Abstract
Chronic obstructive pulmonary disease (COPD) is the third leading cause of death worldwide; smoking is the main risk factor for COPD, but genetic factors are also relevant contributors. Genome-wide association studies (GWAS) of the lung function measures used in the diagnosis of COPD have identified a number of loci, however association signals are often broad and collectively these loci only explain a small proportion of the heritability. In order to examine the association with COPD risk of genetic variants down to low allele frequencies, to aid fine-mapping of association signals and to explain more of the missing heritability, we undertook a targeted sequencing study in 300 COPD cases and 300 smoking controls for 26 loci previously reported to be associated with lung function. We used a pooled sequencing approach, with 12 pools of 25 individuals each, enabling high depth (30x) coverage per sample to be achieved. This pooled design maximised sample size and therefore power, but led to challenges during variant-calling since sequencing error rates and minor allele frequencies for rare variants can be very similar. For this reason we employed a rigorous quality control pipeline for variant detection which included the use of 3 independent calling algorithms. In order to avoid false positive associations we also developed tests to detect variants with potential batch effects and removed them before undertaking association testing. We tested for the effects of single variants and the combined effect of rare variants within a locus. We followed up the top signals with data available (only 67% of collapsing methods signals) in 4,249 COPD cases and 11,916 smoking controls from UK Biobank. We provide suggestive evidence for the combined effect of rare variants on COPD risk in TNXB and in sliding windows within MECOM and upstream of HHIP. These findings can lead to an improved understanding of the molecular pathways involved in the development of COPD.
Introduction
Chronic obstructive pulmonary disease (COPD) is the third cause of death worldwide [1]. Forced expiratory volume in one second (FEV1) and the ratio of FEV1 to forced vital capacity (FVC), measured by spirometry, are used in the diagnosis of COPD. Whilst environmental factors such as air pollution or tobacco smoking have a negative effect on lung function and increase the risk of developing COPD [2, 3], both lung function measures and COPD are heritable [4–6]. Genome-wide association studies (GWAS) of FEV1 and FEV1/FVC have now collectively identified 44 loci that have an effect on lung function [7–12]. This study focuses on the 26 loci that were first identified. Out of these 26 loci, 12 have already shown association with COPD risk, either in GWAS of COPD risk [13–16] or in studies that have only analysed previously-reported lung function associated variants [17, 18]. The known risk variants overall tend to have small effect sizes and only explain a small proportion of the heritability [9]. Most genome-wide association studies of lung function undertaken to date have focused on identifying common variants (minor allele frequency, MAF, >5%), and it is hypothesized that variants with lower allele frequency might have larger effect sizes and therefore might play an important role in explaining the missing heritability [19]. A recent GWAS of lung function [12] using data imputed to the 1000 Genomes Project reference panel [20] identified two low frequency variants with larger effect sizes. In addition, many of the association signals for the loci known to affect lung function are not well localized, and identifying rare (MAF ≤ 1%) or low frequency (1% < MAF ≤ 5%) variants within these regions can aid the identification of the causal variants.
In order to detect genetic associations with COPD risk for variants down to low allele frequencies in 26 loci associated with lung function, we undertook a two-stage study. In stage 1 we sequenced these 26 loci in 300 COPD cases and 300 smoking controls, using a cost-effective pooled design to maximize the sample size. We undertook single variant analysis and applied two collapsing methods in order to increase the power to detect rare variant associations in these regions. In stage 2 we followed up the top signals in 4,249 COPD cases and 11,916 smoking controls within the UK BiLEVE study (a subset of UK Biobank) [11], with genotypes imputed to the joint 1000 Genomes Project [20] and UK10K [21] reference panel.
Results
Study design, sequencing and data processing
Three hundred COPD cases, defined using spirometry (GOLD stage 2 [3] and above), and 300 controls were selected among individuals over 40 years of age who were smokers from three studies: Gedling, Nottingham Smokers and the Leicester COPD cases. Spirometry procedures carried out in these studies can be found in [9, 22] and characteristics of individuals included in the study are presented in S1 Table.
The region sequenced for each of the 26 loci associated with FEV1 or FEV1/FVC was defined using the results from the largest meta-analysis of GWAS for these traits undertaken to date [9]. The parameters used in this definition were: distance from the most significantly associated SNP (sentinel SNP) in each region and strength of association, measured by P-value, (details in the Methods section and in S2 Table). In total 7.7Mb of sequence was covered and the sequencing was undertaken using Illumina HiSeq 2000 with 100 bp paired-end reads and 8 lanes, each with 3 pools. Each pool contained DNA from 25 cases or 25 controls. The data were aligned against 1000 Genomes Project phase 1 data [20] (GRCh37; h19) using BWA.6.2 [23]. Alignments were then sorted and PCR duplicates were removed using SAMtools [24] and indels were locally realigned and quality scores recalibrated using GATK v2.5 [25]. After removing two control pools (50 control individuals) due to lower DNA quality than the rest, the average coverage per individual was 30x.
Variant calling and quality control checks
In order to distinguish true calls from sequencing error, three different calling algorithms specific for pooled data were used (vipR [26], SNVer [27] and Syzygy [28]). vipR was less sensitive than the other two algorithms and it called a much smaller number of variants, 39,211 SNPs and 459 deletions, compared to 62,506 SNPs and 5,811 indels by SNVer and 55,886 SNPs and 5,331 indels by Syzygy. Most of the variants called by vipR had MAF > 1% (85% of SNPs and 99% of indels). SNVer also called mainly variants with MAF > 1% (81% of SNPs and all indels), whereas Syzygy called the largest number of rare variants (43% SNPs and 35% of indels). In terms of specificity, Syzygy and vipR showed the best specificity when assessing the proportion of SNPs with MAF > 1% included in dbSNP137 [29] (97.08% for vipR and 98.2% for Syzygy), compared to a 72% for SNVer. When calling indels all algorithms had lower specificity than when calling SNPs, assessed as the proportion of indels with MAF > 1% included in 1000 Genomes Project phase 1 [20], (67.55% for vipR, 35% for SNVer and 50% for Syzygy).
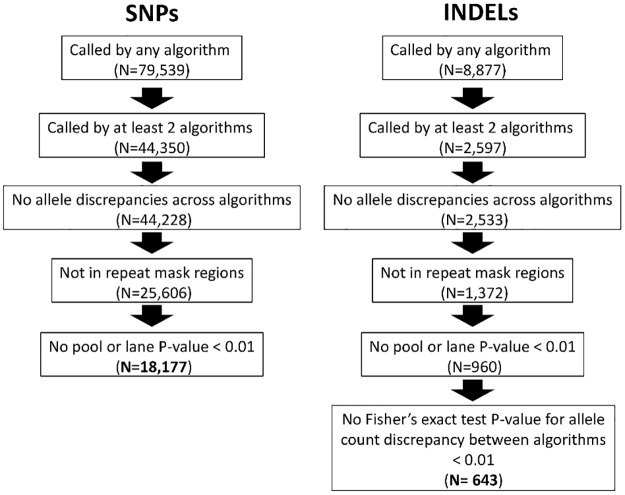
A series of quality control checks and filtering strategies were performed (Fig 1, details in the Methods section). More than 90% of SNPs (> 46% of indels) with MAF > 1% called by at least two algorithms were present in dbSNP137 [29], in contrast to < 70% of SNPs (< 40% for indels) with MAF > 1% called by only one algorithm (S1 Fig). This indicated that the set of variants called by at least two algorithms was more reliable, and therefore variants called only by one algorithm were excluded. Variants were also excluded if the alleles called by different algorithms did not agree or if they were in a repeat masked region. Additional tests were performed in order to remove variants which could generate false associations due to a lane or pool effect (see details in the Methods section). Allele frequencies for SNPs were consistent across algorithms and with those from 1000 Genomes Project phase 1 data [20] (S2a and S2b Fig). There were considerable discrepancies across algorithms for some indels, therefore, a Fisher’s exact test (FET) was performed for indel allele counts in order to exclude those that showed marked discrepancies across algorithms (S2c and S2d Fig). After undertaking quality control checks a total of 18,177 SNPs and 643 indels across the 26 regions were selected for association testing with COPD risk (Fig 1). Some of these variants were novel: 1,429 SNPs (93% with MAF < 1%) were not in dbSNP137 [29] or in the joint 1000 Genomes Project [20] and UK10K [21] reference panel and 216 indels (4% with MAF < 1%) were not in 1000 Genomes Project phase 1 data [20] or reported by Mills et al. [30]. There was a notable difference in the quality of the data for SNPs and indels. Out of all variants called by at least one algorithm, after all quality control checks 23% of SNPs remained, whereas for indels it was only a 7%.
Fig 1. Flow chart of the variant selection process.
Single variant
Given the pooled design of the experiment, no allele counts per individual were available, only allele counts within sets of cases and controls; therefore the association testing was undertaken using Fisher’s exact test. It was performed separately for the allele counts produced by each calling algorithm, for the variants that passed the quality control checks. A total of 8 SNPs and 3 indels, different from the sentinel variants associated with lung function in previous studies [7–10], met a significance threshold adjusted for multiple testing (significance thresholds defined separately for each region, see Methods section and S3 Table) using the allele counts for at least one calling algorithm and showed supportive evidence (P < threshold x 2) when using allele counts from another calling algorithm (S4a Table).
These variants were followed up with data imputed to the joint 1000 Genomes Project [20] and UK10K [21] reference panel in 4,249 COPD cases and 11,916 smoking controls from the UK BiLEVE study [11] (characteristics in S1 Table). Only one variant (rs999741 in the HTR4 region, MAF 26%) had a nominally significant P-value (P = 2x10-3) and had consistent direction of effect in stage 1 and stage 2 (S4 Table). When conditioned on the previously reported variant (rs1985524 [9]) in that region, rs999741 was no longer significant (S5 Table) thereby confirming that this signal was not independent of rs1985524 and confirming the previously reported association with COPD for this SNP.
Association results for the 26 previously reported lung function sentinel SNPs (or SNPs in perfect LD with the sentinel SNPs) are presented in S6 Table. Sentinel lung function SNPs in four regions (MECOM, HHIP, SPATA9, HTR4) reached nominally significant P-values (Table 1) when using allele counts for at least two calling algorithms and their direction of effect agreed with the previously reported [9] effect on lung function (negative effect on FEV1 or FEV1/FVC and increased risk of COPD, or positive effect on FEV1 or FEV1/FVC and reduced risk of COPD). Association with COPD risk for HHIP and HTR4 had already been reported [15, 18], but not for SPATA9 or MECOM. These four variants were also analysed in UK BiLEVE, where again the association of HHIP and HTR4 were confirmed, but no association was found for SPATA9 or MECOM (Table 1).
Table 1. COPD association results in stage 1 and 2 for lung function sentinel variants analysed in stage 2.
| rs number | GWAS gene | Ref allele | Alt allele | Calling algorithm | Stage 1 | Stage 2 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Alt allele freq in cases | Alt allele freq in controls | OR | P-value | OR | P-value | |||||
| rs1344555 | MECOM | C | T | SNVer | 0.238 | 0.154 | 1.72 | 4.93x10−4 | 1.03 | 4.20x10−1 |
| Syzygy | 0.247 | 0.162 | 1.69 | 5.93x10−4 | ||||||
| vipR | 0.260 | 0.180 | 1.6 | 9.29x10−3 | ||||||
| rs11100860 | HHIP | A | G | SNVer | 0.337 | 0.410 | 0.73 | 1.44x10−2 | 0.86 | 3.31x10−9 |
| Syzygy | 0.340 | 0.404 | 0.76 | 3.28x10−2 | ||||||
| vipR | 0.398 | 0.406 | 0.97 | 8.37x10−1 | ||||||
| rs153916 | SPATA9 | C | T | SNVer | 0.600 | 0.530 | 1.33 | 2.03x10−2 | 0.99 | 5.55x10−1 |
| Syzygy | 0.605 | 0.536 | 1.33 | 2.35x10−2 | ||||||
| vipR | 0.576 | 0.526 | 1.09 | 1.33x10−1 | ||||||
| rs1985524 | HTR4 | G | C | SNVer | 0.378 | 0.448 | 0.75 | 2.27x10−2 | 0.88 | 1.08x10−6 |
| Syzygy | 0.383 | 0.446 | 0.77 | 3.67x10−2 | ||||||
| vipR | 0.428 | 0.446 | 0.93 | 5.89x10−1 | ||||||
COPD single variant results for lung function sentinel variants [9] with P < 0.05 when using allele counts for at least two calling algorithms in stage 1 and 2. Full stage 1 results for the 26 lung function loci are presented in S6 Table. OR correspond to the alternative allele.
Abbreviations: Ref = reference, Alt = alternative, freq = frequency, N = number, ac = allele count, OR = odds ratio.
Collapsing methods
In order to increase power to identify associations with rare variants we applied two collapsing methods. A burden test was applied using Fisher’s exact test to assess whether accumulation of rare variants in a locus (number of individuals with at least one rare allele) was associated with COPD risk. The C-alpha test [31] was also applied to test whether a locus was associated with COPD risk allowing for variants to be protective or detrimental. Only rare SNPs (MAF < 1%) were included in these analyses and loci boundaries were defined in three different ways: (i) sliding window: 3kb sliding windows with an overlap of 1.5kb, (ii) gene based: gene coordinates, and (iii) exon based: exons, 5’ UTR and 3’ UTR for each gene.
A total of 59 3kb sliding windows from 18 regions out of the 26 sequenced, and 23 genes (21 from gene based tests, 1 from exon based tests and 1 that was selected for both) from 19 regions out of the 26 sequenced met a significance threshold adjusted for multiple testing (see Methods section and S3 Table) using the allele counts for at least one calling algorithm and showed supportive evidence (P < threshold x 2) when using allele counts from another calling algorithm (S7 Table). Of these, two sliding windows and three genes from the gene based analysis were selected due to their P-values in the burden analysis; all the remaining windows and genes were selected because of their C-alpha test P-values. Only enough variants were available to follow-up 32 sliding windows out of the 59 and the 23 genes in UK BiLEVE. Full stage 2 results are provided in S8 Table.
None of the two sliding windows and three genes selected for their burden test P-values reached nominal significance (P<0.05) in UK BILEVE (S8a Table). The C-alpha test results for sliding windows showed one sliding window in the MECOM region (chr3:169238286–169241286) which met a threshold corrected for multiple testing for that region (P < 8x10-3), and sliding windows in the RARB region (chr3:25633833–25636833) and in the HHIP region (chr4:145293600–145296600) that showed suggestive evidence of association (P = 0.05 and P = 0.04 respectively) (Table 2). For the C-alpha test for gene based analysis, C10orf11 met the significance threshold for that region (P < 0.05, only one gene in the region) and TNXB, in the AGER region, showed near-suggestive evidence of association (P = 0.06) (Table 2). For the C-alpha test for exon based analysis, NPNT in the GSTCD region showed suggestive evidence of association (P = 0.05, Table 2). Full results for the C-alpha test are presented in S8b Table.
Table 2. Collapsing methods results for most significant loci in stage 2.
| Locus, (GWAS gene) | Stage 1 | Stage 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Threshold | P-value | Threshold | All variants | Independent variants | ||||||
| Number of variants | Number of alternative allele counts | P-value | Number of variants | Number of alternative allele counts | P-value | P-value after permutations | ||||
| chr3:25633833–25636833, (RARB) | 5.81x10−4 | 9.43x10−7 | 1.25x10−2 | 3 | 363 | 5.32x10−2 | 3 | 363 | 5.32x10−2 | 6.25x10−2 |
| chr3:169238286–169241286, (MECOM) | 1.87x10−4 | 1.46x10−5 | 8x10−3 | 2 | 573 | 2.94x10−3 | 2 | 573 | 2.94x10−3 | 1.92x10−2 |
| chr4:145293600–145296600, (HHIP) | 2.76x10−4 | 1.11x10−5 | 1.25x10−2 | 2 | 167 | 4.16x10−2 | 2 | 167 | 4.16x10−2 | 6.19x10−2 |
| NPNT (chr4:106816596–106892828), (GSTCD) | 1.25x10−2 | 1.81x10−3 | 2.5x10−2 | 9 | 1400 | 5.25x10−2 | 3 | 422 | 5.1x10−1 | 4.07x10−1 |
| TNXB (chr6:32008931–32077151), (AGER) | 7.14x10−3 | 6.91x10−3 | 5x10−2 | 37 | 8086 | 6.08x10−2 | 11 | 1752 | 4.73x10−2 | 6.69x10−2 |
| C10orf11 (chr10:77542518–78317126) (C10orf11) | 5x10−2 | 3.95x10−18 | 5x10−2 | 304 | 39634 | 4.03x10−2 | 124 | 15529 | 2.77x10−2 | 5x10−1 |
“GWAS gene” presents the gene previously reported for lung function [9] for each of the 26 regions. The most significant stage 1 P-values across the three algorithms for the analysis only including independent variants are presented here; for full results see S7 Table. Stage 2 P-values that meet the threshold are shown in bold.
After repeating the analyses for these six loci keeping only independent variants (r2 < 0.2, see Methods) in each locus, as a sensitivity analysis, NPNT was no longer significant, TNXB and C10orf11 met the significance threshold corrected for multiple testing, and the sliding window results remained the same. The C-alpha test authors (53) recommend undertaking permutations for the top loci. Ten thousand permutations were run for the 6 most significant loci including only independent variants. After permutations, NPNT and C10orf11 were not significant and suggestive evidence was provided for the remaining loci (P = 6.25x10-2 for chr3:25633833–25636833 in RARB, P = 1.92x10-2 for chr3:169238286–169241286 in MECOM, P = 6.19x10-2 chr4:145293600–145296600 upstream of HHIP and P = 6.69x10-2 TNXB).
In order to gain more insights into the 4 loci that showed suggestive evidence of association, single variant results for the variants included in each region were examined and the C-alpha test was undertaken again removing one variant at a time to assess the single variant effect on the results both in stage 1 and stage 2 (S3 Fig). The signal in the sliding window in RARB was driven by the same variant in stage 1 and in stage 2 however, the direction of effect did not agree between stages, indicating that this was probably a false positive association. The remaining signals seemed to be driven by different variants in stage 1 and stage 2.
Discussion
The aim of this study was to identify low frequency and rare variants in genetic regions known to be associated with lung function in order to gain insights into the biological pathways that link these regions with COPD risk. To do this, 26 regions associated with lung function [7–10] were sequenced in 300 COPD cases and 300 controls using a cost-effective pooled design. In order to minimize the occurrence of false positive calls three variant calling algorithms were used in this study. Single and multi (collapsing) variant association analyses were undertaken and the strongest signals were followed up in 4,249 COPD cases and 11,916 controls from the UK BiLEVE study [11]. Suggestive evidence of association with COPD risk was shown for a window in MECOM, one intergenic window upstream of HHIP and for the TNXB gene in the AGER region.
The strongest collapsing signal in stage 2 was for a sliding window (chr3:169238286–169241286) in an intronic region of MECOM, which includes a DNase hypersensitivity site for blood microvascular endothelial cells derived from lung tissue [32]. Variants in MECOM (r2 <0.03 with rs1344555) have been associated with osteoporosis [33], renal function-related traits [34] and nasopharyngeal carcinoma [35] in East Asians and with blood pressure [36] and magnesium levels [37] in Europeans. The MDS1 and EVI1 complex locus protein (MECOM) encodes a number of transcripts that code for nuclear transcription factors [38]. Overexpression of the oncoprotein ecotropic virus integration site 1 protein homolog (EVI1) has been associated with multiple epithelial cancers, such as nasopharyngeal carcinoma, lung and colorectal cancers [35, 39–41]. EVI1 is also involved in embryonic development, through a role in haematopoiesis and mouse knock out models for MECOM have shown embryonic lethality [42–44].
The sliding window ~270kb upstream of HHIP that showed suggestive evidence of association with COPD is located in a region that contains a DNase hypersensitivity site and transcription factors binding sites found in blood cells, renal epithelium cells and embryonic stem cells [32]. This region does not overlap with another region ~85kb upstream of HHIP known to interact with the HHIP promoter and to function as an HHIP enhancer [45].
Single variant association analyses for sentinel variants previously associated with lung function [9] confirmed the previously reported associations with COPD risk for HHIP [15] and HTR4 [18].
Given the pooled nature of the stage 1 design it was not possible to adjust the phenotype for any covariates. Both cases and controls were individuals of over 40 years of age, who smoked between 5 and 100 pack years; and COPD case control status was determined using FEV1 percent predicted, which takes into account age, sex and height. Analyses in stage 2 were undertaken as close as possible to stage 1, and therefore the selection criterion was the same as in stage 1.
A limitation of this study was its reduced sample size. Assuming a COPD prevalence of 30% among smokers, a study with 300 cases and 300 controls would need an OR of 5 in order to detect a variant with MAF ~ 1% and an OR of 2 for a variant with MAF ~ 5% with 80% power at a nominal level of significance (P = 0.05). However, this study was designed to identify genetic associations with rare variants, which are expected to have larger effect sizes [19]. We decided to use a pooled sequencing design in order to maximise stage 1 sample size, however this design also has limitations. It is challenging to determine whether a single variant is homozygous or heterozygous, particularly if the allele frequency of this variant is rare, limiting this way the ability of this approach to discover new variants reliably. In addition, there are a number of factors that can affect the quality of the sequencing data, such as PCR amplification biases, reference allele preferential biases or varying error sequencing rates across sites [46]. This led us to apply three different calling algorithms using different assumptions and statistical models and to apply strict filters. In some instances these filters might have been over conservative.
The three calling algorithms used to minimise the number of false calls used different statistical methods and they also performed differently. vipR and SNVer called mainly variants with MAF > 1%, vipR being the least sensitive of the three algorithms. Syzygy called the largest number of rare variants and along with vipR were the algorithms with best specificity. Calling of indels was more challenging than calling SNPs regardless of which algorithm was used.
The key strength of this study was the ability to identify novel low frequency or rare variants through sequencing. A rigorous analytic pipeline was followed in this study overall. Support from at least two of the three calling algorithms was required for a variant to be called; strict filters were also applied assessing for example potential pool or lane effects. In addition, in order to select the most robust associations with COPD risk, results for at least one calling algorithm had to meet a Bonferroni threshold and support was also required from results using another calling algorithm. Moreover, in order to distinguish real associations from spurious ones, replication was pursued in an independent study. This study despite providing data only for 67% of collapsing methods signals, most of which were imputed rather than genotyped, had a considerably larger sample size and its power was enhanced by the sampling strategy used, selecting individuals from extremes of the lung function distribution in UK Biobank.
Overall, this pooled sequencing study, which implemented strict filtering strategies, identified 18,177 SNPs and 643 indels. It showed suggestive evidence for the association of rare variants with COPD risk in sliding windows in MECOM and upstream of HHIP and in TNXB. These findings will contribute to improve the knowledge of the biological mechanisms underlying the COPD and may lead to the development of new preventive and treatment strategies.
Methods
Samples in stage 1
Individuals from three studies were included in this analysis: Gedling, Nottingham Smokers and the Leicester COPD cases. Spirometry procedures for Gedling and Nottingham Smokers can be found in [9] and for the Leicester COPD cases in [22]. Individuals were excluded if (i) they were younger than 40 years old, (ii) they had pack-years of smoking < 5, or > 100, or (iii) if they had DNA concentration ≤ 20ng/uL. Individuals with asthma were also excluded from the Leicester COPD cases study. This left a sampling frame of 965 individuals (403 from Gedling, 468 from Nottingham Smokers and 96 from the Leicester COPD cases). COPD cases were defined as spirometric GOLD stage 2 [3] and above (percent predicted FEV1 < 80% and FEV1/FVC < 0.7) and controls as individuals with percent predicted FEV1 > 80% and FEV1/FVC > 0.7, based on pre-bronchodilator spirometry. Individuals with percent predicted FEV1 > 80% and FEV1/FVC < 0.7 (GOLD stage 1 [3]) or with percent predicted FEV1 < 80% and FEV1/FVC > 0.7 were excluded from the analysis to minimize misclassification. The calculation of percent predicted FEV1 was undertaken using reference values of FEV1 that take into account age, sex and height according to previously described equations [47, 48]. In order to select the most extreme 300 COPD cases and 300 smoking controls, COPD cases and smoking controls were ranked according to their percent predicted FEV1 and selected from the extremes. In order to remove extremely healthy individuals from the controls, individuals were excluded if (i) they had percent predicted FEV1 > 120.26 (the 99th percentile of percent predicted FEV1) or (ii) if they had FEV1/FVC > 0.85 (the 95th percentile of FEV1/FVC). Individuals were grouped into pools of 25 (separately for cases and controls), following the percent predicted FEV1 ranking, so that individuals with more similar phenotype would be grouped together.
Definition of regions
Region plots produced with data from the largest GWAS to date for FEV1 and FEV1/FVC measures [9] for the 26 loci associated with lung function [7–10] were examined to define the association regions. SNPs with −log10 (P-value) > 2.5 and not further than 50kb away from the next SNP with −log10 (P-value) > 2.5 moving away from the sentinel SNP, were selected. Any gene intersecting the association region was added to the region +/-10kb. If the association region did not include or intersect the closest gene, the association region was enlarged to include the closest gene +/- 10kb. If the enlarged regions also intersected other genes, the regions were not enlarged again, so they included small portions of genes. Regions were selected using the −log10 (P − value) for the most significant trait only, except for CDC123 which was genome-wide significant for FEV1 and FEV1/FVC and the sentinel SNP was the same for both traits. For CDC123 the association region was defined so it included the association regions for both traits. The regions covered a total of 10.3Mb (S2 Table).
Sequencing method
The enrichment kit was produced by Agilent (http://www.agilent.com/) and the sequencing was undertaken by Source BioScience (http://www.sourcebioscience.com/). After applying a correction for GC content and applying repeat-masking filters, a total of 7.7Mb of sequence was covered by probes in the final design. Sequencing was undertaken using Illumina HiSeq2000 with 100 bp paired-end reads and 8 lanes, each with 3 pools. Pools were assigned to lanes sequentially, so that pool1 to pool3 were allocated to lane 1, pool4 to pool6 were allocated to lane 2 and so on; in total there were four case lanes and four control lanes.
Alignment and data processing
The data were aligned against 1000 Genomes Project phase 1 reference panel [20] (GRCh37; h19) using BWA.6.2 [23], with −q 15 for read trimming. Alignments were then sorted and PCR duplicates were removed using SAMtools [24]. After the removal of duplicates, coverage summaries were produced using SAMtools [24] and BEDtools [49]. GATK [25] was used for base quality scores recalibration and local realignment around indels using known indel coordinates from 1000 Genomes Project phase 1 data [20] and Mills et al. data [30] as reference.
Variant calling and quality control checks
In order to distinguish true calls from sequencing error, three different calling algorithms specific for pooled data were used (vipR [26], SNVer [27] and Syzygy [28]) and a series of quality control checks and filtering strategies were performed. Variants were excluded if they were called only by one calling algorithm, if the alleles called by different algorithms differed or if they were in repeat masked regions (extracted from UCSC table browser [50]). A lane test was designed to detect variants affected by lane effects. Given that lanes included only case pools or only control pools, a sequencing artefact in a lane could lead to a false association. A chi-square test with three degrees of freedom was run for the four case lanes and the four control lanes for each variant. A pool test was implemented to detect sequencing artefacts in pools which could lead to false associations and to assess whether the data were consistent within case pools and within control pools. A Binomial test was performed for each pool, to test whether the observed allele count would be expected given the number of chromosomes in the pool and the allele frequency observed across case or control pools (allele frequency was calculated separately for case pools and control pools). Variants with either a P-value < 0.01 in the lane test for case pools or control pools, or a P-value < 0.01 in the pool test for any pool were excluded. Allele frequencies obtained using allele counts from the different calling algorithms were compared across algorithms and with 1000 Genomes Project phase 1 data [20]. A Fisher’s exact test was performed for indel allele counts in order to exclude those that showed discrepancies across algorithms.
Association testing
Variants selected were tested for association with COPD risk using Fisher’s exact tests on allele counts per pool (since no allele accounts per individual were available) using allele counts produced by the different calling algorithms both for SNPs and indels.
Collapsing method tests were only applied to SNPs (with MAF < 1%), due to the lower quality shown for data on indels. Loci boundaries were defined in three different ways, in order to detect associations in three different biological scenarios: (i) sliding window: 3kb sliding windows with an overlap of 1.5kb, (ii) gene based: gene coordinates, and (iii) exon based: exons, 5’ UTR and 3’ UTR for each gene. Gene, UTR and exon coordinates were extracted from UCSC table browser [50] using the RefSeq Genes track. A burden test based on [51] was undertaken using Fisher’s exact test to assess whether the accumulation of rare variants in a locus (number of individuals with at least one rare allele) was associated with COPD risk. In order to infer how many individuals had at least one rare allele it was assumed that individuals with the alternative allele would always be heterozygous (rather than homozygous, since only overall allele count per pool was available). In addition, it was assumed that rare variants within a locus were independent. In order to test whether a locus was associated with COPD risk allowing for variants to be protective or detrimental, the C-alpha test [31] was also applied. This test also assumes that variants within a region are independent. All association testing analyses were performed using R v3.1.0 (https://www.r-project.org/).
In order to assess the effect on the results of the assumption that variants with MAF < 1% in a locus were independent, the collapsing tests were run again for the most significant loci after removing variants in LD (r2 > 0.2) with each other. Within a group of variants in LD the one with the smallest P-value across the three calling algorithms was chosen. LD was calculated using the joint 1000 Genomes Project [20] and UK10K reference panel. The effect on the results of assuming that variants not present in this joint reference panel were independent or were in LD with any of the other variables in the region was assessed by running the collapsing methods with and without variants not in the joint 1000 Genomes Project [20] and UK10K [21] reference panel.
Significance thresholds
Significance thresholds to account for multiple testing were defined for each of the 26 regions separately. As there is already strong prior evidence for the association of the 26 regions with lung function [7–10], no multiple testing adjustment for the number of regions was undertaken. For the single variant analysis the effective number of independent variants tested (equivalent to the number of independent tests) was estimated using the approach developed by Li and Ji [52], and then a Bonferroni correction for the number of independent tests was applied in each region. Data from the joint 1000 Genomes Project [20] and UK10K [21] reference panel were used to estimate the correlation matrix for each region. Variants not included in this reference panel were assumed to be independent. For the collapsing methods a Bonferroni correction was applied for the number of independent tests within each region. For the sliding windows, given the overlap between windows the number of independent tests was defined as half the number of sliding windows, and for the gene based and exon based, the number of independent tests was defined as the number of genes included in each analysis. Significance thresholds for each region and each test are provided in S3 Table.
Selection of signals for follow-up
In order to minimise false positive associations, the criteria to select the top hits required that a variant met the significance threshold using allele counts for one calling algorithm and that it also showed supporting evidence (P-value < threshold x 2) when using allele counts from another calling algorithm. For the collapsing methods, loci had to meet this criterion also after the sensitivity analysis in order to be followed-up. Alignments were visually inspected for all the single variants selected and for a random sample of the variants in the collapsing method selected loci.
Follow-up (stage 2)
Variants and loci selected were followed-up in UK BiLEVE [11], a subset of ~50,000 individuals from UK Biobank (http://www.ukbiobank.ac.uk/) sampled from the extremes of the % predicted FEV1 distribution separately in never smokers and heavy smokers. An Affymetrix Axiom custom array was designed for the genome-wide genotyping of the UK BiLEVE project, including 130K rare missense and loss of function variants, and 642K variants selected for optimal imputation of common variation and improved imputation of low frequency variation (MAF 1–5%), and 9000 variants selected for improved coverage of known and candidate respiratory regions. These data were imputed against the joint 1000 Genomes Project phase 1 [20] and UK10K [21] reference panel.
The sampling frame was made of 20,859 individuals over 40 years old, with no asthma (diagnosed or self-reported) who smoked between 5 and 100 pack years. Case-control status was defined as in the COPD case-control sequencing study. COPD cases defined as in stage 1, but percent predicted FEV1 was obtained using reference values from healthy (no respiratory diseases diagnosed) never smokers (N = 81,719) from UK Biobank. In total there were 4,249 COPD cases and 11,916 smoking controls.
The association of single variants with COPD risk in UK BiLEVE was tested using logistic regression on allele dosages obtained from the imputation output. An adjustment for 5 principal components of ancestry was included. All variants followed up had imputation quality above 0.8 in UK BiLEVE.
The same collapsing methods as in stage 1 were used. The most likely genotype for each individual was used for the analysis, using IMPUTE2 [53] posterior probabilities and including only genotypes with posterior probability > 0.9. Sensitivity analyses were undertaken for the top hits including only independent variants (r2 < 0.2) within each locus. In addition, 10,000 permutations (permuting case control status) were run for the C-alpha top hits, only including independent variants, in order to obtain more accurate P-values. The same loci boundaries as in stage 1 were used for the collapsing method including only variants with MAF < 1%, HWE P-value > 10−6 and imputation quality ≥ 0.8.
Significance thresholds per region were defined by a Bonferroni corrected threshold for the number of independent tests undertaken in each region. For the single variant analysis the number of independent tests were the number of variants followed up. For the gene based and exon based analyses a gene was considered as an independent test. For the sliding window analysis, two overlapping windows were counted as 1.5 tests.
Ethics statement
The Gedling study was approved by the Nottingham City Hospital and Nottingham University Ethics committees (MREC/99/4/01) and written informed consent for genetic study was obtained from participants. The Nottingham Smokers study was approved by Nottingham University Medical School Ethical Committee (GM129901/) and written informed consent for genetic study was obtained from participants. For the Leicester COPD cases ethical approval was obtained from the Leicestershire Research Ethics Committee and written informed consent from all subjects was obtained. UK Biobank has approval from the North West Multi-centre Research Ethics Committee (MREC), which covers the UK. It also sought the approval in England and Wales from the Patient Information Advisory Group (PIAG) for gaining access to information that would allow it to invite people to participate. PIAG has since been replaced by the National Information Governance Board for Health & Social Care (NIGB). In Scotland, UK Biobank has approval from the Community Health Index Advisory Group (CHIAG).
Supporting Information
- a) Venn diagrams of SNPs called by any of the three algorithms with and without a 1% minor allele frequency (MAF) filter. The proportion of SNPs included in dbSNP137 [29] for each section is presented in brackets.
(DOCX)
(DOCX)
A drop one plot for a locus is obtained by undertaking the C-alpha test removing one variant at a time; the P-value plotted for each variant represented on the x-axis is the P-value obtained after removing that variant. Results obtained using calls from different calling algorithms are represented in different colours, according to the legend in each figure. Not all calling algorithms called the same set of variants. When there is no visible P-value represented for a variant for a given calling algorithm, it is because that variant was not called by that calling algorithm. If a region only includes two variants, no drop one plot is produced, since no C-alpha test can be undertaken with only one variant. Asymptotic P-values are presented here for the C-alpha test.
The single variant plots show the P-values obtained for each variant, and they also show the direction of effect by plotting on the y-axis the -log10 (P-value) x direction of effect, with the direction of effect = 1 if OR > 1 and direction of effect = -1 if OR < 1.
For each region the first row shows drop one plots and the second row shows single variant plots for the same variants; the first column presents results from stage 1 excluding variants not in the joint 1000 Genomes Project [20] and UK10K [21] reference panel (UK10K+1000G), the second column presents results from stage 1 including variants not in this reference panel (marked with * on the x-axis) and the third column presents results from stage 2. Variants that are included both in stage 1 and stage 2 are highlighted in bold. The horizontal dashed line represents the collapsing method significance threshold for each region (note that different thresholds were used for stage 1 and stage 2).
- a) chr3:25633833–25636833 (RARB)
- b) chr3:169238286–169241286 (MECOM)
- c) chr4:145293600–145296600 (HHIP)
- d) TNXB (AGER)
(DOCX)
Abbreviations: N = number, sd = standard deviation, y = years, l = litres.
(DOCX)
“GWAS sentinel” and “GWAS gene” present the lung function GWAS sentinel SNP and the closest gene to the sentinel SNP respectively [9]. Abbreviations: Chr = chromosome.
(DOCX)
Significance thresholds for each region are presented for SNPs and indels for the single variant analysis and for SNPs for the collapsing methods (no indels were included in the collapsing methods analyses). The column “GWAS gene” presents the gene reported in the lung function GWAS [9] for each region. Abbreviations: Chr = chromosome, N = number, UK10K+1000G = joint 1000 Genomes Project and UK10K reference panel.
(DOCX)
-
a) Stage 1Single variants results for stage 1 are presented for variants that met the criteria for follow-up. “Threshold” and “Threshold support” present the threshold and the threshold for supporting evidence for each region respectively. “GWAS gene” refers to the gene reported in the lung function GWAS [9] for each region. Abbreviations: chr = chromosome, Ref = reference, Alt = alternative, MAF = minor allele frequency and ac = allele count
-
b) Stage 2The column “GWAS gene” presents the gene reported in the lung function GWAS [9] for each region. The OR correspond to the effect on the alternative allele. Abbreviations: chr = chromosome, Ref = reference, Alt = alternative, MAF = minor allele frequency, OR = odds ratio, SE = standard error.
(DOCX)
The column “GWAS gene” presents the gene reported in the lung function GWAS [9] for each region. Abbreviations: chr = chromosome, Ref = reference, Alt = alternative, MAF = minor allele frequency, OR = odds ratio, SE = standard error.
(DOCX)
Results of COPD risk associations for variants previously associated with lung function [7–10] are presented here ordered by chromosome and position and by P-value significance for each variant. P-values < 0.05 are highlighted in bold. “GWAS gene” presents the closest gene to the lung function sentinel SNP reported in [9]. Abbreviations: MAF = minor allele frequency, N alt ac = number of alternative allele counts, N ref ac = number of reference allele counts, freq = frequency, OR = odds ratio.
(DOCX)
Results are presented for loci (sliding windows or genes) that reach the threshold for follow-up after sensitivity analyses either with (“Independent variants and variants not in UK10K+1000G”) or without (“Independent variants”) including variants not in joint 1000 Genomes Project [20] and UK10K [21] reference panel. Abbreviations: N = number of variants, P = P-value, UK10K+1000G = joint 1000 Genomes Project and UK10K reference panel.
- a) Burden test results in stage 1
- b) C-alpha test results in stage 1
- i) Sliding window
- ii) Gene based
- iii) Exon based
(DOCX)
“GWAS gene” is the gene reported in the lung function GWAS [9] for each region. P-values that reach a Bonferroni corrected threshold as defined in the methods section are highlighted in bold.
- a) Burden test results in stage 2
- b) C-alpha test results in stage 2
- iv) Sliding window
- v) Gene based
- vi) Exon based
(DOCX)
(DOCX)
Acknowledgments
The research undertaken by M.D.T., M.S.A., L.V.W. and N.S. was partly funded by the National Institute for Health Research (NIHR). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. M.D.T. holds a Medical Research Council Senior Clinical Fellowship (G0902313). This research used the ALICE High Performance Computing Facility at the University of Leicester. I.P.H. holds a Medical Research Council programme grant (G1000861). The UK BiLEVE study was funded by a Medical Research Council (MRC) strategic award to M.D.T., I.P.H., L.V.W. and David Strachan (MC_PC_12010). UK BiLEVE members are: Louise V Wain, Nick Shrine, Victoria E Jackson, Ioanna Ntalla, María Soler Artigas, Panos Deloukas, Richard Hubbard, Ian Pavord, Anna Hansell, Neil C Thomson, Eleftheria Zeggini, Andrew P Morris, Jonathan Marchini, David Strachan, Martin D Tobin, and Ian P Hall. This research has been conducted using the UK Biobank Resource under Application Number 648. This study makes use of data generated by the UK10K Consortium (www.UK10K.org). We would like to acknowledge Andrew J Wardlaw’s contribution.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The research undertaken by M.D.T., M.S.A., L.V.W. and N.S. was partly funded by the National Institute for Health Research (NIHR). The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health. M.D.T. holds a Medical Research Council Senior Clinical Fellowship (G0902313). I.P.H. holds a Medical Research Council programme grant (G1000861). The UK BiLEVE study was funded by a Medical Research Council (MRC) strategic award to M.D.T., I.P.H., L.V.W. and David Strachan (MC_PC_12010).
References
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–128. Epub 2012/12/19. 10.1016/S0140-6736(12)61728-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbey DE, Burchette RJ, Knutsen SF, McDonnell WF, Lebowitz MD, Enright PL. Long-term particulate and other air pollutants and lung function in nonsmokers. American journal of respiratory and critical care medicine. 1998;158(1):289–98. Epub 1998/07/09. 10.1164/ajrccm.158.1.9710101 [DOI] [PubMed] [Google Scholar]
- 3.Global Initiative for Chronic Obstructive Lung Disease (GOLD). Global Strategy for the Diagnosis, Management and Prevention of COPD 2014. http://www.goldcopd.org/.
- 4.Ingebrigtsen T, Thomsen SF, Vestbo J, van der Sluis S, Kyvik KO, Silverman EK, et al. Genetic influences on Chronic Obstructive Pulmonary Disease—a twin study. Respiratory medicine. 2010;104(12):1890–5. Epub 2010/06/15. 10.1016/j.rmed.2010.05.004 [DOI] [PubMed] [Google Scholar]
- 5.Palmer LJ, Knuiman MW, Divitini ML, Burton PR, James AL, Bartholomew HC, et al. Familial aggregation and heritability of adult lung function: results from the Busselton Health Study. The European respiratory journal: official journal of the European Society for Clinical Respiratory Physiology. 2001;17(4):696–702. Epub 2001/06/13. [DOI] [PubMed] [Google Scholar]
- 6.Lewitter FI, Tager IB, McGue M, Tishler PV, Speizer FE. Genetic and environmental determinants of level of pulmonary function. American journal of epidemiology. 1984;120(4):518–30. Epub 1984/10/01. [DOI] [PubMed] [Google Scholar]
- 7.Repapi E, Sayers I, Wain LV, Burton PR, Johnson T, Obeidat M, et al. Genome-wide association study identifies five loci associated with lung function. Nature genetics. 2010;42(1):36–44. 10.1038/ng.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilk JB, Chen TH, Gottlieb DJ, Walter RE, Nagle MW, Brandler BJ, et al. A genome-wide association study of pulmonary function measures in the Framingham Heart Study. PLoS Genet. 2009;5(3):e1000429 Epub 2009/03/21. 10.1371/journal.pgen.1000429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Soler Artigas M, Loth DW, Wain LV, Gharib SA, Obeidat M, Tang W, et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nature genetics. 2011;43(11):1082–90. Epub 2011/09/29. 10.1038/ng.941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hancock DB, Eijgelsheim M, Wilk JB, Gharib SA, Loehr LR, Marciante KD, et al. Meta-analyses of genome-wide association studies identify multiple loci associated with pulmonary function. Nature genetics. 2010;42(1):45–52. 10.1038/ng.500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wain LV, Shrine N, Miller S, Jackson VE, Ntalla I, Artigas MS, et al. Novel insights into the genetics of smoking behaviour, lung function, and chronic obstructive pulmonary disease (UK BiLEVE): a genetic association study in UK Biobank. The Lancet Respiratory medicine. 2015;3(10):769–81. Epub 2015/10/02. 10.1016/S2213-2600(15)00283-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Soler Artigas M, Wain LV, Miller S, Kheirallah AK, Huffman JE, Ntalla I, et al. Sixteen new lung function signals identified through 1000 Genomes Project reference panel imputation. Nature communications. 2015;6:8658 Epub 2015/12/05. 10.1038/ncomms9658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho MH, Boutaoui N, Klanderman BJ, Sylvia JS, Ziniti JP, Hersh CP, et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nature genetics. 2010;42(3):200–2. 10.1038/ng.535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho MH, McDonald ML, Zhou X, Mattheisen M, Castaldi PJ, Hersh CP, et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. The Lancet Respiratory medicine. 2014;2(3):214–25. Epub 2014/03/14. 10.1016/S2213-2600(14)70002-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pillai SG, Ge D, Zhu G, Kong X, Shianna KV, Need AC, et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genet. 2009;5(3):e1000421 Epub 2009/03/21. 10.1371/journal.pgen.1000421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilk JB, Shrine NR, Loehr LR, Zhao JH, Manichaikul A, Lopez LM, et al. Genome-wide association studies identify CHRNA5/3 and HTR4 in the development of airflow obstruction. American journal of respiratory and critical care medicine. 2012;186(7):622–32. Epub 2012/07/28. 10.1164/rccm.201202-0366OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castaldi PJ, Cho MH, Litonjua AA, Bakke P, Gulsvik A, Lomas DA, et al. The Association of Genome-Wide Significant Spirometric Loci with COPD Susceptibility. Am J Respir Cell Mol Biol. 2011. Epub 2011/06/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soler Artigas M, Wain LV, Repapi E, Obeidat M, Sayers I, Burton PR, et al. Effect of five genetic variants associated with lung function on the risk of chronic obstructive lung disease, and their joint effects on lung function. American journal of respiratory and critical care medicine. 2011;184(7):786–95. Epub 2011/10/04. 10.1164/rccm.201102-0192OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. Epub 2009/10/09. 10.1038/nature08494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.1000 Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. Epub 2012/11/07. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.UK10K Consortium, Walter K, Min JL, Huang J, Crooks L, Memari Y, et al. The UK10K project identifies rare variants in health and disease. Nature. 2015;526(7571):82–90. Epub 2015/09/15. 10.1038/nature14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stewart CE, Hall IP, Parker SG, Moffat MF, Wardlaw AJ, Connolly MJ, et al. PLAUR polymorphisms and lung function in UK smokers. BMC medical genetics. 2009;10:112 10.1186/1471-2350-10-112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20(9):1297–303. Epub 2010/07/21. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altmann A, Weber P, Quast C, Rex-Haffner M, Binder EB, Muller-Myhsok B. vipR: variant identification in pooled DNA using R. Bioinformatics. 2011;27(13):i77–84. Epub 2011/06/21. 10.1093/bioinformatics/btr205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wei Z, Wang W, Hu P, Lyon GJ, Hakonarson H. SNVer: a statistical tool for variant calling in analysis of pooled or individual next-generation sequencing data. Nucleic acids research. 2011;39(19):e132 Epub 2011/08/05. 10.1093/nar/gkr599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rivas MA, Beaudoin M, Gardet A, Stevens C, Sharma Y, Zhang CK, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nature genetics. 2011;43(11):1066–73. Epub 2011/10/11. 10.1038/ng.952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic acids research. 2001;29(1):308–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, et al. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome research. 2006;16(9):1182–90. 10.1101/gr.4565806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, et al. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7(3):e1001322 10.1371/journal.pgen.1001322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenbloom KR, Sloan CA, Malladi VS, Dreszer TR, Learned K, Kirkup VM, et al. ENCODE data in the UCSC Genome Browser: year 5 update. Nucleic acids research. 2013;41(Database issue):D56–63. 10.1093/nar/gks1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hwang JY, Lee SH, Go MJ, Kim BJ, Kou I, Ikegawa S, et al. Meta-analysis identifies a MECOM gene as a novel predisposing factor of osteoporotic fracture. Journal of medical genetics. 2013;50(4):212–9. 10.1136/jmedgenet-2012-101156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, et al. Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nature genetics. 2012;44(8):904–9. 10.1038/ng.2352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bei JX, Li Y, Jia WH, Feng BJ, Zhou G, Chen LZ, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nature genetics. 2010;42(7):599–603. Epub 2010/06/01. 10.1038/ng.601 [DOI] [PubMed] [Google Scholar]
- 36.Wain LV, Verwoert GC, O'Reilly PF, Shi G, Johnson T, Johnson AD, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nature genetics. 2011;43(10):1005–11. Epub 2011/09/13. 10.1038/ng.922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer TE, Verwoert GC, Hwang SJ, Glazer NL, Smith AV, van Rooij FJ, et al. Genome-wide association studies of serum magnesium, potassium, and sodium concentrations identify six Loci influencing serum magnesium levels. PLoS Genet. 2010;6(8). Epub 2010/08/12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bard-Chapeau EA, Szumska D, Jacob B, Chua BQ, Chatterjee GC, Zhang Y, et al. Mice carrying a hypomorphic Evi1 allele are embryonic viable but exhibit severe congenital heart defects. PloS one. 2014;9(2):e89397 Epub 2014/03/04. 10.1371/journal.pone.0089397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi YW, Choi JS, Zheng LT, Lim YJ, Yoon HK, Kim YH, et al. Comparative genomic hybridization array analysis and real time PCR reveals genomic alterations in squamous cell carcinomas of the lung. Lung cancer. 2007;55(1):43–51. Epub 2006/11/18. 10.1016/j.lungcan.2006.09.018 [DOI] [PubMed] [Google Scholar]
- 40.Starr TK, Allaei R, Silverstein KA, Staggs RA, Sarver AL, Bergemann TL, et al. A transposon-based genetic screen in mice identifies genes altered in colorectal cancer. Science. 2009;323(5922):1747–50. Epub 2009/03/03. 10.1126/science.1163040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yokoi S, Yasui K, Iizasa T, Imoto I, Fujisawa T, Inazawa J. TERC identified as a probable target within the 3q26 amplicon that is detected frequently in non-small cell lung cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2003;9(13):4705–13. Epub 2003/10/29. [PubMed] [Google Scholar]
- 42.Goyama S, Yamamoto G, Shimabe M, Sato T, Ichikawa M, Ogawa S, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell stem cell. 2008;3(2):207–20. Epub 2008/08/07. 10.1016/j.stem.2008.06.002 [DOI] [PubMed] [Google Scholar]
- 43.Yuasa H, Oike Y, Iwama A, Nishikata I, Sugiyama D, Perkins A, et al. Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. The EMBO journal. 2005;24(11):1976–87. Epub 2005/05/13. 10.1038/sj.emboj.7600679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoyt PR, Bartholomew C, Davis AJ, Yutzey K, Gamer LW, Potter SS, et al. The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mechanisms of development. 1997;65(1–2):55–70. Epub 1997/07/01. [DOI] [PubMed] [Google Scholar]
- 45.Zhou X, Baron RM, Hardin M, Cho MH, Zielinski J, Hawrylkiewicz I, et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Human molecular genetics. 2012;21(6):1325–35. 10.1093/hmg/ddr569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen X, Listman JB, Slack FJ, Gelernter J, Zhao H. Biases and errors on allele frequency estimation and disease association tests of next-generation sequencing of pooled samples. Genet Epidemiol. 2012;36(6):549–60. 10.1002/gepi.21648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hankinson JL, Crapo RO, Jensen RL. Spirometric reference values for the 6-s FVC maneuver. Chest. 2003;124(5):1805–11. Epub 2003/11/08. [DOI] [PubMed] [Google Scholar]
- 48.Hankinson JL, Kawut SM, Shahar E, Smith LJ, Stukovsky KH, Barr RG. Performance of American Thoracic Society-recommended spirometry reference values in a multiethnic sample of adults: the multi-ethnic study of atherosclerosis (MESA) lung study. Chest. 2010;137(1):138–45. Epub 2009/09/11. 10.1378/chest.09-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–2. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, et al. The UCSC Table Browser data retrieval tool. Nucleic acids research. 2004;32(Database issue):D493–6. Epub 2003/12/19. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lawrence R, Day-Williams AG, Elliott KS, Morris AP, Zeggini E. CCRaVAT and QuTie-enabling analysis of rare variants in large-scale case control and quantitative trait association studies. BMC bioinformatics. 2010;11:527 Epub 2010/10/23. 10.1186/1471-2105-11-527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity. 2005;95(3):221–7. Epub 2005/08/04. 10.1038/sj.hdy.6800717 [DOI] [PubMed] [Google Scholar]
- 53.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529 Epub 2009/06/23. 10.1371/journal.pgen.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
- a) Venn diagrams of SNPs called by any of the three algorithms with and without a 1% minor allele frequency (MAF) filter. The proportion of SNPs included in dbSNP137 [29] for each section is presented in brackets.
(DOCX)
(DOCX)
A drop one plot for a locus is obtained by undertaking the C-alpha test removing one variant at a time; the P-value plotted for each variant represented on the x-axis is the P-value obtained after removing that variant. Results obtained using calls from different calling algorithms are represented in different colours, according to the legend in each figure. Not all calling algorithms called the same set of variants. When there is no visible P-value represented for a variant for a given calling algorithm, it is because that variant was not called by that calling algorithm. If a region only includes two variants, no drop one plot is produced, since no C-alpha test can be undertaken with only one variant. Asymptotic P-values are presented here for the C-alpha test.
The single variant plots show the P-values obtained for each variant, and they also show the direction of effect by plotting on the y-axis the -log10 (P-value) x direction of effect, with the direction of effect = 1 if OR > 1 and direction of effect = -1 if OR < 1.
For each region the first row shows drop one plots and the second row shows single variant plots for the same variants; the first column presents results from stage 1 excluding variants not in the joint 1000 Genomes Project [20] and UK10K [21] reference panel (UK10K+1000G), the second column presents results from stage 1 including variants not in this reference panel (marked with * on the x-axis) and the third column presents results from stage 2. Variants that are included both in stage 1 and stage 2 are highlighted in bold. The horizontal dashed line represents the collapsing method significance threshold for each region (note that different thresholds were used for stage 1 and stage 2).
- a) chr3:25633833–25636833 (RARB)
- b) chr3:169238286–169241286 (MECOM)
- c) chr4:145293600–145296600 (HHIP)
- d) TNXB (AGER)
(DOCX)
Abbreviations: N = number, sd = standard deviation, y = years, l = litres.
(DOCX)
“GWAS sentinel” and “GWAS gene” present the lung function GWAS sentinel SNP and the closest gene to the sentinel SNP respectively [9]. Abbreviations: Chr = chromosome.
(DOCX)
Significance thresholds for each region are presented for SNPs and indels for the single variant analysis and for SNPs for the collapsing methods (no indels were included in the collapsing methods analyses). The column “GWAS gene” presents the gene reported in the lung function GWAS [9] for each region. Abbreviations: Chr = chromosome, N = number, UK10K+1000G = joint 1000 Genomes Project and UK10K reference panel.
(DOCX)
-
a) Stage 1Single variants results for stage 1 are presented for variants that met the criteria for follow-up. “Threshold” and “Threshold support” present the threshold and the threshold for supporting evidence for each region respectively. “GWAS gene” refers to the gene reported in the lung function GWAS [9] for each region. Abbreviations: chr = chromosome, Ref = reference, Alt = alternative, MAF = minor allele frequency and ac = allele count
-
b) Stage 2The column “GWAS gene” presents the gene reported in the lung function GWAS [9] for each region. The OR correspond to the effect on the alternative allele. Abbreviations: chr = chromosome, Ref = reference, Alt = alternative, MAF = minor allele frequency, OR = odds ratio, SE = standard error.
(DOCX)
The column “GWAS gene” presents the gene reported in the lung function GWAS [9] for each region. Abbreviations: chr = chromosome, Ref = reference, Alt = alternative, MAF = minor allele frequency, OR = odds ratio, SE = standard error.
(DOCX)
Results of COPD risk associations for variants previously associated with lung function [7–10] are presented here ordered by chromosome and position and by P-value significance for each variant. P-values < 0.05 are highlighted in bold. “GWAS gene” presents the closest gene to the lung function sentinel SNP reported in [9]. Abbreviations: MAF = minor allele frequency, N alt ac = number of alternative allele counts, N ref ac = number of reference allele counts, freq = frequency, OR = odds ratio.
(DOCX)
Results are presented for loci (sliding windows or genes) that reach the threshold for follow-up after sensitivity analyses either with (“Independent variants and variants not in UK10K+1000G”) or without (“Independent variants”) including variants not in joint 1000 Genomes Project [20] and UK10K [21] reference panel. Abbreviations: N = number of variants, P = P-value, UK10K+1000G = joint 1000 Genomes Project and UK10K reference panel.
- a) Burden test results in stage 1
- b) C-alpha test results in stage 1
- i) Sliding window
- ii) Gene based
- iii) Exon based
(DOCX)
“GWAS gene” is the gene reported in the lung function GWAS [9] for each region. P-values that reach a Bonferroni corrected threshold as defined in the methods section are highlighted in bold.
- a) Burden test results in stage 2
- b) C-alpha test results in stage 2
- iv) Sliding window
- v) Gene based
- vi) Exon based
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.