Abstract
The genomes of clusters of related eukaryotes are now being sequenced at an increasing rate, creating a need for accurate, low-cost annotation of exon–intron structures. In this paper, we demonstrate that reverse transcription-polymerase chain reaction (RT–PCR) and direct sequencing based on predicted gene structures satisfy this need, at least for single-celled eukaryotes. The TWINSCAN gene prediction algorithm was adapted for the fungal pathogen Cryptococcus neoformans by using a precise model of intron lengths in combination with ungapped alignments between the genome sequences of the two closely related Cryptococcus varieties. This approach resulted in ∼60% of known genes being predicted exactly right at every coding base and splice site. When previously unannotated TWINSCAN predictions were tested by RT–PCR and direct sequencing, 75% of targets spanning two predicted introns were amplified and produced high-quality sequence. When targets spanning the complete predicted open reading frame were tested, 72% of them amplified and produced high-quality sequence. We conclude that sequencing a small number of expressed sequence tags (ESTs) to provide training data, running TWINSCAN on an entire genome, and then performing RT–PCR and direct sequencing on all of its predictions would be a cost-effective method for obtaining an experimentally verified genome annotation.
The first eukaryotic genomes to be sequenced were those of well studied model organisms. Their gene structures were annotated by manual curation of diverse evidence from sequence databases and, to a lesser extent, gene predictions. We have recently entered an era in which clusters of related organisms are being sequenced. Many of these organisms are almost completely unstudied, and their genomes will not be annotated using extensive manual curation. This has created a need for low-cost, high-accuracy automated gene structure annotation. In this paper, we show that reverse transcription-polymerase chain reaction (RT–PCR) and direct sequencing based on predicted gene structures satisfy that need. We develop and test this method using the compact genomes of two varieties of Cryptococcus neoformans.
C. neoformans is an encapsulated yeast of the phylum Basidiomycota. It is the opportunistic pathogen responsible for cryptococcal meningitis, a potentially fatal disease which affects individuals whose immune systems are compromised due to AIDS, chemotherapy, or organ transplant. There are three varieties of C. neoformans, identified by their capsular polysaccharide antigens: Serotype A (C. neoformans var. grubii), Serotype D (C. neoformans var. neoformans), and Serotypes B and C (C. neoformans var. gattii). The genomes of Serotypes A and D have been sequenced to 11-fold redundancy; fourfold shotgun coverage of Serotype B was made public in March 2004 (after the completion of the work reported here).
Cryptococcus is an attractive system for RT–PCR-based annotation because it has relatively complex gene structures, yet it is a single-celled organism, which simplifies the task of obtaining representative mRNA samples. Furthermore, the novel characteristics of the Cryptococcus genome made it an interesting target for de novo gene structure prediction. Unlike those of all previously published fungal genome sequences, the genes of Cryptococcus contain numerous introns (avg. 4.5 per gene). Furthermore, these introns are shorter (avg. 68 base pairs) than those of plants and animals, the only organisms known to have similarly numerous introns.
When we began to study Cryptococcus, the limited number of available expressed sequence tag (EST) and mRNA sequences had been manually curated at The Institute for Genome Research (TIGR), yielding 431 curated gene structures. However, little effort had been devoted to specializing gene structure prediction programs for this clade. Cryptococcus parameters had been estimated for two programs, GlimmerM (Salzberg et al. 1999) and PHAT (Cawley et al. 2001), but accuracy was not as high as had been hoped—both programs predicted less than 20% of known genes exactly (although a recently retrained version of GlimmerHMM approaches 30% gene sensitivity). The unique characteristics of Cryptococcus genes appeared to require innovation rather than simple retraining. For this reason, we set out to adapt TWINSCAN to Cryptococcus. TWINSCAN is a dual genome, de novo gene structure prediction program that we had developed for annotating the mouse and human genomes by comparison to one another (Korf et al. 2001; Flicek et al. 2003).
The application of TWINSCAN to the two Cryptococcus varieties was challenging because their genomes are so different from those of mouse and human. Cryptococcus Serotypes A and D are much more similar (95% identical in aligned coding regions) than mouse and human (86% identical in aligned coding regions). We did not know whether comparison of such similar genomes would be useful for gene structure prediction, as the noncoding sequences would not have diverged much. A second difference from the mammalian genomes is the very short introns of Cryptococcus genes (avg. 68 bp). These posed two problems. First, very short introns may, by chance, lack stop codons that interrupt the open reading frame (ORF) and suggest the presence of an intron. Second, the central regions of the intron that are evolving neutrally (outside of the conserved splice acceptor, splice donor, and branch point regions) are often less than 15 bp long. Thus, they may lack clear signs of unconstrained mutation, especially given the scant divergence between the genomes available for comparison.
The first step we took to overcome these challenges was to perform ungapped BLASTN alignments between the Serotype A and D strains. In general, gaps representing inserted and deleted bases are more common in noncoding alignments than in coding alignments, but mouse and human are so far diverged that gaps occur in coding sequence as well. The two strains of Cryptococcus, in contrast, are so closely related that their coding sequences have essentially no insertions or deletions. Thus, ungapped alignments between the Cryptococcus varieties cover all annotated coding regions, but are frequently broken by a single insertion or deletion in noncoding regions. These breaks in the alignments turned out to provide key evidence about the locations of introns and intergenic regions.
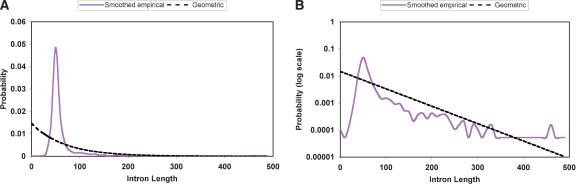
We also took advantage of the short introns and tight intron length distribution in Cryptococcus by constructing a smoothed empirical model of the intron length distribution (Fig. 1). This required a major change to the Hidden Markov Model (HMM) algorithms on which TWINSCAN is based, resulting in a completely new and much more flexible implementation that we identify as TWINSCAN 2.0α (available at http://genes.cse.wustl.edu). The original implementation relied on the partially generalized HMM that GENSCAN uses (Burge and Karlin 1997), which requires the lengths of introns to be modeled by a geometric probability distribution. Although the geometric distribution greatly simplifies and speeds the program, it is far from an accurate model of intron lengths (Fig. 1). By moving to a fully generalized HMM, we were able to turn the short introns of Cryptococcus to advantage.
Figure 1.
Intron length probability distributions used. The smoothed empirical distribution (purple, solid line) closely mirrors the observed intron lengths in the training set (not shown). The geometric distribution (black, dashed line) is the unique member of the geometric family with mean intron length equal to that of the training set (68 bp), but it is clearly a poor fit to the observed distribution. (A) Linear scales; (B) log scale on probability axis.
The result of these efforts is a gene structure prediction program whose accuracy is much greater than has ever been achieved on mammalian genomes. After computational experiments indicated good agreement between TWINSCAN predictions and curated gene structures, we evaluated both the predictions and the curated structures by RT–PCR and direct sequencing. Because experiments aimed at amplifying short segments of cDNAs confirmed the accuracy of the predictions, we went on to amplify and sequence complete ORFs. These experiments constitute a pilot study for experimental annotation of Cryptococcus by amplification and sequencing of all TWINSCAN predictions. The success of the pilot study suggests a new approach to annotation in which gene predictions serve as the hypotheses that drive genome-wide experimental determination of exon–intron structures.
Results
In order to develop an accurate gene finder for Cryptococcus, we experimented with a number of alignment methods and probability model enhancements. Each of these changes was evaluated in computational experiments by training probability model parameters using some of the 431 curated gene structures available at the time, and testing on others. The best results were achieved by using ungapped BLASTN alignments with match score +1 and mismatch score –3. The most productive change to the probability model was replacing the geometric model of intron length with a smoothed empirical model that closely mirrored the observed intron lengths. The RT–PCR and sequencing experiments reported below were obtained using this system, with parameters estimated from the 431 curated genes. Subsequently, TIGR sequenced many additional ESTs and produced a much more comprehensive and more accurate set of curated gene structures, of which 1483 were fully confirmed by ESTs. Retraining and retesting the same system with these more numerous and more accurate curated genes resulted in small but noteworthy improvements in accuracy. The computational results reported below are based on the most recent parameter set, in order to give the most up-to-date picture of the accuracy of gene prediction in Cryptococcus.
Computational experiments
A generalized HMM version of TWINSCAN containing a smoothed empirical intron length model was used to make gene predictions for C. neoformans Serotype D strain JEC21, using ungapped alignments to the genome sequence of Serotype A, strain H99. Both genome model parameters and evolutionary conservation parameters were trained using the 1483 EST confirmed C. neoformans serotype D genes obtained from TIGR. TWINSCAN annotation of the entire genome resulted in 62% of all known genes being predicted exactly right at every base pair. To ensure that this high accuracy was not unduly influenced by training and testing on the same genes, we held out subsets of the genes during training and tested on the held out sets. To enable the estimation of specificity as well as sensitivity, the testing set consisted of the coding and intron regions of the 1483 TIGR genes along with 250 base pairs of flanking sequence. These genes were concatenated into a single sequence with an additional 250 base pairs of randomized sequence between adjacent genes. Prediction accuracy was measured by repeatedly training on 90% of the data and testing on the other 10% (10-fold cross validation). Evaluation of the predictions showed that 60% of genes were predicted exactly right at every base pair, as were 88% of exons.
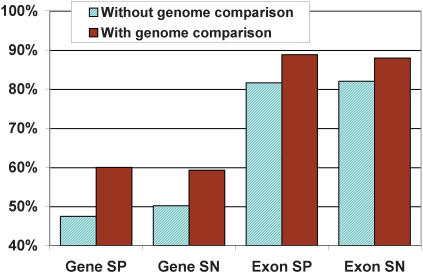
To determine the effect of genome alignment information on prediction accuracy, we ran TWINSCAN on the cross-validation set without genomic alignments but with the empirical intron length distribution. The results show that including a comparison of the Serotype D and Serotype A genomes yields a 10% improvement in exact gene prediction and a 7% improvement in exact exon prediction over the single-genome accuracy, despite the minimal evolutionary divergence between the two genomes (Fig. 2).
Figure 2.
Accuracy of the gene prediction set generated without genome comparison and the final prediction set, which was generated using comparison to the Serotype A genome. The smoothed empirical intron length model was used in generating both sets.
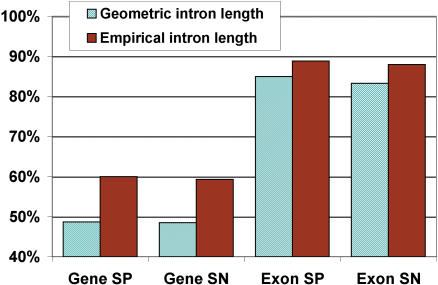
To evaluate how the new empirical intron length model affected prediction accuracy, predictions were made on the cross-validation set using the original geometric intron length distribution (along with the genomic alignments). The results show that the smoothed empirical intron length model provides a striking 12% boost in exact gene prediction and a 4% increase in exact exon prediction (Fig. 3).
Figure 3.
Comparison of TWINSCAN predictions generated using the geometric intron length model to the final TWINSCAN prediction set, which was generated using the smoothed empirical length model. Comparison to the Serotype A genome was used in generating both sets.
Assessment of TWINSCAN predictions and TIGR annotations by RT–PCR
Once comparison of predictions to curated gene structures indicated satisfactory performance, we moved to evaluation by RT–PCR and direct sequencing. The first goal was to experimentally test the predictions that disagreed with curated genes. We therefore designed primers to amplify cDNA regions spanning three categories of introns from the curated genes that disagreed with the TWINSCAN predictions: (1) those with GC/AG splice sites, which are rare but known to exist; (2) those with splice sites that were neither GC/AG nor GT/AG, which are thought not to exist in fungi; and (3) those with standard GT/AG splice sites. RT–PCR was carried out using RNA from Serotype D cells grown under a variety of conditions (see Methods). The products were then sequenced from both PCR primers without gel purification or cloning. The resulting sequences were aligned back to the genome by using the program EST_GENOME (Mott 1997). Only spliced sequences were considered positive results.
The results were as follows. Of the 25 GC/AG introns, we obtained spliced sequences that determined both intron boundaries in 20 cases, all of which agreed with the curated annotations. Of the 38 splice sites in 11 curated genes that did not conform to either the GC/AG or the GT/AG consensus, we obtained spliced sequence that determined the intron boundaries for 27. None of these experimentally determined introns agreed with the nonstandard splice sites in the curated annotations. Alignment of the RT–PCR sequences back to the genome revealed that the correct splice sites all obeyed the standard GT/AG or GC/AG consensus. Furthermore, most of the nonstandard annotated splice sites lay near standard splice sites that we verified. The genes containing the nonstandard splice sites were discarded from the training and testing sets in subsequent experiments, yielding improved TWINSCAN accuracy.
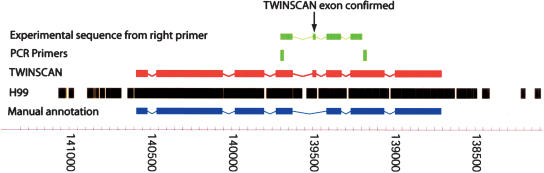
To test the third category, we selected a set of 22 predicted genes at random from among those that overlapped and disagreed with manually annotated genes. Primers were designed to anneal to coding regions of the TIGR annotations and to amplify a region containing an intron on which the two sets disagreed. Targets were successfully amplified and sequenced from 15 of the genes. Of the experimentally determined introns, 12 (80%) agreed with the manual curation, and three (20%) agreed with the TWINSCAN predictions. An example in which the experimental result agrees with the TWINSCAN prediction is shown in Figure 4.
Figure 4.
A curated annotation (blue), blocks of ungapped alignment from the genome sequence of Serotype A Strain H99 (black), the TWINSCAN prediction (red), and the PCR primers and experimental sequence aligned back to the genome (green). TWINSCAN's prediction of the missing exon is influenced by both the long ungapped alignment from H99 and the unusually (though not impossibly) long intron in the curated gene structure.
Amplifying segments of predicted genes
Having assessed the accuracy of manual annotations that disagreed with TWINSCAN predictions, we wanted to estimate the accuracy of TWINSCAN predictions that did not overlap annotated genes. We randomly selected 108 such genes and performed RT–PCR using primers designed to amplify a segment spanning two introns along with the complete exon between them. Alignment of the resulting sequences back to the genome revealed that at least one spliced sequence was obtained from 83 of the predicted genes (75%). For those cases that were determined experimentally, the predicted exons were exactly correct 78% of the time, the predicted introns were exactly correct 87% of the time, and the predicted splice sites were correct 92% of the time (Table 1).
Table 1.
Results for RT-PCR tests of two-intron targets from TWINSCAN predictions
| Targeted | Experimentally determined | Prediction correct | Correct over determined | |
|---|---|---|---|---|
| Intron | 216 | 147 | 128 | 0.87 |
| Exon | 108 | 63 | 49 | 0.78 |
| Splice Site | 432 | 294 | 271 | 0.92 |
“Targeted” is the number of predicted introns, exons, or splice sites between the PCR primers. “Experimentally determined” is the number of each that we could determine unambiguously by aligning high-quality experimental sequence back to the genome. “Prediction correct” is the number of experimentally determined introns, exons, or splice sites that match the prediction exactly. “Correct over determined” is the number of predicted features that were verified as a fraction of the number that could be determined experimentally.
Amplifying complete predicted ORFs
The final goal was to test our predictions of complete gene structures. We picked another 88 predicted genes at random from among those that did not overlap annotated genes. The targeted predictions varied from two to 20 exons. This time we designed primers in the predicted untranslated regions (UTRs) in order to amplify the whole gene. Of the 88 targets, 63 amplified and produced spliced sequences (72%). The other 28% may not have amplified and produced spliced sequences due to incorrect predictions, lack of expression in the limited growth conditions tested, problems with primer design or synthesis, or other experimental failure in PCR or sequencing. In keeping with our goal of developing a low-cost, high-throughput method, we did not revisit failed targets.
The accuracy of the predicted exons and introns in the genes we sequenced was also high (89% and 87%, respectively; Table 2). The experimental sequences were completely consistent with the predictions in 43 cases (68% of those yielding spliced sequence), although some of these experimental sequences did not fully cover the predicted ORF. Nine of the 23 targets that were completely covered by high-quality sequence were exactly as predicted at every splice site (39%). There were no clear trends in the rate of amplification or exactly correct prediction as a function of the targets' lengths or exon counts.
Table 2.
Results of whole-gene RT–PCR tests of TWINSCAN predictions (see Table 1 caption)
| Targeted | Experimentally determined | Prediction correct | Correct/determined ratio | |
|---|---|---|---|---|
| Intron | 374 | 203 | 177 | 0.87 |
| Exon | 286 | 123 | 110 | 0.89 |
| Splice Site | 748 | 406 | 376 | 0.93 |
Methods
Intron length model
GENSCAN and earlier versions of TWINSCAN used a geometric probability distribution to model intron lengths. The probability of an intron having length i was calculated as: (1–p) pi–1, where p is chosen so that the mean of this geometric distribution is the same as the empirical average intron length. In the current implementation, the probability of an intron having length i is calculated directly from the empirical distribution of intron lengths, for all i ≤ 500; introns longer than 500 are given probability 0.
The introns in the training set are grouped by length into 50 bins of 10 nt, up to a maximum length of 500 nt. After adding one to smooth the estimates, the count in each bin is divided by the total count over all bins. This normalizes the probabilities so that they sum to 1.
Genome sequences
Predictions were made on the three strains of C. neoformans for which genome sequence is available, JEC21 (Serotype D), H99 (Serotype A), and B3501 (Serotype D). These predictions are available on our Web site. The JEC21 sequences were obtained from TIGR on January 27th 2003 and represent 9× coverage. H99 sequences were obtained from the Whitehead Institute on March 19th 2003 and June 4th 2003, and represent 9× and 11× coverage, respectively. B3501 sequences were obtained from the Stanford C. neoformans genome project on June 6th 2003. Predictions on the JEC21 and B3501 sequences were made by using H99 as the informant genome, whereas predictions on H99 were made by using JEC21 as informant.
BLAST parameters
As repeat libraries do not yet exist for C. neoformans, no attempt was made to mask the sequences. WU-BLAST 2.0 was used with BLASTn parameters M=1 N=–3 B=100000000 V=100 -nogap.
Primer design
Primer design was done by using Primer3 (Rozen and Skaletsky 2000) with default parameters.
Annotations
The predictions used for the RT–PCR experiments were obtained by training TWINSCAN on 420 of the 431 gene structures obtained from TIGR in March 2002 (11 were excluded due to formatting errors).
cDNA preparation
C. neoformans strain JEC21 was obtained from Joe Heitman (Duke University) and used for all experiments. Two batches of cDNA were used. The first was made from a combination of C. neoformans grown in YPD (1% yeast extract, 1% bacto-peptone, and 2% dextrose) at 30°C, low-iron medium (LIM, Jacobson et al. 1998) at 37°C, YNB pH 7.0 (6.7g/L yeast nitrogen base without amino acids plus 20g/L dextrose and 50 mM MOPS at pH 7.0) at 37°C, YNB pH 7.0 at 25°C, YNB pH 7.0 at 25°C treated with 1 mM H2O2 for 3 h, and YNB pH 4.0 (50 mM succinate, pH 4.0) at 25°C treated with NaNO2 for 3 h. The second batch included RNA from all of the above conditions with the addition of YNB pH 7.0 lacking glucose but containing other carbon sources (2% acetate, 2% succinate, or 2% raffinose). In all cases, cells were grown overnight in the appropriate liquid medium with aeration until midlog phase (107–108/mL). Cells were collected by centrifugation, washed once in PBS, and flash frozen in liquid nitrogen, and the pellets were lyophilized overnight until dry. Cells were disrupted by adding 4 mL of Trizol (Invitrogen) to cells from a 50-mL culture. Following lysis, 0.8 mL of chloroform was added, and the mixture was centrifuged to separate the layers. The upper, aqueous layer was transferred to a fresh tube; 2 mL of isopropanol was added, and the samples were centrifuged to precipitate the RNA. The pellet was washed once in 75% ethanol and the RNA resuspended in RNase free water. One mg of total RNA was used to isolate mRNA using a Qiagen Oligotex kit according to the manufacturer's instructions. The mRNA was treated with DNase (Roche) and repurified with an Oligotex kit (QIAGEN). cDNA was generated from the mRNA using either oligo dT or random oligonucleotides and a first-strand cDNA synthesis kit (Roche) according to the manufacturer's instructions.
PCR, sequencing, and sequence analysis
PCR reactions were performed using REDTaq enzyme with buffer provided by the vendor (Sigma) to amplify cDNA prepared as above, using primers as described above. The amplification program consisted of 94°C for 2 min; 30 cycles of 94°C for 1 min/56°C for 1 min/72°C for 1 min per kb of expected product; and 72°C for 7 min. A portion of each reaction was analyzed by agarose gel electrophoresis (Ready-to-Run gels, Amersham Biosciences), and the remainder was purified using a GFX-96 PCR purification kit (Amersham Biosciences) for DNA sequencing at the Oklahoma University Health Sciences Center (see Web site http://micro-gen.ouhsc.edu for details). Purified products were sequenced twice, once from each PCR primer. Experimental sequences and traces were deposited in the NCBI trace repository (http://www.ncbi.nlm.nih.gov/Traces/) with trace id numbers 513872104-513872538 and 518318058-518318101. Where possible, the two reads were assembled into one sequence by using PHRAP (P. Green, unpubl.). The resulting sequence(s) were aligned back to the genome sequence by using EST_GENOME (Mott 1997), a splice-site aware alignment program.
Discussion
The results presented here suggest a powerful new approach to genome annotation: RT–PCR and direct sequencing of all predicted genes. This approach is complementary to EST sequencing, in that RT–PCR is much more sensitive and cheaper for genes that are expressed at relatively low levels, whereas ESTs provide low-cost data on highly expressed genes without reference to gene predictions. Furthermore, a small set of ESTs would provide training data for the gene prediction program, which plays a critical role in the RT–PCR phase.
We found that 72% of the Cryptococcus genes predicted by TWINSCAN, but not overlapping previously annotated genes, were amplified and end sequenced after trying just one primer pair per target. The estimated cost for RT–PCR is about $11 per predicted ORF, or $56,000 for the entire set of Cryptococcus predictions. A few additional primers and sequencing runs would be needed to fully sequence long ORFs. On the scale of genome sequencing and annotation this is a very modest investment. Indeed, the cost would be very similar to that of the recent project in which TIGR generated 46,000 Cryptococcus ESTs (B. Loftus, pers. comm.). By combining a prediction-driven PCR approach with a modest EST sequencing effort, it would be possible to produce annotation based on native cDNA sequence for most of the genes in Cryptococcus neoformans or any other single-celled eukaryote.
The RT–PCR approach is related to a number of so-called “ORFEOME” projects that aim to provide cDNA clones for the ORFs of all the genes in a genome (Strausberg et al. 2002; Reboul et al. 2003; The MGC Project Team 2004). However, because the goal of these projects is to obtain a physical resource, a significant portion of their effort is often devoted to amplifying and cloning known genes. Our approach, by contrast, is focused on obtaining information about genes whose complete ORFs are not already known. Further, our annotation method does not incur the costs associated with cloning and clone verification. ORFEOME projects do often go beyond known genes to produce information that is invaluable for genome annotation. In fact, a project aimed at obtaining full ORF clones of all Caenorhabditis elegans genes using the predictions of the program Gene Finder (P. Green, unpubl.) has produced good results (Reboul et al. 2003), and TWINSCAN predictions are now being used to fill out the collection (C. Weil, P. Lamesch, M. Arumuga, J. Rosenberg, P. Hu, M. Vidal, and M.R. Brent, in prep.). However, genome-wide RT–PCR and direct sequencing of gene predictions without cloning would be applicable to a wide range of genomes for which the cost of an ORFEOME project is not justified.
There are two primary factors that affect the efficiency of the hypothesis-driven approach to experimental genome annotation: the difficulty of obtaining an RNA pool in which most genes are expressed, and the difficulty of creating a highly accurate gene prediction program. Although we cannot expect to obtain representation of every gene, the completeness of the cDNA sample is affected by the complexity of the organism's body plan, life cycle, and behavior. Thus, a single-celled fungus such as C. neoformans presents a relatively easy case, compared to differentiated plants and animals.
The second factor influencing the efficiency of this approach for Cryptococcus was the high prediction accuracy we achieved using TWINSCAN: about 60% of known genes are predicted exactly right, and the predictions are even able to correct manually curated gene structures (normally thought of as a gold standard) in some cases. This success was enabled, in part, by the fact that Cryptococcus has a relatively compact genome with a small average intron length and a tight intron length distribution. These characteristics contributed to both the computational feasibility and the effectiveness of using a fully generalized hidden Markov model with a smoothed empirical model of intron lengths. Prediction accuracy was also improved by the availability of the genome sequence of a closely related organism. We were able to make effective use of these two genomes by creating ungapped alignments between them. Both of these factors (small introns and sequence from a closely related genome) are also present for Arabidopsis thaliana, on which we have achieved very high prediction accuracy (Allen et al. 2004; P. Hu, unpubl.), and, to a lesser extent, C. elegans. In mammalian genomes, by contrast, prediction accuracy is notably lower (currently about 20%–25% of known genes are predicted exactly right).
Although RT–PCR success rates decrease with increasing developmental and genomic complexity, good results have been obtained with C. elegans (C. Weil, P. Lamesch, M. Aramugam, J. Rosenberg, P. Hu, M. Vidal, and M.R. Brent), Arabidopsis thaliana (P. Hu, unpubl.), and even rat (Wu et al. 2004). Because gene prediction is less accurate and mRNA pools are less complete, mammalian genomes will likely continue to require more RT–PCR reactions per full-ORF cDNA amplicon. On the other hand, the scientific community has been willing to spend more money per gene in annotation of mammalian genomes. Thus, the approach described here is still likely to be a good value, especially compared to sequencing very large numbers of random clones from cDNA libraries. Indeed, the random clone approach appears to reach its limit at about one-half of the human genes (The MGC Project Team 2004).
Currently, most genome annotation efforts rely on some combination of native cDNA sequences obtained by sequencing random clones and homology to cDNAs or proteins from other organisms. The random clone approach results in sequencing hundreds of cDNAs from the same gene, whereas the homology approach draws inferences across species from sequences that may not be a good match for the genome to be annotated. Using the hypothesis-driven approach described here makes it possible, for a relatively modest cost, to obtain native cDNA sequence for most of the genes in compact genomes with numerous small introns.
Acknowledgments
We are grateful to Brendan Loftus and The Institute for Genome Research (TIGR) for providing DNA sequences, annotations, and EST sequences, and for their generous collegiality throughout this project; we are also grateful for Cryptococcus sequence generated by Broad Institute, the Stanford Genome Technology Center, the Duke Center for Genome Technology, and the British Columbia Genome Sequencing Centre. We thank Ian Korf for making available a pre-release version of his Zoe library (which was used in the creation of TWINSCAN 2.0α), Hong Liu (Doering lab) for carrying out laboratory work, Manimozhiyan Arumugam (Brent lab) for helping analyze the PCR sequences, and the Laboratory for Genomics and Bioinformatics at Oklahoma Health Sciences center for working with us on the DNA sequencing. Work in the Brent lab is supported in part by NIH grant R01-AI051209. A.T. was supported in part by T32 grant HG00045. R.H.B. was supported by F33 grant HG002635. Studies in the Doering laboratory are supported by NIH grants R01-GM66303 and R01-AI49173, a Burroughs Wellcome Fund New Investigator Award in Molecular Pathogenic Mycology, and an award from the Edward J. Mallinckrodt Jr., Foundation. Work in the Lodge lab is supported by NIH grants RO1-AI051209 and RO1-AI50184.
Article and publication are at http://www.genome.org/cgi/doi/10.1101/gr.2816704. Article published online before print in October 2004.
Footnotes
[All sequences, predictions, primers, traces, accession numbers, and links to software are available at http://genes.cse.wustl.edu/tenney-04-crypto-data/].
References
- Allen, J.E., Pertea, M., and Salzberg, S.L. 2004. Computational gene prediction using multiple sources of evidence. Genome Res. 14: 142-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burge, C. and Karlin, S. 1997. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268: 78-94. [DOI] [PubMed] [Google Scholar]
- Cawley, S.E., Wirth, A.I., and Speed, T.P. 2001. Phat—A gene finding program for Plasmodium falciparum. Mol. Biochem. Parasitol. 118: 167-174. [DOI] [PubMed] [Google Scholar]
- Flicek, P., Keibler, E., Hu, P., Korf, I., and Brent, M.R. 2003. Leveraging the mouse genome for gene prediction in human: From whole-genome shotgun reads to a global synteny map. Genome Res. 13: 46-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson, E., Goodner, A.P., and Nyhus, K.J. 1998. Ferrous iron uptake in Cryptococcus neoformans. Infect. Immun. 66: 4169-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf, I., Flicek, P., Duan, D., and Brent, M.R. 2001. Integrating genomic homology into gene structure prediction. Bioinformatics (Suppl.) 17: S140-148. [DOI] [PubMed] [Google Scholar]
- Mott, R. 1997. EST_GENOME: A program to align spliced DNA sequences to unspliced genomic DNA. Comput Appl. Biosci. 13: 477-478. [DOI] [PubMed] [Google Scholar]
- Reboul, J., Vaglio, P., Rual, J.F., Lamesch, P., Martinez, M., Armstrong, C.M., Li, S., Jacotot, L., Bertin, N., Janky, R., et al. 2003. C. elegans ORFeome version 1.1: Experimental verification of the genome annotation and resource for proteome-scale protein expression. Nat. Genet. 34: 35-41. [DOI] [PubMed] [Google Scholar]
- Rozen, S. and Skaletsky, H. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365-386. [DOI] [PubMed] [Google Scholar]
- Salzberg, S.L., Pertea, M., Delcher, A.L., Gardner, M.J., and Tettelin, H. 1999. Interpolated Markov models for eukaryotic gene finding. Genomics 59: 24-31. [DOI] [PubMed] [Google Scholar]
- Strausberg, R.L., Feingold, E.A., Grouse, L.H., Derge, J.G., Klausner, R.D., Collins, F.S., Wagner, L., Shenmen, C.M., Schuler, G.D., Altschul, S.F., et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. 99: 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The MGC Project Team. 2004. The status, quality and expansion of the NIH full-length cDNA project: The mammalian gene collection (MGC). Genome Res. 14: 2121-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.Q., Shteynberg, D., Arumugam, M., Gibbs, R.A., and Brent, M.R. 2004. Identification of rat genes by TWINSCAN gene prediction, RT–PCR, and direct sequencing. Genome Res. 14: 665-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
WEB SITE REFERENCES
- http://www.ncbi.nlm.nih.gov/Traces/; NCBI Trace Archive.
- http://genes.cse.wustl.edu/tenney-04-crypto-data/; Supplemental data for this paper.
- http://genes.cse.wustl.edu/; TWINSCAN home page, application, source code, and gene predictions.
- http://micro-gen.ouhsc.edu; Oklahoma University Health Sciences Center.