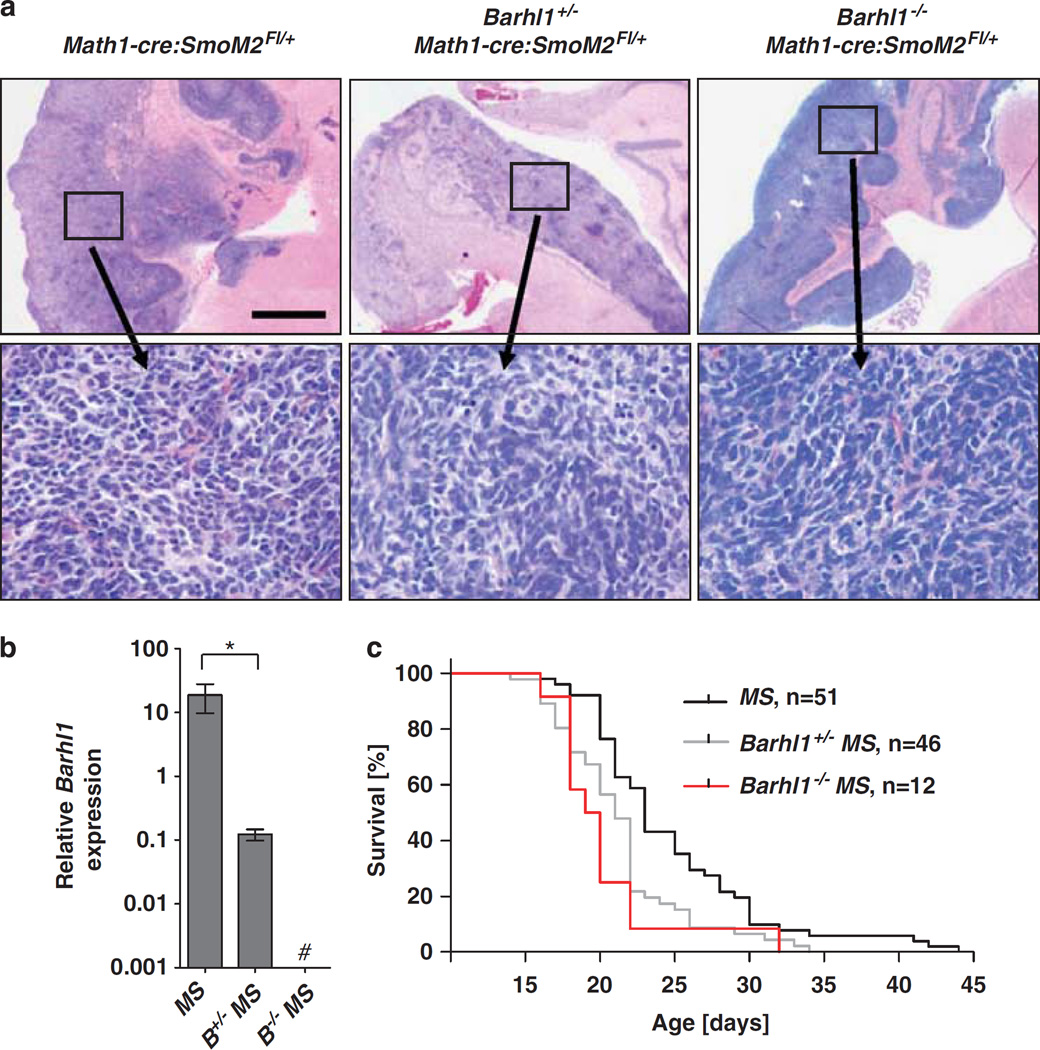
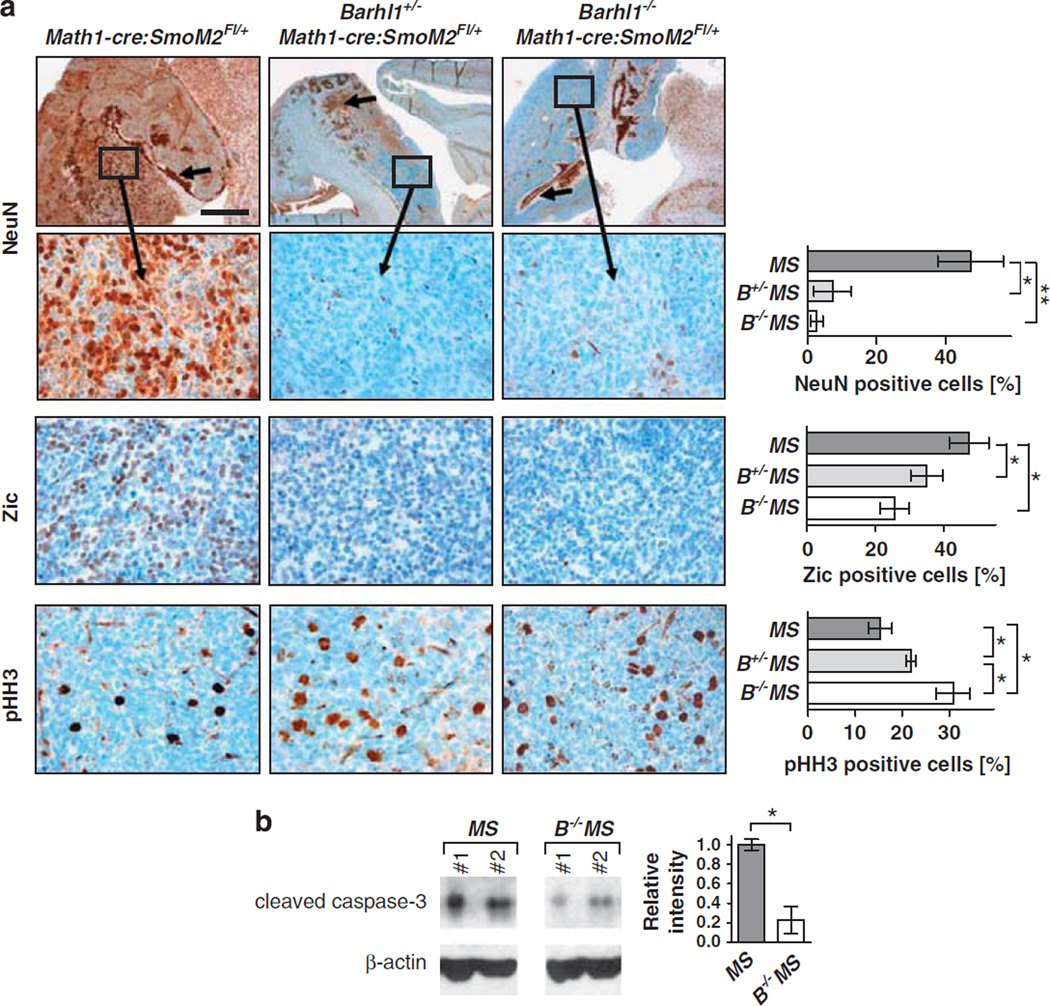
To investigate the functional role of Barhl1 for the development and progression of medulloblastomas, we generated Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/− Math1-cre:SmoM2Fl/+ mice. Similar to the previously described Math1-cre:SmoM2Fl/+ mice, all generated Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice developed medulloblastomas. As seen by hematoxylin and eosin (H&E) staining, all three genotypes displayed a similar morphology with tumors composed of small round blue tumor cells in high density (Figure 3a). Although Barhl1 expression was strongly present in Math1-cre:SmoM2Fl/+ mice, Barhl1 levels were significantly decreased in Barhl1+/−Math1-cre:SmoM2Fl/+ mice (P = 0.029) and were not detectable in Barhl1−/−Math1-cre:SmoM2Fl/+ mice (Figure 3b). To elucidate whether Barhl1 expression in medulloblastomas has an impact on the survival of mice, we established Kaplan–Meier curves of Math1-cre:SmoM2Fl/+ mice (n = 51), Barhl1+/−Math1-cre:SmoM2Fl/+ mice (n = 46) and Barhl1−/−Math1-cre:SmoM2Fl/+ mice (n = 12, Figure 3c). Deletion of Barhl1 resulted in a significantly decreased survival of Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/− Math1-cre:SmoM2Fl/+ mice than in Math1-cre:SmoM2Fl/+ mice (P = 0.003 and P = 0.002, respectively). It is noteworthy that influences of the genetic background on the survival of different groups were excluded by crossing the mice in a manner that all genotypes were produced in one litter. To find out why Barhl1 deletion resulted in a worse prognosis, we further analyzed Math1-cre:SmoM2Fl/+, Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice by immunohistochemistry. As expected, Cre recombinase expression was strong in all three genotypes with a large majority of tumor cells being stained (Supplementary Figure S3). NeuN, a marker for neuronal differentiation, is expressed not only in mature granule neurons of the internal granule cell layer (Weyer and Schilling, 2003) but also in medulloblastomas (Eberhart et al., 2001). Interestingly, Math1-cre:SmoM2Fl/+-derived medulloblastoma cells exhibited a strong expression of NeuN, but significantly fewer NeuN-positive cells were detectable in tumors from Barhl1+/−Math1-cre:SmoM2Fl/+ (P = 0.004) and Barhl1−/−Math1-cre:SmoM2Fl/+ mice (P = 0.012, Figure 4a). Similarly, expression of Zic, a marker for granule neuron differentiation and medulloblastoma (Yokota et al., 1996) was significantly reduced in Barhl1+/− Math1-cre:SmoM2Fl/+ (P = 0.034) and Barhl1−/−Math1-cre:SmoM2Fl/+ mice (P = 0.016, Figure 4a). Although Olig2 and Sox2, both of which label multipotent precursors (Schüller et al., 2008; Sutter et al., 2010), were found to be similarly expressed in all three genotypes (Supplementary Figure S3), Synaptophysin and MAP2, as additional markers for neuronal differentiation, appeared to be more weakly expressed in Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice than in Math1-cre:SmoM2Fl/+ mice (Supplementary Figure S3). In contrast, the percentage of phosphorylated histone H3-positive tumor cells was significantly increased in Barhl1−/−Math1-cre:SmoM2Fl/+ mice as compared to Barhl1+/−Math1-cre:SmoM2Fl/+ mice (P = 0.019) and Math1-cre:SmoM2Fl/+ mice (P = 0.012). Similarly, Barhl1+/−Math1-cre:SmoM2Fl/+ mice showed a higher fraction of phosphorylated histone H3-positive tumor cells than Math1-cre:SmoM2Fl/+ mice (P = 0.045). Furthermore, western blot analysis showed that protein levels of cleaved caspase-3, a marker for apoptosis, were significantly reduced in Barhl1−/−Math1-cre:SmoM2Fl/+ mice when compared to Math1-cre:SmoM2Fl/+ mice (P = 0.036, Figure 4b), and this may indicate some pro-apoptotic effect in medulloblastoma. Hence, medulloblastomas from Barhl1+/− Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice were characterized by a significantly decreased differentiation of tumor cells.
Figure 3.
Deletion of Barhl1 in Math1-cre:SmoM2Fl/+ mice. (a) H&E staining of sagittal sections from medulloblastoma showed a similar morphology in Math1-cre:SmoM2Fl/+, Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice. High magnifications revealed small round blue tumor cells in high density. (b) Barhl1 expression was analyzed using real-time RT–PCR. Target gene level was normalized to B2m mRNA. Barhl1 levels were significantly decreased in Barhl1+/−Math1-cre:SmoM2Fl/+(Barhl1+/−MS) mice than in Math1-cre:SmoM2Fl/+(MS) mice (P = 0.029) and were not detectable in Barhl1−/−Math1-cre:SmoM2Fl/+ (Barhl1−/−MS) mice. Hash indicates no detectable Barhl1 expression. Asterisk: P≤0.05 (c) Survival curves for Math1-cre:SmoM2Fl/+ (MS) mice (n = 51), Barhl1+/−Math1-cre:SmoM2Fl/+ mice (Barhl1+/−MS, n = 46) and Barhl1−/−Math1-cre:SmoM2Fl/+ mice (Barhl1−/−MS n = 12). Deletion of Barhl1 resulted in a significantly decreased survival of Barhl1+/−Math1-cre:SmoM2Fl/+mice and Barhl1−/−Math1-cre:SmoM2Fl/+ mice in comparison with Math1-cre:SmoM2Fl/+ mice (P = 0.003 and P = 0.002, respectively). Scale bar is 1 mm for low-power images and 50 µm for high-power images.
Figure 4.
Immunohistochemical characterization of mouse medulloblastoma. (a) Immunostaining for NeuN, Zic and phosphorylated histone H3 (pHH3). Scale bar is 1 mm for low-power images and 50 µm for high-power images. Immunostaining for NeuN: Arrows point at strongly stained mature granule neurons in the internal granule cell layer (IGL). Strong NeuN expression was also found in tumor cells from Math1-cre:SmoM2Fl/+ mice, but only marginal expression was seen in tumors from Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice. The percentage of NeuN-positive tumor cells was significantly reduced in Barhl1+/−Math1-cre:SmoM2Fl/+(B+/−MS) mice (P = 0.012) and in Barhl1−/−Math1-cre:SmoM2Fl/+ (B−/−MS) mice (P = 0.004) than in Math1-cre:SmoM2Fl/+ (MS) mice. Immunostaining for Zic: The percentage of Zic-positive tumor cells was significantly reduced in B+/−MS mice (P = 0.034) and in B−/−MS mice (P = 0.016) than in MS mice. Immunostaining for pHH3: The percentage of pHH3-positive tumor cells was significantly increased in B−/−MS mice than in B+/−MS mice (P = 0.019) and in MS mice (P = 0.012). B+/−MS mice showed increased pHH3-positive tumor cells when compared with MS mice (P = 0.045). (b) Western blot for cleaved caspase-3. Protein levels of cleaved caspase-3 were significantly reduced in Barhl1−/−Math1-cre:SmoM2Fl/+ mice than in Math1-cre:SmoM2Fl/+ mice (P = 0.036). One asterisk: P≤0.05, two asterisks: P≤0.01.
Expression of BARHL1 in human medulloblastomas correlates with patient survival
Having observed an important impact of Barhl1 on the differentiation and prognosis of murine medulloblastomas, we next aimed to analyze the survival of patients with respect to BARHL1 expression in human medulloblastomas. When separating the patients into two groups by using the median of relative BARHL1 expression in medulloblastomas, we did not see statistically significant differences in survival (data not shown). However, the 13 patients exhibiting the highest relative BARHL1 expression values (high BARHL1) were found to have a significantly better prognosis than the rest of the group (low BARHL1, n = 31, P = 0.049, Figure 5a). In particular, after 5 years of follow-up, none of the high BARHL1 cases had died, whereas the survival rate of patients with low BARHL1 medulloblastomas was only 0.59 (Figure 5a). To validate the prognostic value of BARHL1 expression in a different set of medulloblastoma cases, we re-analyzed the microarray expression data from an Amsterdam series of medulloblastoma patients (Kool et al., 2008) and established Kaplan–Meier curves with respect to BARHL1 expression. To apply a similar threshold for BARHL1 expression, we first used an expression cutoff at third quartile to divide the Amsterdam series into high and low BARHL1 cases. However, the 13 high BARHL1 cases only tended to have a better survival than the low BARHL1 cases, but statistical significance was not reached (n = 36, P = 0.357, Figure 5b). Nevertheless, a significantly prolonged overall survival was found after splitting the series into 41 patients with high BARHL1 expression and 8 patients with low BARHL1 expression (P<0.001, Figure 5c). Therefore, we conclude that analysis of the Amsterdam series did not confirm the results of our series and that Barhl1 does not seem to be suitable as a prognostic indicator for medulloblastoma patients.
To see whether BARHL1 also correlated with differentiation markers in human medulloblastomas, we again re-analyzed the microarray expression data of 62 medulloblastoma samples from the Amsterdam series. Expression levels of NEUN and ZIC1 were significantly increased in the 50 medulloblastoma cases expressing BARHL1 when compared with the 12 medulloblastoma samples with no detectable BARHL1 expression (P = 0.001 and P = 0.021, respectively, see Figures 5d and e). Moreover, MAP2 expression levels were found to be significantly correlated with BARHL1 expression levels (P<0.001, data not shown). These findings suggest that the effects of BARHL1 were similar in mouse and human medulloblastomas, with loss or reduced BARHL1 expression being associated with decreased differentiation of tumor cells.
Discussion
Barhl1 is known to play a key role in cerebellar development having influence on the migration and survival of granule neuron precursors (Li et al., 2004). We observed that Barhl1 expression was maintained in mature granule neurons of several anterior lobes of the cerebellum and in lobe X (Figure 2a). Although the expression of Barhl1 in mature granule neurons per se was confirmed by lacZ expression in Barhl1−/− mice (Li et al., 2004), the regional specificity of Barhl1 expression in the cerebellum has not yet been functionally analyzed. Interestingly, this expression pattern correlates well with the expression of Gli1 and En1 transcription factors, both of which are known to have crucial functions during cerebellar development (Millen et al., 1995; Corrales et al., 2004). Although it is tempting to speculate on the functional role of Barhl1 itself with respect to anteroposterior pattering, it seems possible that Barhl1 functions in cooperation with the abovementioned genes.
We further report that Barhl1 is expressed in medulloblastomas arising in Math1-cre:SmoM2Fl/+ mice and in the majority of human medulloblastoma samples. Interestingly, loss of Barhl1 in Math1-cre:SmoM2Fl/+derived medulloblastomas resulted in a downregulation of neuronal differentiation markers, whereas the mitotic activity of these cells was increased (Figure 4). Hence, apart from its role during normal cerebellar development, Barhl1 might have important roles for cell differentiation in medulloblastomas. Moreover, deletion of Barhl1 in Math1-cre:SmoM2Fl/+ mice led to a significantly decreased survival of these mice. Similarly, patients with low BARHL1 expression had a significantly worse prognosis and died earlier. The observation that a loss of Barhl1 causes migration deficits in neurons of Barhl1 null mice (Li et al., 2004), extenuates the possibility that unfavorable survival of patients with medulloblastomas with low levels of Barhl1 expression might be due to enhanced migration or infiltration of medulloblastoma cells. Therefore, we suggest that the underlying mechanisms leading to a worse prognosis of patients and mice carrying tumors with low or no BARHL1 expression are mainly decreased differentiation and increased proliferation of medulloblastoma cells.
A recent study found medulloblastomas to comprise five distinct molecular variants (Kool et al., 2008). Besides SHH-associated tumors, the WNT group and three other, not yet well-characterized groups have been described. Although our mouse model highlights the role of Barhl1 in SHH-associated medulloblastomas, the functional impact of Barhl1 on tumors belonging to other molecular subgroups seems hard to investigate, mainly because of the lack of appropriate mouse models. Interestingly enough, of the 13 medulloblastoma samples exhibiting the highest BARHL1 expression values in our own series, 4 showed a desmoplastic histology, which is often associated with SHH-associated medulloblastomas and 1 case carried a CTNNB1 mutation, a criterion for the Wnt-associated variant (Kool et al., 2008). However, both desmoplastic histology and CTNNB1 mutations were also found in tumors with low BARHL1 expression. In line with this, the expression of BARHL1 was similar in desmoplastic and in classic medulloblastomas (Figure 1c), suggesting an important role of Barhl1 for several, if not all medulloblastoma subtypes and indicating that the mechanisms revealed by Barhl1 loss in Math1-cre:SmoM2Fl/+ mice might be applied to all medulloblastoma subtypes.
Barhl1 belongs to a family of homeodomain transcription factors, which also includes the homolog Barhl2. Although these proteins are known to display distinct expression patterns in the nervous system, yet overlapping expression was found in granule neuron precursors of the cerebellum (Bulfone et al., 2000; Li et al., 2002; Mo et al., 2004). Moreover, Barhl1 and Barhl2 can transactivate the Barhl1 promoter (Chellappa et al., 2008). This could not only point towards a close interaction between Barhl1 and Barhl2 in granule neuron precursors but also to some compensating mechanisms in a Barhl1-null situation. However, we did not find any upregulation of Barhl2 expression in medulloblastomas from Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice. Neither was Barhl2 upregulated in Barhl1−/− and Barhl1+/− mice (data not shown). Therefore, the function of Barhl2 in medulloblastoma pathology and normal granule cell development seems to be distinct from Barhl1 functions and effects of Barhl1 loss were neither mediated nor extenuated by Barhl2.
We found a statistically significant difference in survival with respect to BARHL1 expression in our group of 44 patients. However, re-analysis of the microarray expression data from the Amsterdam series of medulloblastoma patients (Kool et al., 2008) did not confirm these results when using the same cutoff for BARHL1 expression as an prognostic indicator. This reinforces the fact that molecular markers relying on percentile rankings are not suitable for human clinical trials. Although BARHL1 seems biologically important in medulloblastomas, its prognostic value still has to be defined, ideally prospectively in a large homogenously treated cohort of patients.
Similar to Barhl1, several other genes are known to be associated with a favorable prognosis in medulloblastoma (Pomeroy et al., 2002) Among them, TRKC, the receptor for Neurotrophin-3 (NT-3), is well described (Segal et al., 1994). Interestingly, its ligand, Nt-3, was suggested to be a major downstream gene of Barhl1 during cerebellar development (Li et al., 2004). Further understanding of the molecular interactions of BARHL1 and its partners will be promising to find specific therapeutic strategies for medulloblastoma treatment in the future.
Materials and methods
Patients and tumor samples
A total of 44 surgical tumor samples from patients with medulloblastoma were analyzed. Patients included 25 males and 19 females. They were treated in the University Hospitals of Munich, Göttingen, Bremen, Hannover and Münster (all Germany). The majority of patients were enrolled in the German Society of Pediatric Hematology and Oncology multicenter treatment studies for pediatric malignant brain tumors (HIT). The average age was 11.0 years, and the median age was 7.0 years, ranging from 0.6 to 39.7 years. The follow-up in survivors ranged from 1.1 to 145.2 months with an average of 55.8 months and a median of 46.0 months. In all, 13 patients succumbed to their disease, and 31 were alive as of 1 September 2010. The study included 29 medulloblastomas of classic histology, 11 medulloblastomas of desmoplastic histology and 4 medulloblastomas with extensive nodularity. An overview on clinical data and histology is given in Table 1. Tumor diagnosis was established by standard light-microscopic evaluation of H&E sections and silver stains. Diagnoses were made independently by at least two neuropathologists based on the criteria of the latest WHO brain tumor classification (Louis, 2007). For gene copy number analysis, tumor tissues obtained from six glioblastoma multiforme were included as controls. Autopsy material of normal cerebella was obtained from patients, who gave their informed consent. Patient age ranged between 18 and 67 years for the adult cerebella and between 22 weeks of gestation and 6 months for the developing cerebella. None of these patients died from a central nervous system disorder or showed neuropathological abnormalities on histological examination. All tissue samples were either immediately fixed in formaldehyde and embedded in paraffin or snap frozen and stored at −80 °C until use.
Transgenic mice
SmoM2Fl/Fl (Gt(ROSA)26Sortm1(Smo/EYFP)Amc/J (Mao et al., 2006)) mice were obtained from Jackson Laboratory (Bar Harbor, ME, USA). Generation and characterization of Barhl1−/− and Math1-cre mice have been described previously (Li et al., 2002; Schüller et al., 2007). Genotyping was performed by PCR analysis using genomic DNA from mouse tail biopsies. Primers for Cre, SmoM2 and wild-type Barhl1 have been previously published (Li et al., 2002; Jeong et al., 2004; Heine and Rowitch, 2009). Barhl1 knockout was detected with the following primers: Barhl1 forward, 5′-TCCCATTCCTCCTGACACTC-3′; LacZ reverse, 5′-GACAGTATCGGCCT-CAGGAA-3′. Tumor-prone mice were monitored twice daily for signs of neurological symptoms or general weakness. All mouse procedures were performed according to protocols approved by the local government.
DNA and RNA extraction, real-time PCR
For DNA and RNA extraction, paraffin-embedded tissues containing at least 90% tumor material, as confirmed by H&E sections, were used. For snap-frozen tissues, instantaneous sections were performed to assure a tumor fraction of at least 90%. DNA for real-time PCR analysis was extracted from formalin-fixed paraffin-embedded tissues using the QIAamp DNA FFPE Tissue Kit (Qiagen, Hilden, Germany). Total RNA was extracted from snap-frozen tissues using TRIzol reagent (Invitrogen, Darmstadt, Germany). Total RNA from paraffin-embedded tissues was obtained using the RNeasy FFPE Kit (Qiagen). Total RNA was treated with RNase-Free DNase (Promega, Mannheim, Germany). Random hexamers and Superscript II reverse transcriptase (Invitrogen) were used to run reverse transcription for human samples. Reverse transcription of mouse samples was performed using random hexamers and Transcriptor High-Fidelity Reverse Transcriptase (Roche, Penzberg, Germany). For real-time quantitative RT–PCR, the LightCycler480 system (Roche) and the SYBR Green detection format was used. Beta-2-microglobulin was used as a housekeeper, as it proved to be highly consistently expressed in medulloblastoma cells (data not shown). All analyses were performed in triplicate. Primers were designed using the Primer3 software. Sequences were as follows: B2M (cDNA) forward, 5′-TGTCTTTCAGCAAGGACTGG-3′; B2M (cDNA) reverse, 5′-GATGCTGCTTACATGTATCG-3′; BARHL1 (cDNA) forward, 5′-GAGCGGCAGAAGTACCTGAG-3′; BARHL1 (cDNA) reverse, 5′-AGAAATAAGGCGACGGGAAC-3′; B2M (gDNA) forward, 5′-CAGTAAAGGAGTGGGGGATG-3′; B2M (gDNA) reverse, 5′-TTGTGTTGAGGCAGGAAAAA-3′; BARHL1 (gDNA) forward, 5′-TTCCATTTTCATCCCATCGT-3′; BARHL1 (gDNA) reverse, 5′-TCTTCCCTTTCCCTTCCTTC-3′; B2m forward, 5′-CCTGGTCTTTCTGGTGCTTG-3′; B2m reverse, 5′-TATGTTCGGCTTCCCATTCT-3′; Barhl1 forward 5′-CGCTCAACCTCACCGACA-3′; Barhl1 reverse, 5′-AGAAATAAGGCGACGGGAAC-3′. For each set of primers, post-amplification melting curves were analyzed using the LightCycler480 software, and agarose gel electrophoresis was performed to verify the presence of a single amplification product. Efficiency correction for each set of primers was performed by creating a standard curve.
Histology, in situ hybridization and immunohistochemistry
For H&E staining and for immunohistochemical procedures, brain and tumor tissues were fixed in 4% paraformaldehyde/phosphate-buffered saline overnight at 4 °C. Tissues for frozen sections were equilibrated in 20% glucose/phosphate-buffered saline (pH 7.4) and embedded in optimal compound temperature (OCT). Parasagittal sections of 12-µm thickness were prepared on Superfrost plus slides (Fisher, Schwerte, Germany). Tissues for paraffin wax-embedded sections were dehydrated, embedded and sectioned at 5 µm according to standard protocols. Overall morphology was assessed by staining with H&E. All photomicrographs including those from immunohistochemical experiments were taken digitally using an Olympus Bx50 microscope in combination with the Color view Soft imaging system and Cell software (Olympus, Hamburg, Germany).
In situ hybridization on frozen sections was performed according to standard protocols. In brief, brain sections were hybridized overnight with labeled RNA probes at 65 °C, washed twice in 0.2× SSC pH 4.5, 0.1% Tween20 at 65 °C, washed twice in MBST buffer, pH 7.5, containing 100 mm maleic acid, 150 mm NaCl2, 2 mm levamisole and 0.1% Tween20, blocked in MBST with 2% BM blocking agent (Roche) and 20% lamb serum and incubated with alkaline phosphatase-labeled anti-Digoxigenin antibodies (Roche, 1:2500 in 2% serum) for 2 h. Sections were washed and color was visualized using BM purple (Roche) (detailed protocol available upon request). Immunohistochemistry was performed on frozen or formalin-fixed, paraffin-embedded, rehydrated, 4-µm thick sections using the ABC method on an automated system (BenchMark, Ventana, Tuscon, AZ, USA). Primary antibodies against Synaptophysin (Dako, Glostrup, Denmark, 1:100), MAP2 (Sigma, St Louis, MO, USA, 1:40 000), Cre (Covance, Princeton, NJ, USA, 1:1500), BARHL1 (Chemicon, Billerica, MA, USA, 1:200), NeuN (Chemicon, 1:300), Zic (gift from Dr R Segal, 1:3000), OLIG2 (Chemicon, 1:1000), Sox2 (Chemicon, 1:1000) and phosphorylated histone H3 (Cell Signaling Technology, Danvers, MA, USA, 1:200) were diluted in phosphate-buffered saline and applied on sections for 30 min at 42 °C. Cell nuclei were visualized by hematoxylin.
Western blot analysis
Proteins were extracted from frozen tumor samples and western blots were performed essentially as described before (Hartmann et al., 2005). Filters were incubated with a cleaved caspase-3 (Asp175) (Cell Signaling Technology) and a β-actin antibody (clone AC-15, Sigma) as internal controls according to the instructions of the manufacturers. Secondary antibody labeling and filter development were performed using the ECL kit (Amersham, Buckinghamshire, UK) as described.
Statistical analysis
All obtained results were analyzed using the Prism4 software (GraphPad, La Jolla, CA, USA). Survival of patients and mice was analyzed using Kaplan–Meier survival curves, and the log-rank test was used to examine the significance of results. P-values <0.05 were considered significant. The unpaired t-test was applied to compare the means of two groups with assumed Gaussian distribution and equal variances. If variances were not equal, as confined by the F-test, or Gaussian distribution was not expected, the nonparametric Mann–Whitney test for unpaired data was used to compare the medians of two groups. To compare protein expression patterns of Math1-cre:SmoM2Fl/+, Barhl1+/−Math1-cre:SmoM2Fl/+ and Barhl1−/−Math1-cre:SmoM2Fl/+ mice, at least 3 animals were examined in each group and at least 500 tumor cells were counted in each animal. For phosphorylated histone H3 analysis, 1000 tumor cells were counted. The respective histograms illustrate s.e.m. (error bars). Correlation of two paired data sets was performed with Pearson’s correlation for Gaussian distributions or else with nonparametric Spearman’s correlation, with r as correlation coefficient.
Supplementary Material
Acknowledgments
We are indebted to Michael Schmidt, Veronika Kaltenbrunn, Dagmar Metzger, Philipp Neumann and Silvia Occhionero for excellent technical support, to Dr Mehdi Shakarami and Dr Stefanie Ohlemeyer for animal husbandry. We acknowledge Dr Rosalind Segal for providing Zic antibodies. This work was supported by grants from the Wilhelm-Sander-Stiftung and the Friedrich-Baur-Stiftung (all to US). US is a member of the Max-Eder-junior-research-program of the German Cancer Aid. The HIT-trial-office at the University Medical Center Hamburg-Eppendorf is supported by the German Children’s Cancer Foundation (Deutsche Kinderkrebsstiftung) and by the Fördergemeinschaft Kinderkrebszentrum Hamburg e.V.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Bulfone A, Menguzzato E, Broccoli V, Marchitiello A, Gattuso C, Mariani M, et al. Barhl1, a gene belonging to a new subfamily of mammalian homeobox genes, is expressed in migrating neurons of the CNS. Hum Mol Genet. 2000;9:1443–1452. doi: 10.1093/hmg/9.9.1443. [DOI] [PubMed] [Google Scholar]
- Chellappa R, Li SG, Pauley S, Jahan I, Jin KX, Xiang MQ. Barhl1 regulatory sequences required for cell-specific gene expression and autoregulation in the inner ear and central nervous system. Mol Cell Biol. 2008;28:1905–1914. doi: 10.1128/MCB.01454-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales J, Rocco G, Blaess S, Guo Q, Joyner A. Spatial pattern of sonic hedgehog signaling through Gli genes during cerebellum development. Development. 2004;131:5581–5590. doi: 10.1242/dev.01438. [DOI] [PubMed] [Google Scholar]
- Crawford JR, MacDonald TJ, Packer RJ. Medulloblastoma in childhood: new biological advances. Lancet Neurol. 2007;6:1073–1085. doi: 10.1016/S1474-4422(07)70289-2. [DOI] [PubMed] [Google Scholar]
- Eberhart CG, Kaufman WE, Tihan T, Burger PC. Apoptosis, neuronal maturation, and neurotrophin expression within medulloblastoma nodules. J Neuropathol Exp Neurol. 2001;60:462–469. doi: 10.1093/jnen/60.5.462. [DOI] [PubMed] [Google Scholar]
- Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson RJ. Medulloblastoma: signalling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- Hartmann W, Koch A, Brune H, Waha A, Schüller U, Dani I, et al. Insulin-like growth factor II is involved in the proliferation control of medulloblastoma and its cerebellar precursor cells. Am J Pathol. 2005;166:1153–1162. doi: 10.1016/S0002-9440(10)62335-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine VM, Rowitch DH. Hedgehog signaling has a protective effect in glucocorticoid-induced mouse neonatal brain injury through an 11betaHSD2-dependent mechanism. J Clin Invest. 2009;119:267–277. doi: 10.1172/JCI36376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool M, Koster J, Bunt J, Hasselt NE, Lakeman A, van Sluis P, et al. Integrated genomics identifies five medulloblastoma subtypes with distinct genetic profiles, pathway signatures and clinicopathological features. PLOS One Aug. 2008;3:e3088. doi: 10.1371/journal.pone.0003088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Qiu F, Xu A, Price SM, Xiang M. Barhl1 regulates migration and survival of cerebellar granule cells by controlling expression of the neurotrophin-3 gene. J Neurosci. 2004;24:3104–3114. doi: 10.1523/JNEUROSCI.4444-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SG, Price SM, Cahill H, Ryugo DK, Shen MM, Xiang MQ. Hearing loss caused by progressive degeneration of cochlear hair cells in mice deficient for the Barhl1 homeobox gene. Development. 2002;129:3523–3532. doi: 10.1242/dev.129.14.3523. [DOI] [PubMed] [Google Scholar]
- Louis DN. WHO Classification of Tumours of the Central Nervous System. Lyon: IARC Press; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J, Ligon KL, Rakhlin EY, Thayer SP, Bronson RT, Rowitch D, et al. A novel somatic mouse model to survey tumorigenic potential applied to the Hedgehog pathway. Cancer Res. 2006;66:10171–10178. doi: 10.1158/0008-5472.CAN-06-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millen KJ, Hui CC, Joyner AL. A role for En-2 and other murine homologues of Drosophila segment polarity genes in regulating positional information in the developing cerebellum. Development. 1995;121:3935–3945. doi: 10.1242/dev.121.12.3935. [DOI] [PubMed] [Google Scholar]
- Mo Z, Li S, Yang X, Xiang M. Role of the Barhl2 homeobox gene in the specification of glycinergic amacrine cells. Development. 2004;131:1607–1618. doi: 10.1242/dev.01071. [DOI] [PubMed] [Google Scholar]
- Northcott PA, Korshunov A, Witt H, Hielscher T, Eberhart CG, Mack S, et al. Medulloblastoma comprises four distinct molecular variants. J Clin Oncol. 2011;29:1408–1414. doi: 10.1200/JCO.2009.27.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomeroy SL, Tamayo P, Gaasenbeek M, Sturla LM, Angelo M, McLaughlin ME, et al. Prediction of central nervous system embryonal tumour outcome based on gene expression. Nature. 2002;415:436–442. doi: 10.1038/415436a. [DOI] [PubMed] [Google Scholar]
- Scales SJ, de Sauvage FJ. Mechanisms of Hedgehog pathway activation in cancer and implications for therapy. Trends Pharmacol Sci. 2009;30:303–312. doi: 10.1016/j.tips.2009.03.007. [DOI] [PubMed] [Google Scholar]
- Schüller U, Heine VM, Mao J, Kho AT, Dillon AK, Han YG, et al. Acquisition of granule neuron precursor identity is a critical determinant of progenitor cell competence to form Shh-induced medulloblastoma. Cancer Cell. 2008;14:123–134. doi: 10.1016/j.ccr.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüller U, Zhao Q, Godinho SA, Heine VM, Medema RH, Pellman D, et al. Forkhead transcription factor FoxM1 regulates mitotic entry and prevents spindle defects in cerebellar granule neuron precursors. Mol Cell Biol. 2007;27:8259–8270. doi: 10.1128/MCB.00707-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal RA, Goumnerova LC, Kwon YK, Stiles CD, Pomeroy SL. Expression of the neurotrophin receptor TrkC is linked to a favorable outcome in medulloblastoma. Proc Natl Acad Sci USA. 1994;91:12867–12871. doi: 10.1073/pnas.91.26.12867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter R, Shakhova O, Bhagat H, Behesti H, Sutter C, Penkar S, et al. Cerebellar stem cells act as medulloblastoma-initiating cells in a mouse model and a neural stem cell signature characterizes a subset of human medulloblastomas. Oncogene. 2010;29:1845–1856. doi: 10.1038/onc.2009.472. [DOI] [PubMed] [Google Scholar]
- Weyer A, Schilling K. Developmental and cell type-specific expression of the neuronal marker NeuN in the murine cerebellum. J Neurosci Res. 2003;73:400–409. doi: 10.1002/jnr.10655. [DOI] [PubMed] [Google Scholar]
- Yokota N, Aruga J, Takai S, Yamada K, Hamazaki M, Iwase T, et al. Predominant expression of human Zic in cerebellar granule cell lineage and medulloblastoma. Cancer Res. 1996;56:377–383. [PubMed] [Google Scholar]
- Yokota N, Mainprize TG, Taylor MD, Kohata T, Loreto M, Ueda S, et al. Identification of differentially expressed and developmentally regulated genes in medulloblastoma using suppression subtraction hybridization. Oncogene. 2004;23:3444–3453. doi: 10.1038/sj.onc.1207475. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.