Abstract
The selection-mutation-drift theory of codon usage plays a major role in the theory of molecular evolution by explaining the co-evolution of codon usage bias and tRNA content in the framework of translation optimization. Because most studies have focused only on codon usage, we analyzed the tRNA gene pool of 102 bacterial species. We show that as minimal generation times get shorter, the genomes contain more tRNA genes, but fewer anticodon species. Surprisingly, despite the wide G+C variation of bacterial genomes these anticodons are the same in most genomes. This suggests an optimization of the translation machinery to use a small subset of optimal codons and anticodons in fast-growing bacteria and in highly expressed genes. As a result, the overrepresented codons in highly expressed genes tend to be the same in very different genomes to match the same most-frequent anticodons. This is particularly important in fast-growing bacteria, which have higher codon usage bias in these genes. Three models were tested to understand the choice of codons recognized by the same anticodons, all providing significant fit, but under different classes of genes and genomes. Thus, co-evolution of tRNA gene composition and codon usage bias in genomes seen from tRNA's point of view agrees with the selection-mutation-drift theory. However, it suggests a much more universal trend in the evolution of anticodon and codon choice than previously thought. It also provides new evidence that a selective force for the optimization of the translation machinery is the maximization of growth.
Due to the degeneracy of the genetic code, many codons are synonymous for the same amino acid. Nevertheless, some synonymous codons are more abundant than others. This is the result of mutational biases and selective forces (Grantham et al. 1980; Andersson and Kurland 1990; Bulmer 1991; Sharp et al. 1993). Among the former, G+C content variation is important both in prokaryotes, because their genomes vary from 25% to 75% G+C (Sueoka 1962), and in eukaryotes with intragenomic compositional heterogeneity, for example in the isochore structure of the human genome (Bernardi et al. 1985). The third position of codons is the most neutral to changes and thus shows ampler G+C variations, which in bacteria range from <20% to >90% G+C (Muto and Osawa 1987). As a result, G+C composition is the major factor affecting codon usage variation (Chen et al. 2004). Other compositional biases that affect the relative frequency of synonymous codons include compositional strand bias (Lobry 1996) and transcription-coupled repair-associated biases (Francino and Ochman 2001). With the exception of the latter, which is correlated with gene expression levels, mutational biases affect large groups of functionally unrelated genes. This extends to all genes in the case of G+C genome composition.
Because translation is the most energetically expensive process occurring in exponentially growing cells, its efficiency is under important selective pressure. Under these physiological conditions, a small set of genes accounts for the large majority of transcription and translation taking place in the cell (Andersson and Kurland 1990). This set includes genes related to translation, transcription, and the energy metabolism, and is under strong selective pressure for translation efficiency. The rate-limiting step in the elongation cycle of the polypeptide chains is the diffusion of the cognate ternary complex (tRNA+EfTu+GTP) to the A-site of the ribosome (Varenne et al. 1984). As a result, the most abundant aa-tRNA for a given amino acid is predominantly recruited by the codons of highly expressed genes (Dong et al. 1996). In this context, the most favorable codons are the ones corresponding to the most abundant and efficient cognate aa-tRNA present in the cell (Andersson and Kurland 1990). The selection-mutation-drift theory states that the bias observed in synonymous codon usage is the result of the balance between the forces of selection and mutation in a finite population, with greater intragenomic bias reflecting stronger selection for translation efficiency (Sharp and Li 1986a; Bulmer 1991). Selection can arise for elongation speed, and different codons have different translation rates (Varenne et al. 1984; Sorensen et al. 1989). Selection can also arise for accuracy, if some codons are less prone to mistranslation or drop-off events (Rodnina and Wintermeyer 2001). In Drosophila, highly conserved positions show amino acids coded by the preferred codons more often than nonconserved positions in the same gene, suggesting the importance of accuracy (Akashi 1994). Selection for biased codon usage has also been recently related with thermophily (Lynn et al. 2002) and the metabolic cost of amino acids (Akashi and Gojobori 2002).
There is abundant literature regarding codon usage biases, and sophisticated techniques profit from this information to infer adaptive evolution (Suzuki et al. 2001; Yang and Swanson 2002), horizontal transfer (Médigue et al. 1991), expression levels (Sharp and Li 1987), and cellular localization (Chiapello et al. 1999). However, most works are focused on the analysis of codon frequencies from DNA sequences (Andersson and Kurland 1990; Moszer et al. 1999; Duret 2002). The complementary approach, understanding the distribution of tRNA gene number and anticodon type, has been much less developed in the framework of comparative genomics. Several early studies focused on the correlation between tRNA content and codon usage within one or a few species (Ikemura 1985; Kano et al. 1991; Yamao et al. 1991; Percudani et al. 1997; Kanaya et al. 1999; Duret 2000). From these studies it emerged that tRNA content correlates with codon usage and amino acid composition in many prokaryotic and eukaryotic species. Experimentally, it was found that the correlation is stronger at higher growth rates in Escherichia coli, indicating the importance of translation efficiency in these conditions (Dong et al. 1996). Yet, several questions remain unanswered because they demand the comparative study of many genomes. (1) Why are some codons preferred relative to others recognized by the same anticodon? (2) How do tRNA gene pools evolve in terms of anticodon number and type? (3) How do tRNA gene pools co-evolve with codon usage in relation to the optimization of the translation machinery and the maximization of growth? It is often implicitly assumed that the tRNA pool and its expression levels remain relatively constant in short evolutionary spans. This is not necessarily so, because regulatory sequences evolve quickly (McAdams et al. 2004). As a result, tRNA levels in exponentially growing cells have the potential to change very quickly, and there is no reason to suppose that codon usage adaptation to tRNA pools is much less constrained than tRNA pool adaptation to codon usage bias. Furthermore, at a broader evolutionary level, the tRNA gene composition of genomes also evolves rapidly (Marck and Grosjean 2002). As an example, E. coli O157:H7 strain contains 100 tRNAs, whereas the closely related MG1655 strain contains 88, and the not so distantly related genomes of Buchnera species have 32. Because the number and the type of tRNAs co-evolve with codon usage bias in the framework of the optimization of the translation machinery, it is important to understand how this happens and what the major variables influencing the process are. This should allow a better understanding of codon usage bias, but also of the evolution of genome content and its relation with maximum specific growth rates.
Results
Distribution of tRNAs and their relation with generation time
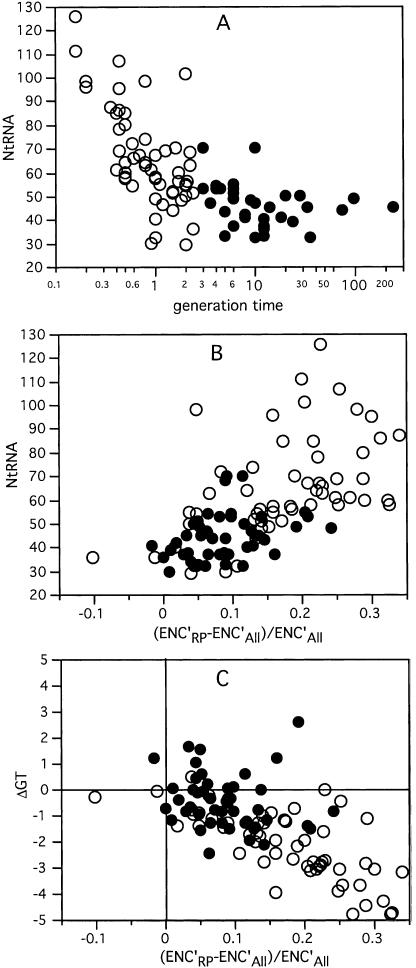
We identified an average of 55.6 tRNA genes per genome, with a maximum of 126 in Vibrio parahaemolyticus, and a minimum of 29 in Mycoplasma pulmonis (see detailed results in Supplemental Tables 1,2). There is a strong negative correlation between the minimal generation times of bacteria and the number of tRNA genes in the genome (Spearman ρ = –0.72, P < 0.001, Fig. 1A). This is not a consequence of genome size, as its correlation with generation time is not significant (ρ = –0.17, P = 0.10). We could not find in the literature minimal generation times for eight obligatory intracellular bacteria such as Chlamydia and Blochmania, although it is clear that they grow slowly. We therefore divided the genomes into slow and fast growers, where the latter have minimal generation times shorter than 2.5 h. Using this categorical data, we found that fast growers have a median of 61 tRNA versus 44 for slow growers. We then computed the number of different anticodons (i.e., tRNA species) present in the tRNA gene sets. This varied from 27 in M. pulmonis to 44 in four: Bradyrhizobium japonicum, Mesorhizobium loti, Rhodopseudomonas palustris, and Thermotoga maritima. As remarked previously (Marck and Grosjean 2002), the maximum number of different tRNAs is always much smaller than 61, the theoretical upper limit. The average number of different anticodons present in the genomes is 37. Surprisingly, fast growers have fewer different tRNAs (median = 34) than slow growers (median = 39, P < 0.05, Wilcoxon test). Thus, fast growers have more tRNA genes that represent a smaller set of the possible anticodons; that is, fast growers have more abundant but less diverse tRNAs. The translation machinery in fast-growing bacteria has thus evolved to enlarge the number of tRNAs and to specialize in the use of a small set of anticodons.
Figure 1.
(A) Number of tRNA genes per genome as a function of the bacterial optimal generation time. (B) Number of tRNA genes per genome as a function of codon usage bias given as the difference in ENC′ between the set of genes coding for ribosomal proteins (ENC′RP) and the whole genome (ENC′All). (C) Enrichment in G and T in the last codon position of twofold-degenerated amino acids in ribosomal proteins compared to the average genome (ΔGT) as a function of codon usage bias. ○, fast growers; •, slow growers (generation time >2.5 h).
This should result in higher codon usage bias in highly expressed genes, because codons cognate for the most abundant tRNAs would tend to be overrepresented in highly expressed genes. We therefore computed the effective number of codons given G+C composition (ENC′) (Novembre 2002), in the entire genome and in the subset of genes coding for ribosomal proteins. The latter are expected to be highly expressed under exponential growth (Jansen et al. 2003). The difference between the two groups (ENCdif = (ENC′RP – ENC′All)/ENC′All) is a good measure of the codon usage bias related to translation optimization. We observe a positive correlation (Spearman ρ = 0.68, P < 0.001) between ENCdif and the number of tRNAs in the genome (Fig. 1B). As expected, generation time is negatively correlated with ENCdif (Spearman ρ = –0.59, P < 0.001). The codon adaptation index (CAI) measures the codon usage bias of a gene towards a set of “optimal” codons determined from a reference set of highly expressed genes, typically ribosomal proteins (Sharp and Li 1987). It has been argued that genomes with translation-associated codon usage bias have a lower average genomic CAI because the codon usage of ribosomal proteins is very different from the average genome in this case. Indeed, the Spearman correlation of average genomic CAI with ENCdif is ρ = –0.81 (P < 0.001), and that with minimal generation time is ρ = 0.54 (Spearman ρ, P < 0.001). About 76% of the genomes with significant codon usage bias (as defined by Rocha and Danchin 2003) have fast growth rates, and 75% of the nonbiased genomes have low growth rates. Thus, fast-growing bacteria have more abundant and similar tRNAs, and this co-evolves with higher codon usage bias in highly expressed genes relative to the rest of the genome. As a result, most fast-growing bacteria have strong codon usage biases, contrary to slow-growing bacteria.
Evolution of anticodon bias with G+C content
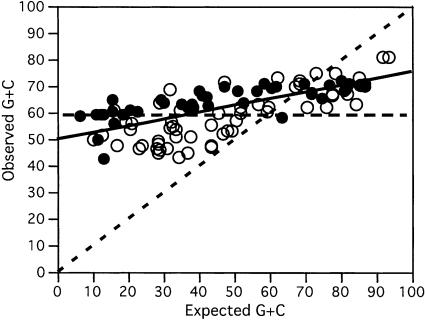
The previous results indicate that for fast growers, only a small set of all possible anticodons is used. Hence, we tried to understand how tRNA sets evolved from the point of view of anticodon usage. Because the G+C contents of third codon positions vary from <20% to >90% (Muto and Osawa 1987), the most frequent codons will dramatically change with G+C content (Chen et al. 2004). Naturally, one would expect the frequency of anticodons in genomes to follow the same trend. Although at the time no systematic multi-genomic study was possible, G+C enrichment was observed in Micrococcus luteus tRNA anticodons, relative to the G+C-poorer E. coli and Mycoplasma (Kano et al. 1991). Therefore, G+C-rich genomes should have G+C-richer first anticodon positions in their set of tRNAs than A+T-rich genomes. To test this hypothesis, we built a linear model for G+C variation at third codon position (%G+C3) relative to genome G+C (%G+CT)inthe 102 genomes, as in Muto and Osawa (1987) (see Supplemental Fig. 1 for details). This provided a regression line %G+C3 = –36.42 + 1.798 × %G+CT (r2 = 0.99, P < 0.001, F-test), which we used to predict the G+C composition of tRNAs at first anticodon position. Unexpectedly, the results show that there is a much smaller variation in anticodon composition (slope = 0.25, Fig. 2) than expected (slope = 1, significant difference P < 0.001, F-test). In fact, a model where the frequency of tRNA anticodons does not change with G+C composition (slope = 0) fits the data better (P < 0.01, F-test). Therefore, tRNA anticodon composition only slightly adapts to the genome G+C composition and to the codon usage that it leads to.
Figure 2.
Observed versus expected G+C composition of tRNA anticodons. Expected was computed using an update of the Muto and Osawa (1987) equations and is indicated by the black dashed main diagonal. The horizontal dashed line indicates the expected value if there was no change in G+C anticodon composition with genome G+C content. The linear regression of the observed data is shown as a solid black line (it has an R2 of 0.50 and a slope of 0.26, significantly different from 1 and 0, P < 0.001).
One can explain these unexpected results, if optimal anticodons are nearly invariant in the bacterial domain. In this case, anticodon variation would be more constrained than codon variation. Therefore, we tried to identify for each amino acid the most frequent anticodon in the set of all genomes. We also checked how often it is the most frequent anticodon and how often it is present in each genome (see Table 1). This clearly demonstrates that in most genomes one given anticodon is almost always present and is also systematically the most frequent. As a general rule, in the first anticodon position of twofold-degenerated amino acids, G is always preferred over A and U is preferred over C. The four- and sixfold-degenerated amino acids show a preference for U, when possible, and then G. The exceptions are Arginine, where ACG is the most frequent anticodon, and Glycine, where GCC outnumbers UCC in several genomes, although it is less ubiquitous. Therefore, a reason for anticodon constancy despite wide variation in the G+C content of genomes may be that the best anticodon is typically the same.
Table 1.
Most frequent anticodon for each amino acid
| N | AC | MF | P | |
|---|---|---|---|---|
| Met | 1 | CAU | 102 | 102 |
| Trp | 1 | CCA | 102 | 102 |
| Ile | 3 | GAU | 97 | 101 |
| Asn | 2 | GUU | 102 | 102 |
| Asp | 2 | GUC | 102 | 102 |
| Cys | 2 | GCA | 102 | 102 |
| His | 2 | GUG | 102 | 102 |
| Phe | 2 | GAA | 102 | 102 |
| Tyr | 2 | GUA | 102 | 102 |
| Glu | 2 | UUC | 98 | 102 |
| Gln | 2 | UUG | 98 | 101 |
| Lys | 2 | UUU | 97 | 102 |
| Ala | 4 | UGC | 98 | 102 |
| Gly | 4 | GCC | 93 | 101 |
| Val | 4 | UAC | 97 | 102 |
| Pro | 4 | UGG | 99 | 102 |
| Thr | 4 | UGU | 95 | 102 |
| Arg2 | 2 | UCU | 60 | 102 |
| Arg4 | 4 | ACG | 87 | 91 |
| Ser2 | 2 | GCU | 77 | 102 |
| Ser4 | 4 | UGA | 90 | 99 |
| Leu2 | 2 | UAA | 70 | 100 |
| Leu4 | 4 | UAG | 71 | 102 |
The anticodon (AC) is the (or among the, if there are ties) most frequent anticodons in MF genomes and present in P genomes. Amino acids with more than N>4 codons were split into codon boxes.
Codon usage invariants
The above results suggest that in most genomes the codons that are favored in highly expressed genes relative to the rest of the genome are the same. We first tested this hypothesis using twofold-degenerated amino acids, where most frequently there is one anticodon that is much more frequent than the others. We then analyzed the difference in codon usage between genes coding for ribosomal proteins and the remaining genes in the genome. If codon usage bias in highly expressed genes evolves to perfectly match the most frequent anticodon, then one would expect C and A richness in the third codon position of these genes (because A and C pair better with G and U anticodons), and conversely G+T poorness. Indeed, we found that third codon position C and A codons are more frequent in twofold-degenerated amino acids of highly expressed genes and especially among fast growers (Fig. 1C).
The analysis of four-codon amino acids (also commonly named quartets) is more complicated, for two reasons. Firstly, there is almost always more than one type of anticodon available in the genome. Although the major one is usually the U-starting anticodon, G-starting and C-starting anticodons are also commonly found. Because there are different overlapping pairing possibilities, it is more difficult to assign the theoretically best codon (see below the analysis of models for codon:anticodon pairing). Secondly, in two-codon amino acids, U is necessarily modified to pair with A and G, whereas in quartets it may be modified to xo5U, which pairs with A, G, and U, or it may not be modified at all, in which case it pairs with any base. Currently, we cannot assign the state of base modification of tRNAs based simply on the genome information. These problems can be illustrated in the following example. E. coli has two anticodons for Alanine (two GGC and three UGC). If there are no modifications, then UGC can read any Alanine codon (although it might pair better with GCA), whereas GGC can only read GCC and GCU. But if U is modified as in Table 2, then it would read only GGA and GGG. Supposing that differences in codon:anticodon pairing are small, in the first case GCC and GCU would be preferred, whereas in the second they would be at a disadvantage.
Table 2.
Assignment of readable codons to tRNA anticodons obtained by tRNAscan
| 1st AP | Nucleoside | Pairs with 3rd CP | For: |
|---|---|---|---|
| A | I | U, C, A | Arg4, Ile |
| A | A | A, C, G, U | aoc |
| C | k2C | A | Ile |
| C | C | G | aoc |
| G | G/Q | C, U | All cases |
| T | mU | A, G | Gln, Glu, Arg2, Lys, Leu2, Trp |
| T | U | A, C, G, U | aoc |
As an example, T at first anticodon position corresponds to mU when coding for twofold degenerate amino acids (pairing with 3rd codon position bases A and G) and to U in other cases (pairing with any 3rd codon position nucleoside). mU, unspecified modification of U (e.g. S2U, Sc2U, Um or xm5U); aoc, “all other cases”; Xxx4 and Xxx2, four- and two-codon boxes of the sixfold degenerate amino acids; CP, codon position; AP, anticodon position.
Despite these difficulties, we tried to test whether highly expressed genes do get enriched in quartet codons ending in the complementary base of the majority anticodon first base. As expected, fast growers showed a very significant enrichment in A (C in Glycine) in ribosomal proteins (+12%), relative to slow growers (–8%, P < 0.001, Wilcoxon test). Thus in quartets of fast-growing bacteria, highly expressed genes also tend to be enriched in the complementary base of the most abundant anticodon first base. However, whereas two-codon amino acids in slow growers also show significantly C+A enrichment, quartets show impoverishment in this group (P < 0.001, signed-rank tests). This exception may be related to the methodological complications of defining the expected best codon in quartets in genomes with a more diverse tRNA gene pool, as discussed above. Nevertheless, although the detailed analysis of quartets requires further work, these results clearly indicate that there are more similar trends of codon usage optimization in highly expressed genes of different fast-growing bacteria than previously thought. We then tried to investigate the reasons behind the common preference for particular anticodons.
Models explaining codon:anticodon preference
We set up the correspondence tables that associate each anticodon to a set of readable codons. The wobble at the anticodon first position (pairing with the codon third position) is particularly permissive in the bacterial context, allowing one tRNA to decode several codons (Table 2). Under the aa-tRNA-demand theory for codon usage bias, codons recognized by the same tRNAs should be equally frequent, apart from mutational biases. The frequency model is a generalization of this, where the favored codon is the one that can be read by the largest number of tRNAs (see the text entitled “Models” in the Methods section). Because some codons can be read by several different anticodons, they should be preferred. Indeed, the codon choice follows significantly the frequency model in fast- and slow-growing bacteria, more so among the highly expressed genes (Table 3). However, a preliminary inspection of codon usage in E. coli (∼50%G+C) shows that this model is not enough to explain codon usage biases. In E. coli there are six tRNA genes for Lys, all with anticodon UUU. Although the frequency model would predict similar codon frequencies of AAA and AAG, in ribosomal protein genes 71% of codons are AAA. In E. coli there are six tRNA genes for Glycine, one with the anticodon CCC, one with UCC, and four with GCC. However, the most frequent codon in highly expressed genes is GGU (60%). Thus, some codons are strongly preferred relative to others that are recognized by the same set of aa-tRNAs. This has been experimentally confirmed for several aa-tRNAs in E. coli (Thomas et al. 1988; Curran and Yarus 1989).
Table 3.
Average observed/expected (O/E) values for the three models explaining codon usage in slow growers and fast growers
|
Fast growers
|
Slow growers
|
|||
|---|---|---|---|---|
| dif | All | dif | All | |
| Frequency | 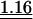 |
1.04 | 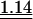 |
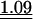 |
| Perfect match | 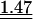 |
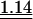 |
1.03 | 0.99 |
| Stability | 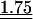 |
0.93 | 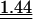 |
0.94 |
Note that because expected values are generally different in the different models, O/E values between different models cannot be compared in a straightforward manner. “All” corresponds to the fit when the most frequent codon per amino acid is taken from the codon usage of all genes. “Dif” corresponds to the fit when the most frequent codon per amino acid is the one increasing the most in frequency in ribosomal proteins compared to the rest of the genome. Wilcoxon tests (Ha: O/E > 1), underlined values (P < 0.05), double underlined values (P < 0.001).
Two other models have been proposed to explain biased codon choice (see “Models” in the Methods section). In the perfect match model, the most frequent codon should make the optimal codon-anticodon interaction, that is, it should perfectly match the most abundant anticodon. This should increase the specificity and the sensitivity of the ribosome (Ikemura 1981). The stability model maintains that very strong and very weak codon-anticodon interactions should be avoided (Grosjean and Fiers 1982), the former because they slow tRNA turnover in the ribosome and the latter because they might lead to frequent mistranslation and/or to higher rates of incorrect rejection of aa-tRNAs by the ribosome. Under this model, the best codons starting with two strong (S={G,C}) bases are the ones with a weak (W={A,U}) third base. Inversely, the best codons starting with two Ws should have a third base S.
We tested the two models using two sets: the codon usage of all genes and the difference in codon usage between ribosomal proteins and the remaining genes (Table 3). The latter analysis aims at finding patterns that become dominant only in highly expressed genes relative to the entire genome. The perfect match model shows significant fit in fast-growing bacteria, especially among highly expressed genes. The stability model shows significant fit among highly expressed genes. Therefore, both models explain part of the observed co-evolution of tRNAs and codon usage, but under different circumstances and with different overall fit. Although these heterogeneous results strongly suggest that the significance of the models is not due to mutual dependency, we tested whether by keeping one model constant, the other would still provide significant fit. This was done for fast-growing highly expressed genes (first data column of Table 3), because it is the only case where the significant fit of the two models coincides. When the perfect match model is fitted to codons starting with SW or WS (for which the stability model has no expectations), the observed/expected (O/E) value is almost unchanged (O/E = 1.40). When the perfect-match codons were removed from the set of fourfold and sixfold degenerate codons, the stability model still significantly fitted the data of this subset (O/E = 1.46). Therefore, both models show significant fit, when the other is controlled for.
Discussion
The literature on codon usage bias has often insisted on the correlation between the concentration of aa-tRNA, the cell generation time, and codon usage bias (Andersson and Kurland 1990; Sharp et al. 1993; Moszer et al. 1999). Here, we demonstrate this for the first time using data on minimal generation times and genome data on tRNA and codon usage bias for 102 bacterial species. We observed that tRNA genes are more numerous in fast-growing bacteria, and that these tRNAs are less diverse. This strongly suggests that the optimization of the translation machinery to high growth rates is achieved by having more tRNAs, but of fewer different types, which allows near-saturation of ribosomes with the smaller pool of ternary complexes (Ehrenberg and Kurland 1984). At high growth rates, the concentration of rRNA in cells increases faster than the concentration of aa-tRNA (Dong et al. 1996). As a result, the number of aa-tRNA per ribosome decreases, and elongation rates depend more dramatically on fast aa-tRNA diffusion to the decoding site. It is thus more favorable to have more tRNAs of the same type, because this allows the co-evolution of codon usage bias in highly expressed genes, which then creates a strong demand for these smaller sets of tRNAs (Curran and Yarus 1989; Berg and Kurland 1997). If this view turns out to be correct, codon usage bias in highly expressed genes results from the selection of optimal codons associated with the most frequent tRNA genes, but the increase in frequency of these tRNA genes also results from codon usage bias. This co-evolution renders the expression of highly expressed genes more efficient, as these genes overrepresent the codons corresponding to the overabundant tRNAs.
Codon usage bias in highly expressed genes relative to the rest of the genome is expected to be under stronger selection in organisms for which growth rate is an important element of the overall fitness, that is, fast-growing bacteria. There are some exceptions to this rule. For example, three Mycoplasma (M. pulmonis, M. gallisepticum, and Ureaplasma urealyticum) have short generation times with few tRNAs and low codon usage bias. However, these bacteria are very small (about 1000 × smaller than E. coli), and the metabolic effort necessary to duplicate the entire cell is certainly much smaller. Indeed, the characteristic enrichment of tRNA in exponential growth phase associated with codon usage bias in E. coli (Dong et al. 1996) is absent from M. capricolum (Yamao et al. 1991). For comparison, Buchnera has a volume close to that of E. coli and grows slowly with its minimal translation apparatus (Baumann et al. 1995). This suggests that models aiming at explaining codon usage bias from selection to translation optimization should take into account the cost and kinetics of cell doubling. It also puts forwards a problem with the use of minimal generation times. Here, we consider these to represent the maximal growth capacity of the cell. However, optimal growth conditions are probably not known with equal accuracy for all species, which introduces a certain error in the analysis.
Given the close association between codon usage bias, tRNA abundance, and generation time, one would expect anticodon usage to follow the same compositional trends as codon usage with respect to G+C variation (Muto and Osawa 1987; Osawa et al. 1988; Kano et al. 1991). However, we find a much smaller variation than expected. This is probably because some anticodons are preferably chosen in most genomes, which results in constraining anticodon evolution in function of G+C composition. As such, directional selection of anticodon usage to adapt to codon usage is partially compensated by selection for a more universally efficient tRNA. The reasons why such tRNAs are so widely preferred are not entirely clear. In the 102 genomes there are only 200 tRNAs with an A(I) in the first anticodon position (out of a total of 5670 tRNAs), and 188 of these correspond to the anticodon ACG for Arginine. There is thus a clear counterselection for A at first anticodon position. In two-codon amino acids this may be related to the fact that both A and Inosine (I) pair with A at the third codon position, leading to mistranslation (Osawa et al. 1992). However, Inosine in a hypothetical IAU anticodon would allow the recognition of the three codons for Isoleucine, avoiding the systematic modification of CAU (for Methionine) to k2CAU to recognize (AUA) (Muramatsu et al. 1988). Yet, IAU was never found in this work. A-avoidance may be related to stereochemical destabilization of the codon:anticodon duplex (Lim and Curran 2001), but this does not explain why it is favored for coding Arginine. In any case, A-avoidance may partially explain the preference for G at twofold degenerate amino acids.
As a general rule, U is preferred at the first anticodon position when possible. This includes amino acids that are coded by more than two codons. For most cognate amino acids, U or some modified nucleosides derived from U can pair with all synonymous codons. This is an advantageous anticodon sparing strategy (Marck and Grosjean 2002), particularly for small genomes and for fast-growing bacteria. For small genomes, the choice of tRNA genes with U-starting anticodons allows a reduction of the number of tRNA genes to a minimum. For fast-growing bacteria, it allows the allocation of one single anticodon to each amino acid, which may then be amplified in multiple copies, resulting in optimized translation of highly expressed genes.
Surprisingly, when the tRNA composition is matched with codon usage, different genomes show similar patterns of codon usage bias in highly expressed genes. Although this finding will require further analysis, it strongly suggests enrichment in these genes of codons with optimal pairing with the most frequent ubiquitous anticodons. Interestingly, enrichment of A-ending codons was found in mitochondria of organisms with a high metabolic rate (Xia 1996), suggesting that these results may also apply to them. Naturally, codon usage bias is also constrained by the genome average sequence composition (Muto and Osawa 1987), which reflects mutational biases and tends to increase the differences between highly expressed genes in genomes with very different average compositions.
We have tried to unravel the constraints imposed by codon:anticodon interaction on the definition of optimal codons, both in the genome and in the set of highly expressed genes, by applying three previously proposed models. As expected, the frequency model fits the data well, especially among highly expressed genes. This model is a generalization of the aa-tRNA demand model and thus confirms the importance of using the codons corresponding to the most frequent aa-tRNAs in the cell. If the concentration of aa-tRNA species is the major determinant of translation efficiency, then one would expect codons recognized by the same tRNA to be equally frequent apart from compositional biases. This does not seem to be the case, because of the different possible codon:anticodon interactions (Thomas et al. 1988; Curran and Yarus 1989). There are multiple possible reasons why some interactions could be more efficient than others, and many involve selection for accuracy. These can take the form of avoiding the erroneous incorporation of an amino acid or translation accidents such as frame-shift or ribosomal drop-off events (Rodnina and Wintermeyer 2001). Translation slow-down may also result from incorrectly refusing a tRNA. For example, hyperaccurate ribosomes inhibit growth (Ruusala et al. 1984). A codon:anticodon interaction optimizing recognition would reduce this problem. It has been proposed that such interaction corresponds to the canonical Watson–Crick base-pairing (Ikemura 1981), and this may be why the perfect match model better fits the data for fast-growing bacteria and especially for their highly expressed genes. The reason for a higher bias related to accuracy in highly expressed genes stems from their higher conservation (Rocha and Danchin 2004), which suggests a stronger selective pressure for accurate translation.
The initial formulation of the stability model proposed that average bonding energy would be selected in the codon usage of highly expressed genes, and inversely, counterselected in weakly expressed genes (Grosjean and Fiers 1982). Counterselection in lowly expressed genes has been questioned on the basis of population genetics models, as selection is not expected to operate at the very low adaptive value of selecting less-efficient codons in weakly expressed genes (Sharp and Li 1986b). However, we found that the majority of genes do have O/E values smaller than 1, confirming the previous observations for low-expressed genes of E. coli, MS2 coliphage (Grosjean and Fiers 1982), and S. cerevisiae (Percudani and Ottonello 1999). These opposite observations can be reconciled by the recent model of supply and demand for genes expressed under starvation conditions, which predict an overrepresentation of rare codons in these genes (Elf et al. 2003). It may also be a fortuitous consequence of the interaction between selective and mutational effects, associated with biased oligonucleotide usage (Burge et al. 1992), or contextual effects (Chen et al. 2004). The stability model seems to apply to highly expressed genes and especially in fast-growing bacteria. This is in agreement with the observation that the most demanded aatRNAs in highly expressed genes, via codon usage bias, have intermediate values of intrinsic translation rate (Curran and Yarus 1989). Very weak or very strong interactions can pose problems to the proofreading system and slow down tRNA turnover at the ribosome. Both are likely to retard elongation. Therefore the stability model applies essentially to highly expressed genes.
It is as yet unclear what the overlaps or conflicts between these models are. Further analysis must take more precisely into account the concentration of the different tRNA in cells and their precise nucleoside modifications at the anticodons. The latter may significantly change codon:anticodon interaction rules even if outside the wobble base (Yarian et al. 2002), and thus change the parameters of the models we analyzed (especially the frequency model, which depends on the decoding rules). An interesting consequence of the anticodon choices that we unraveled is that highly expressed genes tend to be richer in A and C at third codon positions, which opposes several mutational biases. Compositional strand bias leads to higher G+T gene composition for leading strand genes, and the leading strand tends to overrepresent highly expressed genes due to their frequent essential character (Rocha 2004). Comparative analysis, by allowing the study of many diverse sets of tRNAs, genome compositions, and bacterial ecologies, will allow a better understanding of the functioning and evolution of the translation machinery. This will be fundamental to disentangle the selective from the many mutational bases of codon usage and gene composition, as well as essential to a better understanding of codon:anticodon interactions and their role in translation.
Methods
Genome and tRNA data
One hundred and two genomes, corresponding to 102 bacterial species, were retrieved from GenBank (see Supplemental Table 1 for a comprehensive listing). Minimal generation times were taken from the literature or obtained by personal communication with researchers in the field. The tRNA genes were searched with tRNAscanSE (Lowe and Eddy 1997), using the default parameters for bacteria. Each anticodon was assigned a set of readable codons using the wobble rules for bacteria (Yokohama and Nishimura 1995; Lim and Curran 2001). Wobble allows noncanonical nucleoside pairing of the first anticodon position with the third position of codons, depending on the chemical modifications of nucleosides at the first anticodon position. Because such modifications are difficult to predict without further information, we proceeded by parsimony: the first anticodon nucleoside was a priori regarded as unmodified. Modifications were then introduced when required to avoid mistranslation (Table 2). For example, U in the first anticodon position can pair with any nucleotide, which in twofold degenerate amino acids poses a mistranslation problem. Therefore, it is assumed that in this case U is modified to pair only with A and G (Yokohama and Nishimura 1995; Lim and Curran 2001). The bacterial species, their tRNA numbers, and other information are available as Supplemental material.
Codon usage bias
To measure how codon usage deviates from random values we used ENC′, a variant of the Effective Number of Codons index (ENC) (Wright 1990) that takes nucleotide composition into account (Novembre 2002). ENC′ varies between 20 (only one codon used per amino acid) and 61 (all synonymous codons used at the same frequency, given G+C composition). To avoid problems associated with small sequences, ENC′ was computed on the concatenation of all sequences for a given genome and for the genes coding for its ribosomal proteins separately. Qualitatively similar results were obtained using Karlin and Mrazek's B*(a) (Karlin and Mrazek 1996). We computed CAI values for all genes in each genome (Sharp and Li 1987), using ribosomal proteins as a reference for highly expressed genes, which correlates well with expression data (Coghlan and Wolfe 2000; Jansen et al. 2003).
Models
We considered three models to explain the association between the frequency of anticodons and codon usage bias. A given anticodon is present in Na copies in the genome, which corresponds to the Na tRNA genes in the genome harboring such an anticodon. A given codon is present in a relative frequency Fc,all in all genes and Fc,rp in ribosomal proteins. Fc,all and Fc,rp vary between 0 and 1 (respectively, absent codon and only codon used for a given amino acid). Fc,diff is the difference of the relative frequency of one codon in ribosomal proteins and all genes (Fc,diff = Fc,rp – Fc,all). Fc,diff is close to –1 if the codon is very frequent in most genes and nearly absent in the ribosomal genes, and is close to +1 in the inverse situation. It is important to notice that an anticodon can read several codons (see Table 2) and that a codon can be read by several different anticodons. This complicates the association between tRNA concentration and codon usage bias in genes. For example, suppose that there are four tRNA genes for Glycine in a genome, one for each anticodon. A GGU codon can be recognized by anticodons ACC, GCC, and UCC, but a GGA codon can only be recognized by anticodons ACC and UCC. The readability of a codon (Rc) is the number of anticodons than can read it given the tRNA gene pool (e.g., three and two in the preceding example). Naturally, if there is only one anticodon that can read all codons, then all codons have similar Rc values.
In the frequency model, the most frequent codon is the one that can be decoded by the largest number of aa-tRNAs in the cell. Because tRNA concentrations are unavailable in most cases, we consider that the aa-tRNA concentration is proportional to the number of each tRNA in the genome. This is in reasonable agreement with experimental data (Dong et al. 1996; Percudani et al. 1997; Kanaya et al. 1999). The most frequent codon for an amino acid matches the model if it corresponds to the codon that maximizes the number of anticodons with which it can interact. That is, the model is matched for a given amino acid in a genome if the codon having the highest Fc,all (or Fc,diff, depending on the analysis) for the amino acid corresponds to the codon which is most frequently read by the anticodons available, that is, to the most readable codon (highest Rc). The observed value is the sum of the amino acids that match the model (ranging from two to 20, because Met and Trp have only one codon). The expected value for the model under random codon usage is the sum of the number of most readable codons (if there is more than one) divided by the number of codons for each amino acid. The significance of the model is given by the ratio observed/expected (O/E), and is tested with a Wilcoxon test on the set of 102 genomes.
The perfect match model predicts that the most abundant codon (highest Fc,all or Fc,diff, depending on the analysis) is the one making a perfect codon:anticodon interaction with the most abundant anticodon (highest Na for a set of synonymous tRNA genes) (Ikemura 1981). The significance of the model is given by the sum of the number of amino acids for which this is verified divided by the expected value (O/E). For this, we assume that the perfect match is always the canonical Watson–Crick pairing, and that modified residues do not change the perfect match. This is in agreement with the literature for both Inosine (best match U) and modified U nucleosides (best match A) and Q (best match C) (Yokohama and Nishimura 1995).
The stability model predicts that for S = {G,C} and W = {A,U}, codons starting with S1S2 should have a W3 base and inversely, codons starting with W1W2 should be followed by S3 (Grosjean and Fiers 1982). For the other cases, the model has no predictions. The observed value is the difference between the numbers of amino acids with the most frequent codons respecting the model (i.e., S1S2W3 or W1W2S3) against those that do not (i.e., S1S2S3 and W1W2W3). Because in this case the expected value is 0, O/E stands for 2fWWS,SSW/(fWWS,SSW + fWWW,SSS) and varies between 0 and 2, and the model has a significant fit if O/E > 1, as for the other models.
Acknowledgments
I thank all the researchers who have shared references or unpublished data on optimal generation times of bacteria, and Antoine Danchin, Isabelle Gonçalves, Hugo Naya, and David Ardell for discussions and criticisms.
E-mail erocha@pasteur.fr; fax 33 1 44 27 6312.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2896904. Article published online before print in October 2004.
Footnotes
[Supplemental material is available online at www.genome.org.]
References
- Akashi, H. 1994. Synonymous codon usage in Drosophila melanogaster: Natural selection and translational accuracy. Genetics 136: 927-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi, H. and Gojobori, T. 2002. Metabolic efficiency and amino acid composition in the proteomes of Escherichia coli and Bacillus subtilis. Proc. Natl. Acad. Sci. 99: 3695-3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson, S.G.E. and Kurland, C.G. 1990. Codon preferences in free-living microorganisms. Microbiol. Rev. 54: 198-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, P., Baumann, L., Lai, C.-H., and Rouhbakhsh, D. 1995. Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular symbionts of aphids. Annu. Rev. Microbiol. 49: 55-94. [DOI] [PubMed] [Google Scholar]
- Berg, O.G. and Kurland, C.G. 1997. Growth rate-optimised tRNA abundance and codon usage. J. Mol. Biol. 270: 544-550. [DOI] [PubMed] [Google Scholar]
- Bernardi, G., Olofsson, B., Filipski, J., Zerial, M., Salinas, J., Cuny, G., Meunier-Rotival, M., and Rodier, F. 1985. The mosaic genome of warm-blooded vertebrates. Science 228: 953-958. [DOI] [PubMed] [Google Scholar]
- Bulmer, M. 1991. The selection-mutation-drift theory of synonymous codon usage. Genetics 129: 897-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C., Campbell, A.M., and Karlin, S. 1992. Over- and under-representation of short oligonucleotides in DNA sequences. Proc. Natl. Acad. Sci. 89: 1358-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, S.L., Lee, W., Hottes, A.K., Shapiro, L., and McAdams, H.H. 2004. Codon usage between genomes is constrained by genome-wide mutational processes. Proc. Natl. Acad. Sci. 101: 3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiapello, H., Olivier, E., Landès-Devauchelle, C., Nitschké, P., and Risler, J.-L. 1999. Codon usage as a tool to predict the cellular location of eukaryotic ribosomal proteins and aminoacyl-tRNA synthetases. Nucleic Acids Res. 27: 2848-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan, A. and Wolfe, K.H. 2000. Relationship of codon bias to mRNA concentration and protein length in Saccharomyces cerevisiae. Yeast 16: 1131-1145. [DOI] [PubMed] [Google Scholar]
- Curran, J.F. and Yarus, M.Y. 1989. Rates of aa-tRNA selection at 29 sense codons in vivo. J. Mol. Biol. 209: 65-77. [DOI] [PubMed] [Google Scholar]
- Dong, H., Nilsson, L., and Kurland, C.G. 1996. Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol. 260: 649-663. [DOI] [PubMed] [Google Scholar]
- Duret, L. 2000. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 16: 287-289. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 12: 640-649. [DOI] [PubMed] [Google Scholar]
- Ehrenberg, M. and Kurland, C.G. 1984. Cost of accuracy determined by a maximal growth rate constraint. Quart. Rev. Biophys. 17: 45-82. [DOI] [PubMed] [Google Scholar]
- Elf, J., Nilsson, D., Tenson, T., and Ehrenberg, M. 2003. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science 300: 1718-1722. [DOI] [PubMed] [Google Scholar]
- Francino, M.P. and Ochman, H. 2001. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol. Biol. Evol. 18: 1147-1150. [DOI] [PubMed] [Google Scholar]
- Grantham, R., Gautier, C., Gouy, M., Mercier, R., and Pavé, A. 1980. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 8: r49-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean, H. and Fiers, W. 1982. Preferential codon usage in prokaryotic genes: The optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene 18: 199-209. [DOI] [PubMed] [Google Scholar]
- Ikemura, T. 1981. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J. Mol. Biol. 146: 1-21. [DOI] [PubMed] [Google Scholar]
- ———. 1985. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 2: 13-34. [DOI] [PubMed] [Google Scholar]
- Jansen, R., Bussemaker, H.J., and Gerstein, M. 2003. Revisiting the codon adaptation index from a whole-genome perspective: Analyzing the relationship between gene expression and codon occurrence in yeast using a variety of models. Nucleic Acids Res. 31: 2242-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya, S., Yamada, Y., Kudo, Y., and Ikemura, T. 1999. Studies of codon usage and tRNA genes of 18 unicellular organisms and quantification of Bacillus subtilis tRNAs: Gene expression level and species-specific diversity of codon usage based on multivariate analysis. Gene 238: 143-155. [DOI] [PubMed] [Google Scholar]
- Kano, A., Andachi, Y., Ohama, T., and Osawa, S. 1991. Novel anticodon composition of transfer RNAs in Micrococcus luteus, a bacterium with a high genomic G+C content. J. Mol. Biol. 221: 387-401. [DOI] [PubMed] [Google Scholar]
- Karlin, S. and Mrazek, J. 1996. What drives codon choices in human genes? J. Mol. Biol. 262: 459-472. [DOI] [PubMed] [Google Scholar]
- Lim, V.I. and Curran, J.F. 2001. Analysis of codon:anticodon interactions within the ribosome provides new insights into codon reading and the genetic code structure. RNA 7: 942-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobry, J.R. 1996. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol. Biol. Evol. 13: 660-665. [DOI] [PubMed] [Google Scholar]
- Lowe, T. and Eddy, S. 1997. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25: 955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn, D.J., Singer, G.A., and Hickey, D.A. 2002. Synonymous codon usage is subject to selection in thermophilic bacteria. Nucleic Acids Res. 30: 4272-4277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marck, C. and Grosjean, H. 2002. tRNomics: Analysis of tRNA genes from 50 genomes of Eukarya, Archaea and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA 8: 1189-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAdams, H.H., Srinivasan, B., and Arkin, A.P. 2004. The evolution of genetic regulatory systems in bacteria. Nat. Rev. Genet. 5: 169-178. [DOI] [PubMed] [Google Scholar]
- Médigue, C., Rouxel, T., Vigier, P., Henaut, A., and Danchin, A. 1991. Evidence for horizontal gene transfer in E. coli speciation. J. Mol. Biol. 222: 851-856. [DOI] [PubMed] [Google Scholar]
- Moszer, I., Rocha, E.P.C., and Danchin, A. 1999. Codon usage and lateral gene transfer in Bacillus subtilis. Curr. Opin. Microbiol. 2: 524-528. [DOI] [PubMed] [Google Scholar]
- Muramatsu, T., Nishikawa, K., Nemoto, F., Kuchino, Y., Nishimura, S., Miyazawa, T., and Yokoyama, S. 1988. Codon and amino-acid specificities of a transfer RNA are both converted by a single post-transcriptional modification. Nature 336: 179-182. [DOI] [PubMed] [Google Scholar]
- Muto, A. and Osawa, S. 1987. The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. 84: 166-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre, J.A. 2002. Accounting for background nucleotide composition when measuring codon usage bias. Mol. Biol. Evol. 19: 1390-1394. [DOI] [PubMed] [Google Scholar]
- Osawa, S., Ohama, T., Yamao, F., Muto, A., Jukes, T.H., Ozeki, H., and Umesono, K. 1988. Directional mutation pressure and transfer RNA in choice of the third nucleotide of synonymous two-codon sets. Proc. Natl. Acad. Sci. 85: 1124-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa, S., Jukes, T.H., Watanabe, K., and Muto, A. 1992. Recent evidence for evolution of the genetic code. Microbiol. Rev. 56: 229-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percudani, R. and Ottonello, S. 1999. Selection at the wobble position of codons read by the same tRNA in Saccharomyces cerevisiae. Mol. Biol. Evol. 16: 1752-1762. [DOI] [PubMed] [Google Scholar]
- Percudani, R., Pavesi, A., and Ottonello, S. 1997. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 268: 322-330. [DOI] [PubMed] [Google Scholar]
- Rocha, E.P.C. 2004. The replication-related organisation of the bacterial chromosome. Microbiology 150: 1609-1627. [DOI] [PubMed] [Google Scholar]
- Rocha, E.P.C. and Danchin, A. 2003. Gene essentiality as a determinant of chromosomal organization in bacteria. Nucleic Acids Res. 31: 6570-6577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2004. An analysis of determinants of protein substitution rates in bacteria. Mol. Biol. Evol. 21: 108-116. [DOI] [PubMed] [Google Scholar]
- Rodnina, M.V. and Wintermeyer, W. 2001. Fidelity of aminoacyl-tRNA selection on the ribosome: Kinetic and structural mechanisms. Annu. Rev. BioChem. 70: 415-435. [DOI] [PubMed] [Google Scholar]
- Ruusala, T., Andersson, D., Ehrenberg, M., and Kurland, C.G. 1984. Hyper-accurate ribosomes inhibit growth. EMBO J. 3: 2575-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.M. and Li, W.H. 1986a. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 24: 28-38. [DOI] [PubMed] [Google Scholar]
- ———. 1986b. Codon usage in regulatory genes in Escherichia coli does not reflect selection for “rare” codons. Nucleic Acids Res. 14: 7737-7749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 1987. The codon Adaptation Index—A measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 15: 1281-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.M., Stenico, M., Peden, J.F., and Lloyd, A.T. 1993. Codon usage: Mutational bias, translational selection, or both? Biochem. Soc. Trans. 21: 835-841. [DOI] [PubMed] [Google Scholar]
- Sorensen, M.A., Kurland, C.G., and Pedersen, S. 1989. Codon usage determines translation rate in Escherichia coli. J. Mol. Biol. 207: 365-377. [DOI] [PubMed] [Google Scholar]
- Sueoka, N. 1962. On the genetic basis of variation and heterogeneity of DNA base composition. Proc. Natl. Acad. Sci. 48: 582-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, Y., Gojobori, T., and Nei, M. 2001. ADAPTSITE: Detecting natural selection at single amino acid sites. Bioinformatics 17: 660-661. [DOI] [PubMed] [Google Scholar]
- Thomas, L.K., Dix, D.B., and Thompson, R.C. 1988. Codon choice and gene expression: Synonymous codons differ in their ability to direct aminoacylated-transfer RNA binding to ribosomes in vitro. Proc. Natl. Acad. Sci. 85: 4242-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varenne, S., Buc, J., Lloures, R., and Ladzunski, C. 1984. Translation is a non-uniform process: Effect of tRNA availability on the rate of elongation of the nascent polypeptide chains. J. Mol. Biol. 180: 549-576. [DOI] [PubMed] [Google Scholar]
- Wright, F. 1990. The “effective number of codons” used in a gene. Gene 87: 23-29. [DOI] [PubMed] [Google Scholar]
- Xia, X. 1996. Maximizing transcription efficiency causes codon usage bias. Genetics 144: 1309-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamao, F., Andachi, Y., Muto, A., Ikemura, T., and Osawa, S. 1991. Levels of tRNAs in bacterial cells as affected by amino acid usage in proteins. Nucleic Acids Res. 19: 6119-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. and Swanson, W.J. 2002. Codon-substitution models to detect adaptive evolution that account for heterogeneous selective pressures among site classes. Mol. Biol. Evol. 19: 49-57. [DOI] [PubMed] [Google Scholar]
- Yarian, C., Townsend, H., Czestkowski, W., Sochacka, E., Malkiewicz, A.J., Guenther, R., Miskiewicz, A., and Agris, P.F. 2002. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 277: 16391-16395. [DOI] [PubMed] [Google Scholar]
- Yokohama, S. and Nishimura, S. 1995. Modified nucleosides and codon recognition. In tRNA: Structure, modification and function (eds. D. Soll and U. RajBhandary), pp. 207-223. ASM, Washington.