Abstract
Trypanosoma brucei switches between variant surface glycoproteins (VSGs) allowing immune escape. The active VSG is in one of many telomeric bloodstream form VSG expression sites (BESs), also containing expression site-associated genes (ESAGs) involved in host adaptation. The role of BES sequence diversity in parasite virulence can best be understood through analysis of the full repertoire of BESs from a given T. brucei strain. However, few BESs have been cloned, as telomeres are highly underrepresented in standard libraries. We devised a strategy for isolating the repertoire of T. brucei 427 BES-containing telomeres in Saccaromyces cerevisiae by using transformation-associated recombination (TAR). We isolated 182 T. brucei 427 BES TAR clones, 167 of which could be subdivided into minimally 17 BES groups. This set gives us the first view of the breadth and diversity of BESs from one T. brucei strain. Most BESs ranged between 40 and 70 kb (average, 57 ± 17 kb) and contained most identified ESAGs. Phylogenetic comparison of the cohort of BES promoter and ESAG6 sequences did not show similar trees, indicating rapid evolution most likely mediated by sequence exchange between BESs. This cloning strategy could be used for any T. brucei strain, facilitating research on the biodiversity of telomeric gene families and host-pathogen interactions.
The African trypanosome Trypanosoma brucei has a haploid genome size about threefold that of Saccaromyces cerevisiae distributed over 11 megabase chromosomes and up to 100 intermediate and minichromosomes (Borst et al. 1982; Melville et al. 1998, 2000). African trypanosomes cause African sleeping sickness, whereby the parasites evade antibody-mediated destruction by using a sophisticated system of antigenic variation (for review, see Barry and McCulloch 2001; Vanhamme et al. 2001; Donelson 2003). Each parasite is shielded by a variant surface glycoprotein (VSG) coat encoded by a VSG transcribed from one of ∼20 telomeric bloodstream form VSG expression sites (BES). VSG switching can involve DNA rearrangements inserting a new VSG into the active BES, or transcriptional switching between different BESs (Borst and Ulbert 2001).
BESs are large polycistronic transcription units containing an assortment of expression site-associated genes (ESAGs) in addition to the VSG. Many ESAGs appear to encode surface molecules, and at least some of these genes play a role in host adaptation (Bitter et al. 1998; Pays et al. 2001). T. brucei has an unusually large host range, infecting a broad range of African mammals (Ashcroft 1958; Geigy et al. 1971). Some of the ESAGs encode products that allow the parasite to adapt to a variable bloodstream environment. The serum resistance-associated (SRA) gene confers resistance to lysis by human serum (Vanhamme et al. 2003) and is present in a truncated BES (Xong et al. 1998). In addition, the trypanosome transferrin receptor subunits are encoded by ESAG6 and ESAG7 (Schell et al. 1991; Ligtenberg et al. 1994; Steverding et al. 1995). T. brucei appears to have adapted to the difficulties of internalizing polymorphic transferrins present in different species of host by having nonidentical transferrin receptors encoded in different BESs (Bitter et al. 1998). Switching between BESs allows the trypanosome to alternate between transferrin receptors with varying affinities for the different transferrins present in a range of host species (Salmon et al. 1997; Bitter et al. 1998; Gerrits et al. 2002).
The role of BES sequence diversity in parasite–host interaction can only be understood in the context of the complete cohort of BES sequences from a given T. brucei strain. However, as BESs are all located at telomeres, they have proven difficult to characterize. Most of the ESAGs that they contain are members of highly similar multicopy gene families, making chromosome walking from the telomeric VSG an extremely laborious means of cloning an entire BES (Kooter et al. 1987; Pays et al. 1989). BES sequences are underrepresented in large insert T. brucei bacterial artificial chromosome (BAC) libraries, again presumably because of the difficulties of cloning sequences located at chromosome ends. Screening a highly redundant T. brucei 427 BAC library (RPCI-97 from the BACPAC Resources Centre at http://bacpac.chori.org) with single copy sequences from the 221 or VO2 BESs showed that these clones were more than 10-fold underrepresented. In addition, virtually all of the BES BAC clones isolated contained only partial BESs (Berriman et al. 2002; G. Rudenko, unpubl.).
Despite the importance of BESs for both antigenic variation and host adaptation in T. brucei, only four complete BESs from four different T. brucei strains have been analyzed in detail (Berriman et al. 2002). Although T. brucei 427 is not the genome strain, it is the widest used laboratory strain, and powerful experimental systems have been developed in it, including tetracyclin inducible expression (Wirtz et al. 1999). Tetracyclin inducible RNAi is allowing the systematic analysis of gene function in this strain (LaCount et al. 2002; Morris et al. 2002).
To obtain a complete set of telomeric T. brucei 427 BESs, we have devised a modified transformation-associated recombination (TAR) procedure to construct a telomere specific library in S. cerevisiae. TAR cloning in yeast relies on recombination between a target fragment present in a TAR cloning vector and the same sequences present in heterologous genomic DNA (Larionov et al. 1996a,b). We cotransformed S. cerevisiae with a TAR vector containing a BES promoter sequence, yeast telomere repeats, as well as yeast selectable markers, a centromere, and origin of replication together with T. brucei 427 genomic DNA. Homologous recombination resulted in stable linear half YACs with a yeast telomere on one end, and a T. brucei BES containing telomere on the other. This allowed us to construct a T. brucei 427 telomere-specific library of 182 T. brucei BES TAR clones. Sequence determination of regions from the promoter and ESAG6 allowed typing and sorting of these clones into minimally 17 BES groups. These BES TAR clones were analyzed for size, ESAG composition, and presence of known VSGs. Strikingly, most BESs appeared to contain most known ESAGs. Phylogenetic comparisons of BES promoter and ESAG6 sequences showed no evidence of linkage in the evolution of these sequences within any given BES. This is indicative of rapid sequence exchange between BESs.
Results
Construction of a T. brucei 427 BES containing telomere library
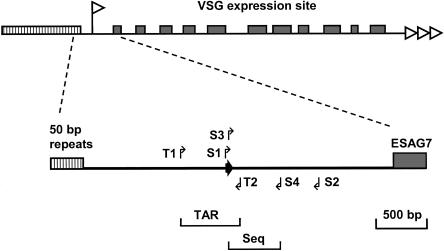
TAR cloning was adapted to facilitate the construction of a library of T. brucei 427 bloodstream form VSG expression site (BES) containing telomeres in S. cerevisiae. The TAR vector pEB4 contains a yeast telomere, centromere, origin of replication (ARS), and positive and negative selectable markers, and uses the T. brucei 427 dominant expression site (DES) promoter (Zomerdijk et al. 1991) as recombinational target (Fig. 1A). Yeast cotransformation of vector pEB4 containing the target and a single yeast telomere located at one end of the linearized vector, and T. brucei 427 genomic DNA resulted in cloning of trypanosome telomeres as linear half YACs (Fig. 1B). In three independent experiments, a total of 2000 transformants were screened by colony-PCR using diagnostic primers for ESAG7 (primers ESAG7a, see Table 1). ESAG7 is thought to be an essential gene present in all T. brucei BESs (Ansorge et al. 1999; Berriman et al. 2002; G. Rudenko, unpubl.). In this primary screen, 182 colonies (9%) were positive for ESAG7.
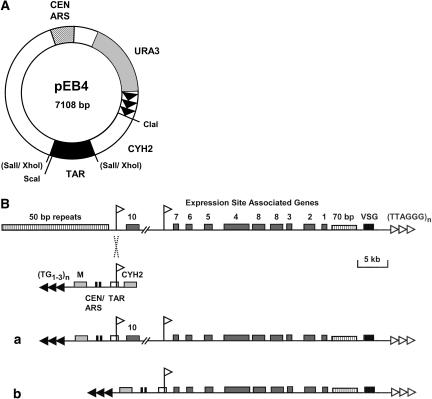
Figure 1.
(A) Schematic of T. brucei BES-specific TAR cloning vector pEB4 constructed from the general TAR vector pEB2 (M. Becker and E.J. Louis, unpubl.). The 560-bp T. brucei TAR target including the BES promoter is indicated as the black box labeled TAR. S. cerevisiae telomere repeats are indicated as black arrows. Further indicated are the yeast centromere and the origin of replication (striped box labeled CEN, ARS), the positive selectable marker gene URA3 (grey box labeled URA3) and the negative selectable marker CYH2 (white box labeled CYH2). Restriction enzyme sites ClaI, used for linearization of the vector prior to transformation; ScaI, used for proper orientation of the TAR fragment; and XhoI/SalI, used for insertion of the TAR fragment, are indicated. The latter were destroyed upon insertion. (B) Schematic for selective isolation of telomeric T. brucei bloodstream form VSG expression sites (BES) via TAR cloning in yeast. At the top of the figure is a schematic of a telomeric BES with duplicated promoters indicated with white flags, various expression site-associated genes (ESAGs) indicated with numbered grey boxes, and the VSG with a black box. Characteristic 50-bp and 70-bp repeat arrays are indicated with hatched boxes, and the T. brucei telomere repeats are indicated with white triangles. The linear S. cerevisiae TAR cloning vector pEB4 is indicated below, with S. cerevisiae telomere repeats indicated with black triangles. The 560-bp T. brucei TAR target fragment, including the BES promoter, is indicated with a white box marked TAR. The TAR vector further includes yeast centromere and ARS sequences (black boxes marked CEN/ARS), a positive marker gene URA3 (grey box labeled M), and a negative marker gene CYH2 (grey box labeled CYH). (a) Product of a recombination between the T. brucei target and an upstream BES promoter, producing a linear telomere clone as half YAC. (b) Product of a recombination between the T. brucei target and a downstream BES promoter, producing an ∼13-kb-shorter linear telomere clone.
Table 1.
Primers used in VSG expression site analysis.
| Name | Sense primer | Antisense primer | Acc. no. | Coordinates | |
|---|---|---|---|---|---|
| TAR T1/T2 | TACGCGTCGACTGAGGTAAGGAATATCGACG | TACGCGTCGACATAAATCGAATACGG | AL671259 | 71184-71742 | |
| ESAG7a | TTGGTTTGTGCTGTTGGCTC | CACTTCATGGAGTTGCAAGC | AL671259 | 74776-75346 | |
| ESP S1/S2 | ATATCCCTATTACCACACCA | CAATACCCACCCATACACAG | AL671259 | 71589-74112 | |
| ESP S3/S4 | CAGGGGAGGTTATTACAGAA | CCGTTTGGGCCATTTACACA | AL671259 | 71619-73768 | |
| ESAG6amp | CAGTATTGAGGAATGAGTTTA | GTTCACTCACTCTCTTTGACA | AL671259 | 77401-78159 | |
| ESAG6seq | CCAGCAGGAGTTGGAGGAAAT | CTCTGTAGCATCACCGTATTC | AL671259 | 77426-77918 | |
| ESAG1-1 | TTGAAATTGTGGAATTGGTTG | GACATCCTCCCACACTGTCC | AL671259 | 118926-119416 | |
| ESAG1-2 | ATGCGGAAGTCAAAAGGTTG | CCTCGGTCTTGAGTTTAGTGG | AL671259 | 119449-119882 | |
| ESAG1-3 | TCTCCGTTGTCTTTCGGATG | GAAATTGCTATGGACTTCGTTG | AL671259 | 119053-119845 | |
| ESAG2-1 | GCGAGTTACGAATGGTAGGC | GTCTCCGTAAGAACCGCAAC | AL671259 | 116570-117071 | |
| ESAG2-2 | CCGCTGGGGACTATAACCTAAC | ATGACTGTCAGCCTCCTTGG | AL671259 | 117205-117689 | |
| ESAG2-3 | CCTTTAAGAATCGTGAGAAGGA | TAATCACATCCCATGCCTCA | AL671259 | 116707-117390 | |
| ESAG3-1 | TGAAATTTGGTACCTCGAGAGC | CCAACTGAGGGGCATAACAC | AL671259 | 99689-100189 | |
| ESAG3-2 | ATCACAAGGAGCATGTGCAG | CGAAATTCCCACTTCTCTCC | AL671259 | 100208-100730 | |
| ESAG3-3 | GCAGAATACTGCGGAAAATGA | TCACCCCACTACTGGGAATAG | AL671259 | 100077-100555 | |
| ESAG4-1 | TCCACTTTTATCCGACAATGG | GGATTGGTGCCTGTGAAAAG | AL671259 | 88548-89025 | |
| ESAG4-2 | TTGAGGGAGAGTTGATGGTG | GTTCGCTGCCTCAGCTATG | AL671259 | 89167-89663 | |
| ESAG4-3 | GGCTTCAACGCCTCACTG | GTCCTTCGCAAGCGGATTG | AL671259 | 88308-89057 | |
| ESAG5-1 | TGTTGTATCATCGGCTCTGG | GGCATCTTTGATTTCCCTTC | AL671259 | 81975-82478 | |
| ESAG5-2 | GATAAGGATGTGCTTCCCAAG | CAGGTCCACGGCAACTTC | AL671259 | 82513-83001 | |
| ESAG5-3 | ATTCAAGAGGCGGCTGTTAC | AGCACGACCAGTTCAAGTCTCAT | AL671259 | 82056-82670 | |
| ESAG6-1 | ACCGCCGCTAATAAAGTGTG | GGAGTTGCAAGCTTTTCTGC | AL671259 | 77205-77691 | |
| ESAG6-2 | GCAGAAAAGCTTGCAACTCC | AACCTCTTTCTGCCCTATGC | AL671259 | 77672-78077 | |
| ESAG6-3 | CATTCCAGCAGGAGTTGGAGG | TTGTTCACTCACTCTCTTTGACAG | AL671259 | 77422-78161 | |
| ESAG7-3 | CATTCCAGCAGGAGTTGGAGG | TTAGGTTCCCCGCCACAC | AL671259 | 75069-75527 | |
| ESAG8-1 | CAGATGGCGAGGAAACTAGG | CACCTTCAGCTTCCCATCAG | AL671259 | 97088-97612 | |
| ESAG8-2 | ACTGCTATTGGGGGTGTGAG | CAATCCACTCAGGCTTGTTATTC | AL671259 | 97649-97143 | |
| ESAG8-3 | GTATCGGAGGGAGACGGAAG | CTGCTTCCCATCAGGATGAA | AL671259 | 96981-97604 | |
| ESAG10-2 | CACAAGCTCACGAAGCTGAG | ACCAAACCCAACTGACAAGC | AL670322 | 25506-26093 | |
| ESAG10-3 | TATGATTGGGGCTGTTGGTGGTGT | TAGACGCGCCGCTGGTATCA | AL670322 | 26752-27249 | |
| ESAG11-1 | GGGGCTGAAACATTATGTAACC | GTCGTCATCGACACTTCTCTTG | AJ239059 | 141-1055 | |
| ESAG11-2 | AATAGTGCAGGGTTTGGAGAA | CCATTTGCAACCTCTTCCGTA | AJ239059 | 287-766 | |
| VSG 221 | GCATGCCTTCCAATCAGGAGGC | GCTGTATCGGCGACAACTGCAG | X56762 | 19-558 | |
| VSG 121 | CAATAGCAGCCGATCTAGCC | TGATGGTCGCTAAGTTGTCG | X56764 | 317-808 | |
| VSG V02 | CAGCGGCTGTACATACTAAC | TAGCGTGCACATTTTCGCG | G. Rudenko, and P. Borst, unpubl. | ||
| VSG 1.8 | AAGGAGACACAATCGCCAAG | CCGTCTTTGACGCAATAGTTC | N. Aitcheson, P. Borst, and G. Rudenko, unpubl. | ||
| VSG 800 | ACTACAGACCGCCGACAGTATC | GGGCTACTAGCAGATTGCATTT | S. Hajduk, pers. comm. | ||
| VSG bR-2 | GTGAAGTCCACAAAAGACAGTGG | CGCGAGCTTGTATTCTTTTAGC | G. Cross, pers. comm. | ||
| VSG T3 | GGCACAACTTTGCTACACAATG | GGGGTCATTTATTGTCTCAAGC | G. Cross, pers. comm. | ||
| VSG 224 | CGATGACGTCAATCCAGATG | CCGTTGGTGTCGTGTCTTC | G. Cross, pers. comm. | ||
| VSG JS1 | CCATAAAGCACGAAGCCTGGAAG | TAGTCCGCATCTGCCTAGTCCTG | N. Aitcheson and G. Rudenko, unpubl. | ||
Analysis of T. brucei telomere clones
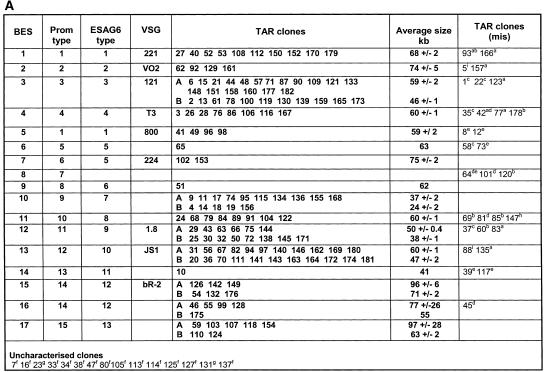
The 182 ESAG7–positive telomere clones were subjected to several analyses in order to categorize them. A combination of size and number of bands on pulsed field gels, along with sequences of the promoter regions adjacent to the target sequence and ESAG6, as well as PCR assessment of ESAG and VSG composition allowed us to categorize 167 of these clones into potentially 17 BES types (Fig. 2A). Thirteen clones had either had no visible, faint, multiple bands, or a <20-kb YAC and for a further two clones promoter and ESAG6 sequence could not be obtained (category “uncharacterized clones”) (Fig. 2A). Thirty of the clones either had missing information or were consistent with some form of rearrangement occurring during the cloning procedure (category TAR clones [mis]) (Fig. 2A). The 137 clones with complete and consistent information fall into 17 BES classes (category “TAR clones”) (Fig. 2A). Details of these analyses follow.
Figure 2.
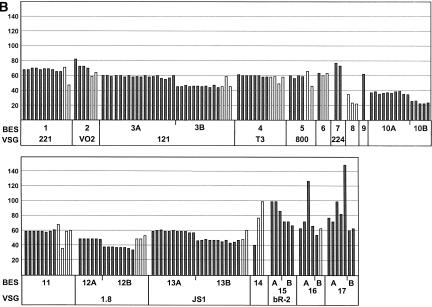
(A) Analysis of 182 T. brucei telomere clones, 167 of which are categorized into BES sets by insert size, BES promoter, ESAG6, and VSG type. TAR clones were categorized into BES sets (first column) primarily according to their BES promoter sequence type (second column) (for sequence alignment, see Fig. 5A). A variable region of ESAG6 was also sequenced, and the ESAG6 sequence type is indicated (for sequence alignments, see Fig. 5B). Presence of several known VSGs (indicated in column 4) was analyzed by PCR (for primers, see Table 1). Clones marked in bold have the correct promoter, ESAG6 and, where indicated, VSG type for the BES set (column marked TAR clones). The average size in kilobases with standard deviant (±SD) for the different TAR clones is indicated. Different size classes form BES subsets, marked with A and B, and correspond to the presence or absence of duplicated BES promoters (see Fig. 1B). Clones marked with A and B were checked by PCR for ESAG10 to confirm that they indeed had a duplicated BES promoter as expected. Clones in the column marked TAR clones (mis) have either missing or different BES promoter, ESAG6 or VSG types or combinations of mismatches indicated. These clones were not included in the size estimations. Clones in the last row marked “uncharacterized clones”, could not be categorized due to the problems indicated. aNo VSG if expected; bESAG6 type different; cmissing promoter type; dmissing ESAG6 type; edifferent known VSG; fmissing, faint or too small (<20 kb) or multiple band on PFGE;, gpromoter, ESAG6 and VSG type not obtained; hpromoter sequence with one mismatch; and ipromoter sequence with multiple mismatches. (B) Size distribution of the T. brucei BES TAR clones analyzed. The different BES sets and VSG type if known are indicated below. The size of the individual TAR clones described in A is indicated in kilobases. Clones that have the correct promoter sequence, ESAG6 and VSG type (TAR clones marked in bold in panel A) are indicated with dark bars. TAR clones with mismatched sequences or VSG type (TAR clones [mis] in A) are indicated with white bars. Some BES sets fell into size classes differing by ∼13 kb, which are indicated with A and B. PCR for ESAG10 confirmed its presence in sets A and its absence in sets B which is compatible with the presence of a duplicated promoter in these BESs (Fig. 1B).
Insert size analysis
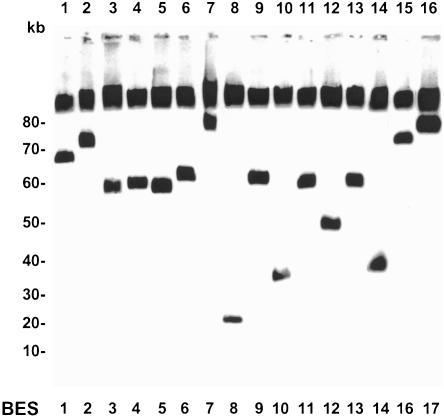
The sizes of all 182 telomere clones were determined. Genomic DNA embedded in agarose plugs was subjected to pulsed field gel electrophoresis (FIGE for clones up 80 kb or CHEF for clones <80 kb), and analyzed by Southern blotting using vector pEB2 as a probe. An example for this is shown in Figure 3. Insert sizes were found to range from ∼10 to 150 kb (data not shown) with an average size of 57 ± 17 kb (Fig. 2A,B). Eighty-four percent of the telomere clones in this library had inserts of 40–80 kb, which is consistent with the size of most T. brucei BESs (Berriman et al. 2002). Insert sizes >80 kb were detected in nine clones, seven of which fell into just three BES sets (BES 15, 16, and 17). Analysis of T. brucei TAR clones after prolonged propagation indicated that they were stably maintained in yeast (data not shown; M. Becker and E.J. Louis, unpubl.).
Figure 3.
Size determination of the T. brucei telomere clones. Genomic DNA of telomere clones was separated by FIGE gel electrophoresis. A representative telomere clone for the different ES types (only A sets) is shown, with the ES type indicated underneath. The gel was blotted and hybridized with vector pEB2 as probe. In addition to the telomere clone, this probe also hybridizes with yeast chromosomal DNA that is not separated under these conditions. Lanes are shown above, and size markers are indicated on the left in kilobases.
Some T. brucei 427 BESs like the DES (Gottesdiener et al. 1991, 1992) are known to contain duplicated BES promoters. In accordance with that, 98 telomere clones fell into two size categories (indicated with A and B in Fig. 2) differing by ∼13 kb, the size of the segment containing the duplicated BES promoter. This is to be expected, depending on whether the upstream or the downstream promoter recombines with the target in vector pEB4 (Fig. 1B). In addition to the DES (BES 3), six additional BES sets also contained clones falling into two size classes (BES10, 12, 13, 15–17). By using PCR to detect the presence of ESAG10, which is expected to be located between duplicated BES promoters (Gottesdiener et al. 1991), clones in all BES sets A were all found to contain ESAG10, and clones in all BES sets B did not. This is consistent with the presence of a duplicated BES promoter in the larger clones (Fig. 2B).
BES promoter, ESAG6, and VSG type analysis
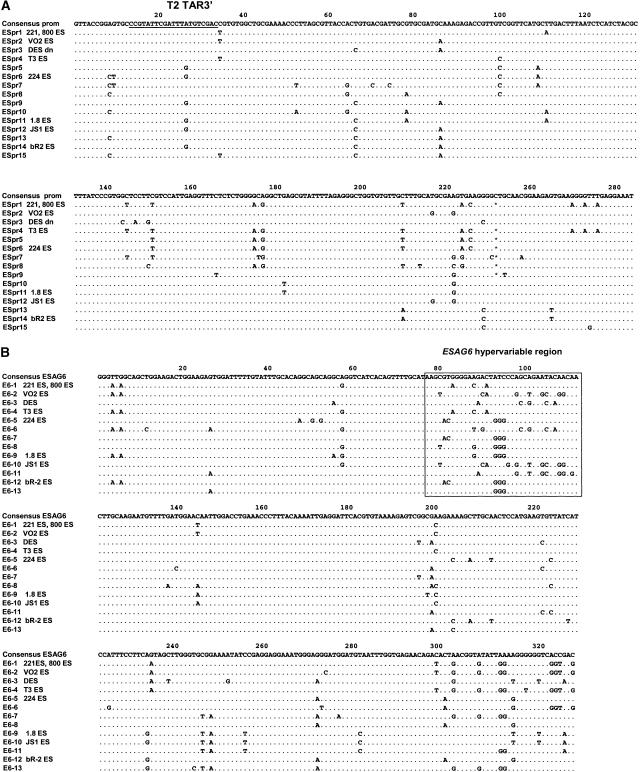
To categorize the telomere clones into sets corresponding to a particular BES, we determined their BES promoter type and their ESAG6 type, and tested for a range of known VSGs of which nine were identified (Fig. 2A). The downstream BES promoter was selectively PCR amplified by using primers S1 and S2 (Fig. 4). This strategy would only generate sequence from a downstream BES promoter, even if the telomere clones had duplicated promoters. Sequence was obtained from each strand using the nested primers S3 and S4 (for primers used, see Fig. 4; Table 1). Single nucleotide polymorphisms (SNPs) allowed the categorization of these clones into 15 different BES promoter types (Figs. 2A, 5A). The telomere clones were also sequenced over a variable region of ESAG6, including the hypervariable region identified by Zomerdijk et al. (1990; Fig. 5B). Alignment of the ESAG6 sequences showed thirteen different ESAG6 types in T. brucei 427 (Figs. 2A, 5B). The number and sequence of ESAG6 variants found is compatible with those found in an independent analysis of T. brucei 427 ESAG6 containing BAC clones (H. van Luenen and P. Borst, pers. comm.). We have identified one ESAG6 variant (ESAG6 type 6) that was not picked up in their study. The analysis of van Luenen and Borst picked up one ESAG6 variant, which was not found in our analysis. However, the ESAG6 region analyzed in the two studies was not completely overlapping, making the comparison only indicative. Most of the multiple independent clones from the same BES with consistent sequence and VSG composition are similar in size, indicating general stability during the cloning procedure (Fig. 2B).
Figure 4.
Schematic of the T. brucei BES promoter region between the 50-bp repeat arrays (hatched box) and the most upstream of the expression site associated genes (ESAG7). The top schematic of a VSG expression site is labeled according to Figure 1B, with the promoter indicated with a white flag. In the expanded section between the 50-bp repeat arrays and ESAG7, the primers used to PCR amplify the 560-bp TAR target fragment are indicated with arrows T1 and T2. The BES promoter is indicated with a dark arrow. Initial sequence characterization of the telomere clones obtained involved PCR amplification between S1 and S2, and subsequent sequencing of each strand of the PCR product using nested primers S3 and S4 (for primer used, see Table 1).
Figure 5.
(A) Sequence alignment of different polymorphic BES promoter types found in the different T. brucei telomere clones. The number of the sequence type is indicated on the left. The consensus sequence is shown at the top. Similar nucleotides are indicated with dots. The T2 TAR3′ primer indicated is the downstream end of the TAR target fragment. (B) Sequence alignment of the variable region of ESAG6 in the different T. brucei telomere clones. The number of the ESAG6 type is indicated with numbers on the left. The ESAG6 hypervariable region as identified by Zomerdijk et al. (1990) is indicated with a box.
Clone integrity
The sequences of a number of T. brucei 427 VSGs are known. We tested telomere clones across the different BES types with T. brucei 427 VSG specific primers (see Table 1) for the presence of VSGs 221, VO2, 121, T3, 800, 224, 1.8, JS1, and bR-2. The analysis (Fig. 2A) showed that 99 clones had a known VSG. Two BES types with known promoter sequences (221 BES and VO2 BES) indeed had the expected VSG (either VSG 221 in 10 out of 12 clones or VSG VO2 in four out of six). In the other BES sets, we were able to link a given VSG with a pair of promoter and ESAG6 sequence types. The majority of telomere clones within each identified set had the same VSG, and ranged from 29 out of the 32 clones for the VSG 121 (BES 3) expected to be found in the DES, to four out of six clones for the set found to have VSG 800 (BES 5). Based on the number of clones in the known BES sets that had the expected promoter, ESAG6 and VSG type, we estimated that at a minimum 85% (99/117) of all clones are correct, namely, have not undergone undesired recombination. As, in general, all of the clones within a set have the same BES promoter and ESAG6 sequence type, differences in the telomeric VSG are presumably due to recombination between T. brucei telomere ends during the TAR cloning procedure. The eight clones in Figure 2A marked superscript “e” for different known VSG are presumably products of such rearrangement. As VSG is the most telomeric gene, the presence of the correct VSG within a telomere clone is a good indication that the clone has not exchanged sequences with other T. brucei telomeres. However, in some cases, the possibility that a rare combination of promoter type, ESAG6 type, and VSG represents a different BES type cannot be excluded. Sequence analysis of several clones from these sets is required to decide if these clones contain rearranged or separate BESs.
Cloning bias of T. brucei BESs
The T. brucei 427 BES library is comprised of multiple independent clones representing at least 17 different T. brucei chromosome ends. The representation of chromosome ends is fairly well distributed with 15 BES promoter types targeted several times. There is some selectivity toward the sequence used as a target. The DES BES type BES3 is particularly well represented with 30 clones, presumably as sequences from the DES promoter were used for the TAR target fragment. However, these are <20% of the clones. Despite the ∼5% divergence at non-DES promoters (Fig. 5A), there was not a great shortfall in recovery of other BES recombinants. Based on mitotic recombination studies in yeast, >95% of the clones would be expected to be BES3 if TAR cloning was susceptible to the mismatch repression of recombination (Datta et al. 1996). Similar requirement for homology is seen in gene targeting in T. brucei (Blundell et al. 1996). Recombination during TAR cloning, however, is relatively insensitive to mismatches, as demonstrated by using Alu repeats, which are shorter (∼300 bp) than the target used here (560 bp) and more divergent (85% identity on average) (Larionov et al. 1996b; Noskov et al. 2003b). The distribution of the non-BES3 clones is similar to that of random telomere targeting in yeast, in which there are imperfect repeats (Louis and Borts 1995).
ESAG composition
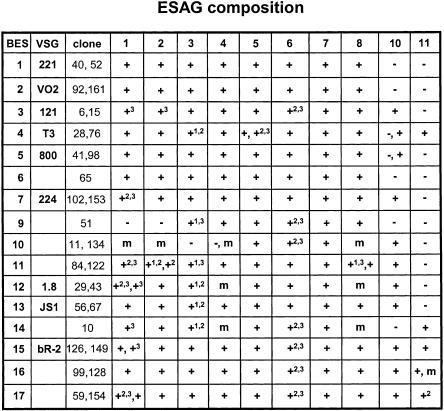
T. brucei BESs contain a range of different genes known as expression site-associated genes (or ESAGs) in addition to the VSG. Different ESAGs are present as similar polymorphic variants in different BESs, and most of the BESs contain most of the ESAGs (Berriman et al. 2002). We performed diagnostic PCR reactions for ESAGs1–8, 10, and 11 using two or three different primer sets per ESAG corresponding to different regions of the gene (for primers used, see Fig. 6; Table 1). We were not able to design diagnostic primers for ESAG9 due to the degree of sequence divergence in this family (K. Matthews, pers. comm.). ESAG1 to 8 were found in most of the BES types. Variability in the PCR results between different ESAG primer sets was presumably due to difficulties in PCR amplifying polymorphic sequence types. Some BES sets (BES10, 12, 14) gave inconclusive results (indicated with “m” in Fig. 6) Occasionally there was variability between the clones analyzed per set. Several clones in BES4, 5, 7, and 11 tested positive for ESAG10 without indication for promoter duplication (similar sized clones in each set), as was observed for other BES sets that tested ESAG10 positive. In these cases ESAG10 may occur in other BES locations. Analysis of the complete repertoire of BESs within the T. brucei 427 strain showed that, surprisingly, most BES types contained most of the ESAGs that have been identified until now. This could indicate a selective advantage for the maintenance of the complete cohort of ESAG types within the individual BES transcription units.
Figure 6.
ESAG composition of selected telomere clones per BES type. Representative telomere clones (indicated in the third column) were analyzed by PCR using three primer sets per ESAG to minimize problems associated with analyzing polymorphic gene families with the exception of ESAG10 and ESAG11, in which two primer sets were used (for ESAG primers used, see Table 1). Results are indicated with + if all ESAG primer sets gave a positive result. If a subset of the primer sets gave positive results, these are indicated with a superscript number (1 indicates first primer set; 2, second primer set; 3, third primer set) (see Table 1). If the result was inconclusive, that is, after three repetitions of the PCR, mixed positive, negative, or weak positive results were obtained, this is indicated with m. Negative PCR results for all three primer sets are indicated with –. If the TAR clones analyzed for a given BES set differed, a comma separates the two results.
Expression site sequence evolution
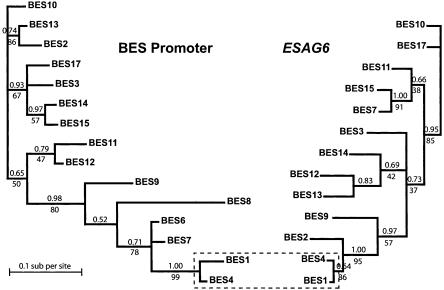
We compared the relative degrees of relatedness of the BES promoter and ESAG6 sequence types within the T. brucei 427 BES cohort by using Bayesian analysis (Fig. 7). The phylogenetic trees inferred by using these two sequence types were not similar in topology, indicating separate evolution of these two sequence types within T. brucei BESs. Only one node was common to BES promoter and ESAG6 trees (BES1 and BES4). This loss of linkage between promoter and ESAG6 evolution within a given BES reflects rapid sequence exchange between BESs. This degree of dissimilarity between tree topologies is particularly striking with regard to the proximity between these two sequence types within BESs. Interestingly, trees inferred by using either the 5′ or 3′ halves of the ESAG6 sequences were also dissimilar in topology (three of 11 nodes common to both trees, although with expected lower support for most nodes) (data not shown), suggesting that sequence exchange is rapid compared with accumulation of point mutations, even when considering directly abutting sequences.
Figure 7.
Phylogenetic analysis of BES promoter and ESAG6 sequences. The trees shown were calculated by Bayesian inference. Posterior probabilities (the proportion of time any given topology was visited during tree inference) are shown above branches. Support for the Bayesian topology from maximum parsimony methods is indicated by bootstrap values (%) under branches. Only one node of the overall topology is common to both trees (boxed).
Discussion
We have devised a means of selectively isolating what we believe is almost the entire set of polymorphic BES-containing telomeres from a given T. brucei strain as linear YACs using TAR cloning in yeast. We have constructed a T. brucei 427 BES library of 182 TAR clones that were characterized by size and 167 which were subdivided in sets according to BES promoter and ESAG6 sequence type. We found 15 BES promoter sequence variants and 13 ESAG6 sequence variants, resulting in a conservative estimate of 17 BESs in the T. brucei 427 strain. Most of the BES TAR clones had insert sizes of between 40 and 80 kb, which is compatible with the size of most known BESs (Berriman et al. 2002). In the cases in which the telomeric VSG was known, PCR across the BES sets with VSG-specific primers showed that most of the clones of a given set indeed had the correct VSG. Once cloned, the TAR clones appeared stable over hundreds of generations in yeast.
The frequency of artifacts such as chimeras and deletions is low in YACs generated by TAR cloning compared with standard methods (Zeng et al. 2001). Based on the information on BES promoter type, ESAG, and VSG content for each clone, we estimate that at least 85% of clones in this library represent chromosome ends that were not rearranged during the TAR cloning procedure. However, there are multiple examples of trimolecular TAR recombinants with the TAR vector recombined with a particular BES promoter and ESAG6 sequence type, but with a different VSG than expected at the other end of the YAC. The most likely explanation for this is a double recombination event between multiple T. brucei telomeres in the same transformant. However, in most cases the majority of the telomere clones in each BES set were the same size and had the proper VSG when this was known. This indicates that although multiple recombination events are a problem, this can be overcome by choosing the predominant type for further analysis. We did not obtain evidence for posttransformation rearrangements or deletions occurring, as the telomere clones remained stable in size and ESAG composition even during amplification in yeast. Sequence analysis of duplicate clones from different BES sets (project in collaboration with the Sanger Institute) will give us further information about the relative integrity of the different telomere clones.
Although most of the BESs appeared to be 40–80 kb, two BES types were much smaller in size: BES8 and BES10. The BES8 set (with an average size of 27 ± 6 kb) contains at least one rearranged clone, and the clones each contained different ESAG6 sequences. They may all be rearranged, or perhaps there is more than one truncated BES with this unique promoter sequence in 427. BES10 clones fall into two sets that differ by 13 kb, depending on the presence of a duplicated promoter as confirmed by ESAG10 PCR. BES10 appears to be severely truncated, and the shorter version is only ∼24 kb. A truncated BES has also been found in the T. brucei ETat1 strain, in which the ETat1.2CR BES was found to contain only ESAG5, 6, and 7 in addition to the SRA gene conferring human serum resistance. However, truncated BESs appear to be an unusual situation in T. brucei.
Judging from ESAG composition as determined by PCR using ESAG specific primers across the different telomere clones, most BESs within the cohort of T. brucei 427 BESs contain most of the ESAGs. This could indicate that a full complement of ESAGs is useful for T. brucei 427 even under laboratory culturing conditions in vitro or in laboratory rodents. Alternatively, the relatively short period that these parasites have been in culture has not been long enough to lead to loss of nonessential BES sequences. There is now a need to compare BES diversity between the cohort of T. brucei 427 BESs and BESs present in T. brucei strains that have not undergone extensive culturing in the laboratory. The experimental procedures described here present a firm basis for doing so.
Have we cloned all of the T. brucei 427 BESs? We have found BES TAR clones corresponding to all known T. brucei 427 BESs with the exception of the Bn-2 BES. The BES 13 TAR clones have the Bn-2 promoter sequence but a different ESAG6 type. Either we lack this BES in our set, or gene conversion between the ESAG6 sequences in different BESs has introduced variability between different T. brucei 427 isolates. This issue still needs to be resolved. Quantitation of Southern blot hybridization experiments using a BES promoter probe estimated ∼30 BES promoters in T. brucei 427 (Navarro and Cross 1996). As about half of BESs are estimated to have duplicated BES promoters in this strain, this is compatible with a total of ∼20 telomeres with BESs (Gottesdiener et al. 1991, 1992). Most, but possibly not all, BES promoters are linked to ESAG7 and part of a functional BES. Our analysis of 167 TAR clones yields an estimate of 17 BESs in T. brucei 427.
At least seven of the BES sets determined here have duplicated promoters, with clones corresponding to two size classes with ESAG10 present in the larger set. It is possible that some of the unique promoter and ESAG6 combinations are found at more than one telomere, making the total number of BESs possibly higher. For example, the BES14–17 clones have a wide range of sizes, and some of these could represent different telomeres with the same promoter. Some of these are BESs cloned as minor variants, and only sequence analysis of multiple versions will tell us if these are correct. However, despite the complication of unwanted extra recombination events, this TAR cloning technique represents a major step forward in our ability to isolate large numbers of complete BES-containing telomeres in a selective fashion. Sequencing in a redundant fashion, for example, two clones per BES set, might be the most cost-effective way of establishing that the clones that we analyze in detail are indeed representative.
Phylogenetic comparisons of the T. brucei BES promoter and ESAG6 sequences allowed inference of trees, showing the likely relatedness of these sequences. The different structure of the trees generated indicate that pairs of sequences within individual BESs do not appear to have coevolved. There was also only poor linkage between two proximal regions of the ESAG6 gene itself. This indicates that BESs are exchanging sequences at a very high rate, and that gene conversion may be the major mechanism of BES evolution. Moreover, loss of linkage suggests that the occurrence of a similar cohort of ESAGs in most BESs is not the result of a very recent amplification of a particular BES in strain T. brucei 427, as considerable shuffling of sequences has occurred since individual BES genesis.
The sequence of the T. brucei 927 genome is nearing completion. However, it is expected that sequences from the polymorphic subtelomeres will be highly underrepresented. Completing the T. brucei 927 genome will require a concerted effort aimed at cloning the different types of chromosome ends in addition to those of BESs. The conserved nature of BESs, including the promoter elements, has made this BES TAR cloning project possible. Isolating telomeres containing T. brucei metacyclic ESs (MESs) will prove to be a much greater challenge, as these monocistronic transcription units contain only VSGs and very divergent promoter elements (Ginger et al. 2002).
Sequence of the complete T. brucei genome containing all of the chromosome internal genes will provide us with a scaffold that is expected to be relatively similar across a range of T. brucei strains. However, telomeres of parasitic protozoa are particularly polymorphic areas where sequence diversity is generated at a high rate (for review, see Rudenko 1999; Scherf et al. 2001). To assess the degree of genetic variability across T. brucei strains and subspecies, we will need to clone and sequence the telomeric regions of the genome at a greater level of redundancy across different trypanosome isolates. This analysis will allow us to compare different BES sets across T. brucei strains. How does the distribution of ESAG types vary, and can we discover new ESAG families? As ESAGs are expected to play a role in host adaptation, can we discern any difference in their relative distribution and diversity across T. brucei strains that may have adapted to a different repertoire of mammalian host species?
Although our approach should be applicable for cloning BESs from a wide range of T. brucei strains, it will also be more widely applicable over a range of parasitic protozoa that have telomeres with sequence elements that are conserved enough to allow telomere trapping via the TAR method described here. This approach will allow a much more rapid and focused analysis of the subset of the genome particularly relevant for infection, and will greatly facilitate studies of antigenic variation.
Methods
T. brucei strain
The trypanosome strain used was T. brucei 427 clone HNI (221+). T. brucei HNI transformants are described in Rudenko et al. (1998) and contain a hygromycin-resistance gene inserted immediately behind the promoter of the active 221 VSG expression site, and a neomycin-resistance gene behind the promoter of the silent VO2 VSG expression site. Bloodstream form trypanosomes were amplified in ICR mice to sub maximal parasitaemia. Trypanosomes were isolated from whole blood by using DEAE column chromatography, and genomic DNA was isolated according to the method of Bernards et al. (1981).
Construction of TAR cloning vectors.
The T. brucei BES-specific TAR vector pEB4 was constructed from the basic TAR cloning vector pEB2 (M. Becker and E.J. Louis, unpubl.), which contains the yeast selectable marker URA3, the counter-selectable marker CYH2 (Raymond et al. 2002), as well as a yeast centromere, origin of replication, and one telomere (Fig. 1A). Alternative counter-selectable systems have been used in TAR cloning, which could be incorporated to improve the efficiency of the vectors used here (Noskov et al. 2003a). The T. brucei TAR target fragment was a 560-bp PCR product containing a BES promoter from the T. brucei 427 DES (accession no. X56598). This was amplified from plasmid subclone pDES2 (Zomerdijk et al. 1991) by using primers TAR T1/T2 (for sequences, see Table 1). This BES promoter region was chosen as it is highly similar in an alignment of different T. brucei BES promoters and was therefore likely to be highly conserved at many core bloodstream form promoters. T. brucei BES promoter regions compared were from the T. brucei 427 DES upstream (X67472) and downstream (X56598, L46868) promoters, 221 VSG BES (acc no.: AL671259), VO2 VSG BES (AL671256), Bn-2 VSG BES (both upstream and downstream promoters (AL670322), and the T. brucei EATRO 1125 AnTat 1.3A BES (L20156), and the T. brucei TREU 927/4 GUTat 10.1 BES (AC087700). This PCR fragment incorporated SalI linkers and was cloned into the XhoI-site of pEB2. Proper orientation of the fragment was confirmed by using an asymmetric ScaI site within the sequence amplified (Fig. 1A). The sequences presented here have GenBank accession nos. AY681978–AY682005.
Yeast strain and transformation.
The S. cerevisiae strain TYC1 was used as the host for transformation. This strain is an isogenic derivative of S288C (Winston et al. 1995) with the following genotype: MATα, ura3-52, leu2Δ1, cyh2r was isolated as a segregant from a cross between FY833 (Winston et al. 1995) and a spontaneous cyh2r mutation of S288Cα. This strain was chosen as it is the sequenced strain and is efficiently transformed (M. Becker and E.J. Louis, unpubl.). In addition, the DNL4gene (encoding DNA ligase IV used in nonhomologous end-joining) was disrupted by using KANMX (Wach et al. 1994) in order to reduce the possibility of spurious ligations during transformation (M. Becker and E.J. Louis, unpubl.). Spheroplasts were prepared by using modifications of previously described methods (M. Becker and E.J. Louis, unpubl.). Transformants are plated onto synthetic 1 M sorbitol and 200 μgL–1 cyclohexamide containing media lacking uracil in 15 mL premolten top agar of the same recipe (M. Becker and E.J. Louis, unpubl.). This media selects positively for the TAR vector that provides URA3 function and negatively against vectors that have not recombined, as the CYH2 gene confers dominant sensitivity to cycloheximide (Fig. 1A).
PCR identification and sequence analysis of T. brucei TAR transformants
Positive T. brucei transformants were identified by screening yeast colonies by PCR using ESAG7-specific primers (ESAG7a, see Table 1) and the following standard PCR conditions: 45 mM Tris-HCl (pH 8.8), 11 mM ammoniumsulphate, 4.5 mM MgCl2, 6.7 mM 2-mercaptothanol, 4.4 mM EDTA (pH 8.0), 113 μg/mL BSA, 1 mM dATP, dTTP, dCTP, and dGTP in a final volume of 10 μL. Thermocycling conditions consisted of initial 5 min at 95°C, 40 cycles of 30 sec at 95°C, 30 sec at 55°C, 2 min at 72°C, followed by a final 5 min at 75°C.
Yeast clones were streaked on URA drop-out plates to produce single colonies, and yeast genomic DNA from positive T. brucei TAR clone transformants was isolated by using standard techniques (Borts et al 1986). All of the 180 ESAG7-positive T. brucei TAR clones that were isolated were sequenced over a region of the BES promoter and ESAG6. Polymorphisms in the sequences allowed the further categorization of the TAR clones. A BES promoter fragment was PCR amplified by using primers ESP S1 and ESP S2. This PCR fragment was subsequently sequenced on both strands by using the nested primers ESP S3 and S4 (for primers used, see Table 1). Similarly, a variable region of ESAG6 was PCR amplified by using primers ESAG6amp and was sequenced on both strands by using both of the nested primers ESAG6 seq (Table 1). Each 50 μL reaction contained 2.5 U Taq DNA polymerase and 1× PCR buffer (Qiagen), 10 pmol each of forward and reverse primers, 10 nmol dNTPs, and 200 ng template DNA. The thermal cycle comprised a hot start for 3 min at 94°C, then 35 cycles for 30 sec at 94°C, 30 sec at 55°C, and 1 min at 72°C; this was followed by a 10-min extension at 72°C. The PCR products were precipitated by the addition of 60 μL 20%PEG-8000/2.5 M NaCl, allowing the removal of residual dNTPs and oligonucleotides. DNA sequencing was carried out by using the BigDye Terminator version 3.1 Cycle Sequencing kit (Applied Biosystems) according to the manufacturer's instructions. Sequencing reaction products were analyzed on an ABI PRISM 3700 DNA Analyzer or ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Sequences were assembled by using ContigExpress and aligned with AlignX from the VectorNTI suite of programs (Informax). The various BES TAR clones were grouped according to shared SNPs. To avoid over prediction of BES groups, only SNPs found in more than one BES clone, or where a clone varied by multiple SNPs, were considered.
PCR analysis of telomere clones
Sequence analysis allowed the categorization of the different telomere clones into different BES sets. PCR was performed on two telomere clones from each BES set by using primers for different T. brucei 427 VSGs. If one or more clones reacted positive with a given primer set, all of the telomere clones in the set were tested (for VSG primers used, see Table 1). A characterization of the ESAG composition in the different telomere clones was performed by using three different primer sets for each ESAG (see Table 1). PCR reactions were carried out in a volume of 10 μL containing 100 ng template DNA, 4 pmol each of the forward and reverse primers, 2 nmol dNTPs, 1.5 mM MgCl2, and 0.4 U Taq DNA polymerase (Invitrogen). The typical thermal cycling conditions were a hot start for 3 min at 94°C; followed by 35 cycles of 30 sec at 94°C, 30 sec at 55°Cto65°C (depending on the primer pair used), and 1 min at 72°C; followed by a 10-min final extension at 72°C.
CHEF and FIGE analysis of telomere clones
To estimate insert size, genomic DNA from yeast transformants was prepared in agarose plugs (Louis 1998) and separated in 1% agarose gels in 0.5× TBE by using a Field Inversion Gel Electrophoresis (FIGE) mapper system (BIORAD) and stored protocol P7 (size range, 25–75 kb; switch time linear ramp, 0.4 to 1.5 sec; 20 h; 180 V forward; 120 V reverse). A 5-kb DNA ladder (GIBCO) was used as a DNA standard. Clones with inserts >80 kb were analyzed in 1% agarose, 0.5× TBE gels on a CHEF-DRIII system (BIORAD) at 6 V/cm, 120° separation angle and switch times from 3.0 s (initial) to 48 sec (final) for 16 h. Subsequent Southern blot, hybridization, and detection procedures were carried out by using standard methods (Sambrook and Russell 2001). pEB2 was fluorescein-labeled by using Random-prime labeling (Amersham) and used as a probe.
The few clones with multiple bands on pulsed field gels could be due to instability during culturing in yeast or to multiple YACs within a culture, both of which have been seen in conventional YAC cloning (Nemeth et al. 1993; Larionov et al. 1994). We have been able to segregate single clones from mixed cultures which maintain their size after prolonged propagation (data not shown; M. Becker and E.J. Louis, unpubl.). This is consistent with multiple transformants being a complication rather than inherent instability.
Phylogenetic analyses
Bayesian trees were inferred from DNA alignments by using the Metropolois-coupled Markov chain Monte Carlo method implemented by the programme MrBayes 3.0b4 (Hulsenbeck and Ronquist 2001). During inference, BES promoter and ESAG6 sequences were treated as entirely independent. Among-site variation in substitution rates were modelled by γ-distributions approximated to one invariable and four variable categories. Rates of transitions and transversions were allowed to differ and their prior ratios were estimated from the sequence alignments, as were base frequencies. The calculations were run for 500,000 generations on four MCM chains. These chains were sampled every 100 generations with swapping of states every generation and a “temperature” of 0.2. The topology inferred by Bayesian methods was nearly identical to that inferred by much simpler maximum parsimony methods implemented by the program PAUP4.0b10 (Swofford 1998)
Acknowledgments
We thank George Cross, Piet Borst, and Steve Hajduk for communicating unpublished results and for providing sequences of unpublished T. brucei 427 VSGs. We are grateful to Jesse Shapiro for help in identifying new T. brucei 427 VSGs and to Keith Jolley for help with sequencing protocols. We thank Henri van Luenen and Piet Borst for unpublished sequences of T. brucei 427 ESAG6 variants, and to Keith Matthews for information on ESAG9. We thank Emma Carlin for technical assistance, Cleria Inglis for constructing the dnl4 knockout strain TYC1, and the masters students of the MSc in Molecular Genetics University of Leicester class of 2002 to 2003 for isolating new clones in their laboratory practical. We are grateful to Keith Gull, Piet Borst, and Katie Hughes for stimulating discussions and comments on the manuscript. M.B. was funded by a Wellcome Trust Value In People award, and E.B. was funded by a Wellcome Trust Programme grant to E.J.L. G.R. is a Wellcome Senior Fellow in the Basic Biomedical Sciences.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2955304.
References
- Ansorge, I., Steverding, D., Melville, S., Hartmann, C., and Clayton, C. 1999. Transcription of “inactive” expression sites in African trypanosomes leads to expression of multiple transferrin receptor RNAs in bloodstream forms. Mol. Biochem. Parasitol. 101: 81-94. [DOI] [PubMed] [Google Scholar]
- Ashcroft, M.T. 1958. The importance of African wild animals as reservoirs of trypanosomiasis. East African Med. J. 36: 289-297. [PubMed] [Google Scholar]
- Barry, J.D. and McCulloch, R. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49: 1-70. [DOI] [PubMed] [Google Scholar]
- Bernards, A., Van der Ploeg, L.H., Frasch, A.C., Borst, P., Boothroyd, J.C., Coleman, S., and Cross, G.A. 1981. Activation of trypanosome surface glycoprotein genes involves a duplication-transposition leading to an altered 3′ end. Cell 27: 497-505. [DOI] [PubMed] [Google Scholar]
- Berriman, M., Hall, N., Sheader, K., Bringaud, F., Tiwari, B., Isobe, T., Bowman, S., Corton, C., Clark, L., Cross, G.A., et al. 2002. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122: 131-140. [DOI] [PubMed] [Google Scholar]
- Bitter, W., Gerrits, H., Kieft, R., and Borst, P. 1998. The role of transferrin-receptor variation in the host range of Trypanosoma brucei. Nature 391: 499-502. [DOI] [PubMed] [Google Scholar]
- Blundell, P., Rudenko, G., and Borst, P. 1996. Targeting of exogenous DNA into Trypanosoma brucei requires a high degree of homology between donor and target DNA. Mol. Biochem. Parasitol. 76: 215-229. [DOI] [PubMed] [Google Scholar]
- Borst, P. and Ulbert, S. 2001. Control of VSG gene expression sites. Cell 114: 17-27. [DOI] [PubMed] [Google Scholar]
- Borst, P., van der Ploeg, M., van Hoek, J.F., Tas, J., and James, J. 1982. On the DNA content and ploidy of trypanosomes. Mol. Biochem. Parasitol. 6: 13-23. [DOI] [PubMed] [Google Scholar]
- Borts, R.H., Lichten, M., and Haber, J.E. 1986. Analysis of meiosis-defective mutations in yeast by physical monitoring of recombination. Genetics 113: 551-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta, A., Adjiri, A., New, L., Crouse, G.F., and Jinks Robertson, S. 1996. Mitotic crossovers between diverged sequences are regulated by mismatch repair proteins in Saccaromyces cerevisiae. Mol. Cell. Biol. 16: 1085-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson, J.E. 2003. Antigenic variation and the African trypanosome genome. Acta Trop. 85: 391-404. [DOI] [PubMed] [Google Scholar]
- Geigy, R., Mwambu, P.M., and Kauffmann, M. 1971. Sleeping sickness survey in Musoma district, Tanzania, IV: Examination of wild mammals as a potential reservoir for T. rhodesiense. Acta Trop. 28: 211-220. [PubMed] [Google Scholar]
- Gerrits, H., MuBmann, R., Bitter, W., Kieft, R., and Borst, P. 2002. The physiological significance of transferrin receptor variations in Trypanosoma brucei. Mol. Biochem. Parasitol. 119: 237-247. [DOI] [PubMed] [Google Scholar]
- Ginger, M.L., Blundell, P.A., Lewis, A.M., Browitt, A., Gunzl, A., and Barry, J.D. 2002. Ex vivo and in vitro identification of a consensus promoter for VSG genes expressed by metacyclic-stage trypanosomes in the tsetse fly. Eukaryot Cell 1: 1000-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesdiener, K., Chung, H.M., Brown, S.D., Lee, M.G., and Van der Ploeg, L.H. 1991. Characterization of VSG gene expression site promoters and promoter-associated DNA rearrangement events. Mol. Cell. Biol. 11: 2467-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesdiener, K.M., Goriparthi, L., Masucci, J.P., and Van der Ploeg, L.H. 1992. A proposed mechanism for promoter-associated DNA rearrangement events at a variant surface glycoprotein gene expression site. Mol. Cell. Biol. 12: 4784-4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huelsenbeck, J.P. and Ronquist, F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754-755. [DOI] [PubMed] [Google Scholar]
- Kooter, J.M., van der Spek, H.J., Wagter, R., d'Oliveira, C.E., van der Hoeven, F., Johnson, P.J., and Borst, P. 1987. The anatomy and transcription of a telomeric expression site for variant-specific surface antigens in T. brucei. Cell 51: 261-272. [DOI] [PubMed] [Google Scholar]
- LaCount, D.J., Barrett, B., and Donelson, J.E. 2002. Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277: 17580-17588. [DOI] [PubMed] [Google Scholar]
- Larionov, V., Kouprina, N., Nikolaishvili, N., and Resnick M.A. 1994. Recombination during transformation as a source of chimeric mammalian artificial chromosomes in yeast (YACs). Nucl. Acids Res. 22: 4154-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov, V., Kouprina, N., Graves, J., Chen, X.N., Korenberg, J.R., and Resnick, M.A. 1996a. Specific cloning of human DNA as yeast artificial chromosomes by transformation-associated recombination. Proc. Natl. Acad. Sci. 93: 491-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larionov, V., Kouprina, N., Graves, J., and Resnick, M.A. 1996b. Highly selective isolation of human DNAs from rodent–human hybrid cells as circular yeast artificial chromosomes by transformation-associated recombination cloning. Proc. Natl. Acad. Sci. 93: 13925-13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligtenberg, M.J., Bitter, W., Kieft, R., Steverding, D., Janssen, H., Calafat, J., and Borst, P. 1994. Reconstitution of a surface transferrin-binding complex in insect form Trypanosoma brucei. EMBO J. 13: 2565-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis, E.J. 1998. Whole chromosome analysis. In: Methods in microbiology: Yeast genome analysis (eds. M.F. Tuite and A.J.P. Brown), pp. 15-31. Academic Press, London.
- Louis, E.J. and Borts, R.H. 1995. A complete set of marked telomeres in Saccharomyces cerevisiae for physical mapping and cloning. Genetics 139: 125-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melville, S.E., Leech, V., Gerrard, C.S., Tait, A., and Blackwell, J.M. 1998. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei and the assignment of chromosome markers. Mol. Biochem. Parasitol. 94: 155-173. [DOI] [PubMed] [Google Scholar]
- Melville, S.E., Leech, V., Navarro, M., and Cross, G.A. 2000. The molecular karyotype of the megabase chromosomes of Trypanosoma brucei stock 427. Mol. Biochem. Parasitol. 111: 261-273. [DOI] [PubMed] [Google Scholar]
- Morris, J.C., Wang, Z., Drew, M.E., and Englund, P.T. 2002. Glycolysis modulates trypanosome glycoprotein expression as revealed by an RNAi library. EMBO J. 21: 4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, M. and Cross, G.A. 1996. DNA rearrangements associated with multiple consecutive directed antigenic switches in Trypanosoma brucei. Mol. Cell Biol. 16: 3615-3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeth, A.H., Hunter, N., Coleman, M.P., Borts, R.H., Louis, E.J., and Davies, K.E. 1993. Rescue of a single yeast artificial chromosome from a cotransformation event utilizing segregation at meiosis. Genet. Anal. Tech. Appl. 10: 123-127. [DOI] [PubMed] [Google Scholar]
- Noskov, V.N., Kouprina, N., Leem, S.H., Ouspenski, I., Barrett, J.C., and Larionov, V. 2003a. A general cloning system to selectively isolate any eukaryotic or prokaryotic genomic region in yeast. BMC Genomics 4: 1-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noskov, V.N., Leem, S.H., Solomon, G., Mullokandov, M., Chae, J.Y., Yoon, Y.H., Shin, Y.S., Kouprina, N., and Larionov, V. 2003b. A novel strategy for analysis of gene homologues and segmental genome duplications. J. Mol. Evol. 56: 702-710. [DOI] [PubMed] [Google Scholar]
- Pays, E., Tebabi, P., Pays, A., Coquelet, H., Revelard, P., Salmon, D. and Steinert, M. 1989. The genes and transcripts of an antigen gene expression site from T. brucei. Cell 57: 835-845. [DOI] [PubMed] [Google Scholar]
- Pays, E., Lips, S., Nolan, D., Vanhamme, L., and Perez-Morga, D. 2001. The VSG expression sites of Trypanosoma brucei: Multipurpose tools for the adaptation of the parasite to mammalian hosts. Mol. Biochem. Parasitol. 114: 1-16. [DOI] [PubMed] [Google Scholar]
- Raymond, C.K., Sims, E.H., and Olson, M.V. 2002. Linker-mediated recombinational subcloning of large DNA fragments using yeast. Genome Res. 12: 190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudenko, G. 1999. Genes involved in phenotypic and antigenic variation in African trypanosomes and malaria. Curr. Opin. Microbiol. 2: 651-656. [DOI] [PubMed] [Google Scholar]
- Rudenko, G., Chaves, I., Dirks-Mulder, A., and Borst, P. 1998. Selection for activation of a new variant surface glycoprotein gene expression site in Trypanosoma brucei can result in deletion of the old one. Mol. Biochem. Parasitol. 95: 97-109. [DOI] [PubMed] [Google Scholar]
- Salmon, D., Hanocq-Quertier, J., Paturiaux-Hanocq, F., Pays, A., Tebabi, P., Nolan, D.P., Michel, A., and Pays, E. 1997. Characterization of the ligand-binding site of the transferrin receptor in Trypanosoma brucei demonstrates a structural relationship with the N-terminal domain of the variant surface glycoprotein. EMBO J. 16: 7272-7278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J. and Russell, D.W. 2001. Molecular cloning: A laboratory manual. Cold Spring Harbor Press, New York.
- Schell, D., Evers, R., Preis, D., Ziegelbauer, K., Kiefer, H., Lottspeich, F., Cornelissen, A.W., and Overath, P. 1991. A transferrin-binding protein of Trypanosoma brucei is encoded by one of the genes in the variant surface glycoprotein gene expression site. EMBO J. 10: 1061-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf, A., Figueiredo, L.M., and Freitas-Junior, L.H. 2001. Plasmodium telomeres: A pathogen's perspective. Curr. Opin. Microbiol. 4: 409-414. [DOI] [PubMed] [Google Scholar]
- Steverding, D., Stierhof, Y.D., Fuchs, H., Tauber, R., and Overath, P. 1995. Transferrin-binding protein complex is the receptor for transferrin uptake in Trypanosoma brucei. J. Cell. Biol. 131: 1173-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. 1998. Phlyogenetic analysis using parsimony (and other methods). Sinauer, Sunderland, MA.
- Vanhamme, L., Pays, E., McCulloch, R., and Barry, J.D. 2001. An update on antigenic variation in African trypanosomes. Trends Parasitol. 17: 338-343. [DOI] [PubMed] [Google Scholar]
- Vanhamme, L., Paturiaux-Hanocq, F., Poelvoorde, P., Nolan, D.P., Lins, L., Van Den Abbeele, J., Pays, A., Tebabi, P., Van Xong, H., Jacquet, A., et al. 2003. Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422: 83-87. [DOI] [PubMed] [Google Scholar]
- Wach, A., Brachat, A., Pohlmann, R., and Phillippsen, P. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10: 1793-1808. [DOI] [PubMed] [Google Scholar]
- Winston, F., Dollard, C., and Ricupero-Hovasse, S.L. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53-55. [DOI] [PubMed] [Google Scholar]
- Wirtz, E., Leal, S., Ochatt, C., and Cross, G.A. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99: 89-101. [DOI] [PubMed] [Google Scholar]
- Xong, H.V., Vanhamme, L., Chamekh, M., Chimfwembe, C.E., Van Den Abbeele, J., Pays, A., Van Meirvenne, N., Hamers, R., De Baetselier, P., and Pays, E. 1998. A VSG expression site-associated gene confers resistance to human serum in T. rhodesiense. Cell 95: 839-846. [DOI] [PubMed] [Google Scholar]
- Zeng, C., Kouprina, N., Zhu, B., Cairo, A., Hoek, M., Cross, G., Osoegawa, K., Larionov, V., and de Jong, P. 2001. Large-insert bac/yac libraries for selective re-isolation of genomic regions by homologous recombination in yeast. Genomics 77: 27-34. [DOI] [PubMed] [Google Scholar]
- Zomerdijk, J.C., Ouellette, M., ten Asbroek, A.L., Kieft, R., Bommer, A.M., Clayton, C.E., and Borst, P. 1990. The promoter for a variant surface glycoprotein gene expression site in Trypanosoma brucei. EMBO J. 9: 2791-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zomerdijk, J.C., Kieft, R., Duyndam, M., Shiels, P.G., and Borst, P. 1991. Antigenic variation in Trypanosoma brucei: A telomeric expression site for variant-specific surface glycoprotein genes with novel features. Nucleic Acids Res. 19: 1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://bacpac.chori.org; BACPAC Resource Center with information about BAC libraries.