Abstract
Differentiating acute pyelonephritis (APN) from acute rejection (AR) in renal allograft biopsies can sometimes be difficult because of overlapping clinical and histologic features, lack of positive urine cultures, and variable response to antibiotics. We wanted to study differential gene expression between AR and APN using biopsy tissue. Thirty-three biopsies were analyzed using NanoString multiplex platform and PCR (6 transplant baseline biopsies, 8 AR, 15 APN [8 culture positive, 7 culture negative], and 4 native pyelonephritis [NP]). Additional 22 biopsies were tested by PCR to validate the results. CXCL9, CXCL10, CXCL11, and IDO1 were the top differentially expressed genes, upregulated in AR. Lactoferrin (LTF) and CXCL1 were higher in APN and NP. No statistically significant difference in transcript levels was seen between culture-positive and culture-negative APN biopsies. Comparing the overall mRNA signature using Ingenuity pathway analysis, interferon-gamma emerged as the dominant upstream regulator in AR and allograft APN, but not in NP (which clustered separately). Our study suggests that chemokine pathways in graft APN may differ from NP and in fact resemble AR, due to a component of alloreactivity, resulting in variable response to antibiotic treatment. Therefore, cautious addition of steroids might help in resistant cases of graft APN.
Keywords: acute pyelonephritis, acute rejection, CXCL1, CXCL10, CXCL11, renal allograft
1 | INTRODUCTION
Urinary tract infection complicated by acute pyelonephritis (APN) is a known complication in renal transplant recipients, especially in the early post-transplant period.1–4 Differentiating between renal allograft APN and acute rejection (AR) based on clinical symptoms alone can be difficult. The classic tetrad of costovertebral angle tenderness, fever, elevated white blood cell count, positive urine culture used to diagnose acute pyelonephritis in the native kidney, is frequently not useful for renal allografts. Fevers and leukocytosis can be attenuated by immunosuppressive therapy. Graft tenderness may be present even in acute rejection. Urine cultures can be frequently negative in renal allograft patients with APN, because these immunosuppressed patients receive long-term prophylactic antibiotics. Lack of positive urine cultures has also been reported in a high percentage of patients with native kidney pyelonephritis in a large case series of 223 patients by Rollino et al.5 in which they report that only 23.5% of their patients had positive urine cultures. In our previous study6 of 49 kidney transplant recipients with APN in first two years post-transplant, we showed that only 32% (16/49) had concomitant positive urine cultures at biopsy, and in 8 of these 16 patients, colony count was less than 105 CFU/mL. In 14 of 49 patients, positive urine culture did not coincide with the biopsy (had positive culture beyond 10 days before or after biopsy), and in 19 of 49 patients, urine cultures were negative. Urinalysis findings in patients with APN and AR can also overlap and may not be specific.
Renal allograft biopsy therefore plays an important role in diagnosis. Even on biopsy, APN vs AR can pose a differential diagnostic dilemma.7,8 Both conditions are associated with tubulointerstitial inflammation.9,10 Characteristic histologic features described for APN such as tubular microabscesses are usually focal in distribution and therefore may not get sampled in the biopsy specimen. Tubular apoptotic cell debris accompanying severe acute tubular necrosis (ATN) can mimic tubular microabscesses. Neutrophil-rich inflammatory infiltrate usually associated with infection and can occasionally be seen in the inflammatory infiltrate of AR as well. Although there are well-defined Banff criteria for the diagnosis and grading of AR (based on interstitial inflammation and tubulitis),11 tubulitis also tends to be focal. All these factors can complicate diagnosis of APN in renal allografts. APN in renal allografts can potentially affect long-term graft function by causing focal scars and reduction in functional parenchyma;3,4 therefore, early diagnosis and treatment are important.
Ours is a preliminary exploratory study aimed at assessing differential gene expression pattern of allograft rejection (AR) and allograft pyelonephritis (APN), using archived biopsy material. We included small number of biopsies of native kidney pyelonephritis as a comparison group. Additionally, we studied patients whose biopsy shows features of APN but have negative urine culture results. These biopsies pose a diagnostic dilemma, and therefore, our second aim was to see whether these biopsies show gene expression pattern of infection or rejection. The hope was to be able to avoid unnecessary immunosuppression in such patients. Validation of gene expression by PCR, immunostaining for selected genes, and in silico functional pathway analysis using Ingenuity software were also performed.
2 | METHODS
Institutional review board (IRB) approval was obtained (Protocol number 2011H0364, 2012H0382).
2.1 | Patient cohort and biopsy samples for gene expression study by Nanostring assay
This is a retrospective study using biopsy tissue remaining in the paraffin blocks after routine pathology workup. We previously published a case series describing biopsy findings and urine culture results in 49 renal allograft recipients with APN.6 The histologic criteria that were used for biopsy diagnosis of APN include—interstitial inflammation with predominance of neutrophils, tubular infiltration with neutrophils, and formation of tubular microabscesses.9,10 For the present study, we used the APN biopsies from the same cohort. Biopsies with AR, native pyelonephritis (NP), and living donor baseline post-perfusion transplant biopsies were used for comparison and were selected from our pathology archival material. Selection was mainly based on adequate tissue availability for RNA isolation. All the biopsies for mRNA testing were carefully reviewed (by AS and TN). We did not include biopsies with moderate-to-extensive chronic allograft injury, because that may alter the mRNA profile.
We had a total of 33 biopsy samples, depicted in Tables 1 and 2. These were divided into four groups as shown below:
Group I—Six post-perfusion baseline transplant biopsies designated as “B” for baseline (B2, B3, B4, B5, B7, and B8), serving as normal kidney controls.
Group II—Eight allograft biopsies with unequivocal acute rejection designated as “R” for rejection (R1, R2, R3, R4, R5, R6, R7, and R8). These were performed within one year post-transplant (Banff Grade IB to Banff Grade IIA). These were predominantly acute cellular rejections. One of the biopsies had mixed cellular and humoral rejection.
Group III—Three native kidney biopsies and one native kidney nephrectomy specimen with acute pyelonephritis designated as “NP” for native kidney pyelonephritis (NP1, NP2, NP3 and NP4).
Group IV—Fifteen allograft biopsies with histologic features of acute pyelonephritis (APN) designated as “I” for infection (I1 to I15).
TABLE 1.
Baseline transplant biopsies (n=6), biopsies with acute rejection, AR (n=8), native kidneys with acute pyelonephritis (n=4). Biopsy diagnosis and follow-up serum creatinine levels at 1 month and 1 year after biopsy are shown
| Biopsy | Age | Race | Sex | Type of transplant |
Duration between transplant and biopsy |
Biopsy diagnosis | PRA Class I; II |
Duration between biopsy and urine culture result |
Urine culture result | Colony count (CFU)/mL |
Baseline S. Cr. before biopsy |
S. cr. at time of biopsy |
S. cr. 1 month post- biopsy |
S. cr. 1 year post- biopsy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B2 | 48 | C | F | LUD | 0 days | Unremarkable renal cortex | N/A | Day of biopsy | Negative | N/A | N/A | N/A | N/A | N/A |
| B3 | 61 | C | M | LRD | 0 days | Unremarkable renal cortex | N/A | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| B4 | 58 | C | M | LUD | 0 days | Unremarkable renal cortex | N/A | 5 days after biopsy |
Negative | N/A | N/A | N/A | N/A | N/A |
| B5 | 47 | AA | M | LRD | 0 days | Unremarkable renal cortex | N/A | Negative | N/A | N/A | N/A | N/A | N/A | N/A |
| B7 | 41 | C | M | LUD | 0 days | Unremarkable renal cortex | Not done | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| B8 | 47 | C | M | LUD | 0 days | Unremarkable renal cortex | Not done | N/A | N/A | N/A | N/A | N/A | N/A | N/A |
| R1 | 44 | C | F | Cad | 0 days | Acute cellular rejection Banff grade 2B; C4d negative |
0; 0 | On day of biopsy |
Negative | N/A | 2.4 | 8.8 | 1.3 | 1.1 |
| R2 | 29 | C | F | Cad | 6 months | Acute cellular rejection Banff grade IIA, mild diffuse C4d |
0; 0 | 4 days before biopsy |
Positive for Enterococcus faecalis (UTI) |
10 000– 50 000 |
0.7 | 2.1 | 1.5 | Dialysis |
| R3 | 23 | C | F | LUD | 4 months | Acute cellular rejection Banff grade 1B, C4d mild diffuse |
0; 9 | 2 days before biopsy |
Negative | N/A | 0.9 | 2.4 | 1.2 | 0.9 |
| R4 | 61 | AA | F | Cad | 19 days | Acute cellular rejection Banff grade IIB, C4d negative |
95; 70 | 6 days before biopsy |
Negative | N/A | DGF | 1.3 | 1 | 1.4 |
| R5 | 51 | C | F | Cad | 1 month, 9 days |
Acute cellular rejection Banff grade 1B, C4d negative |
81; 0 | 7 days before biopsy |
Positive for MRSE; E. coli (lower UTI) |
100 000 | DGF | 5.8 | 1.8 | Dialysis |
| R6 | 45 | AA | F | Cad | 7 months | Combined cellular and humoral rejection (Banff grade 1B), strong diffuse C4d staining |
24; 92 | N/A | Negative | N/A | 2.5 | 4.6 | 4.3 | 4.2 |
| R7 | 34 | AA | M | Cad | 11 days | Acute cellular rejection (Banff grade 1B), C4d negative |
0; 0 | N/A | Negative | N/A | 2.9 | 5.8 | 6.1 | Dialysis |
| R8 | 44 | C | F | LRD | 4 days | Acute cellular rejection, Banff grade 2B; C4d negative |
Not done | N/A | N/A | N/A | 1.3 | 2 | 1.4 | Not available |
| NP1 | 52 | C | M | Native kidney biopsy |
N/A | Acute pyelonephritis (neuro- genic bladder, ureteral strictures) |
N/A | N/A | Positive with pyuria, esterase positive |
Details not available |
1.5 | 3.8 | Not available |
Not available |
| NP2 | 75 | AA | F | Native kidney biopsy |
N/A | Acute pyelonephritis (resected urinary bladder cancer) |
N/A | N/A | Cultures performed after starting antibiotics. Negative |
Details not available |
1.2 | 4.6 | Not available |
Not available |
| NP3 | 48 | C | F | Native nephrectomy |
N/A | Acute pyelonephritis (hemipel- vectomy for chondrosarcoma) |
N/A | N/A |
Enterococcus faecalis |
Details not available |
0.7 | 1.1 | 0.5 | 0.5 |
| NP4 | 79 | C | M | Native kidney biopsy |
N/A | Acute pyelonephritis (cystic- tomy for bladder cancer) |
N/A | 7 days before biopsy |
Klebsiella pneumonia |
Not available | 1.4 | 4 (dialysis) |
Not available |
Not available |
B, baseline; R, rejection; NP, native kidney pyelonephritis; Ph, Phillipino; LUD, living-unrelated donor; LRD, living-related donor; Cad, cadaveric donor; S. cr., serum creatinine in mg/dL; MRSE, methicillin-resistant Staphylococcus epidermidis; UTI, urinary tract infection.
TABLE 2.
Fifteen patients with diagnosis of acute pyelonephritis (APN) on allograft biopsy. Culture results, biopsy features, and serum creatinine levels are shown
| Biopsy sample |
Age | Race | Sex | Transplant type |
Duration from transplant to biopsy |
Biopsy diagnosis |
C4d staining |
PRA Class I; Class II |
Duration between biopsy and urine culture |
Microorganisms identified on urine culture |
Colony count on culture |
Treatment | Baseline S. cr. before biopsy |
S. cr. at biopsy |
S. cr. 10 days post- biopsy |
S. cr. 1 month post- biopsy |
S. cr. 1 year post- biopsy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I1 | 65 | C | F | Cad | 12 days | Acute pyelonephritis |
Negative | 0; 68 | 10 days before biopsy |
Klebsiella pneumonia |
>100 000 | Antibiotics | DGF | 4.3 | 3.5 | 1.1 | 1.49 |
| I2 | 53 | AA | M | Cad | 1 year, 4 months |
Acute pyelonephritis |
Negative | 18; 59 | 1 day after biopsy |
Enterococcus faecalis |
10 000– 50 000 |
Antibiotics | 2.5 | 4.3 | 2.6 | 2.6 | 3.73 |
| I3 | 51 | Ph | F | Cad | 3 years, 3 months |
Acute pyelonephritis |
Negative | 0;0 | On day of biopsy |
Klebsiella oxytoca | >100 000 | Antibiotics | 1.4 | 2.3 | 2.4 | 2.2 | 2.4 |
| I4 | 21 | AA | M | Cad | 3 weeks | Acute pyelonephritis |
Negative | 0; 0 | Negative at biopsy |
n/a | 13 days before biopsy; Candida lusitanae 1–5000 |
Antibiotics | DGF | DGF 12.25 |
9.3 | 3.5 | 2.6 |
| I5 | 36 | C | F | Cad | 8 days | Acute pyelonephritis |
Negative | 0; 0 | 10 days before biopsy |
Enterococcus faecalis |
>100 000 | Antibiotics | DGF | DGF 8.2 |
6.9 | Lost | Lost |
| I6 | 57 | C | F | LRD | 6 months | Acute pyelonephritis |
Negative | 0; 0 | 4 days after biopsy |
Candida glabrata; Mycoplasma |
<50 000 | Antibiotics | 1.4 | 4.5 | 6.8 | 4.2 | 2.4 |
| I13 | 32 | C | F | Cad | 15 days | Acute pyelonephritis |
Negative | 19; 0 | 5 days before biopsy |
MRSE | <50 000 | Antibiotics | 1.1 | 3.6 | 1.5 | 1 | 1 |
| I14 | 52 | C | F | LUD | 5 years, 1 months |
Acute pyelonephritis |
Negative | 0; 0 | 2 days before biopsy |
E. coli | >100 000 | Antibiotics | 2 | 3.3 | 2.9 | 2.1 | 1.6 |
| I15 | 52 | AA | F | Cad | 2 years, 3 months |
Acute pyelonephritis |
Negative | 0; 0 | 10 days before biopsy |
E. coli, also sepsis | >100 000 | Antibiotics | 6.8 | 3.7 | 3.3 | 3.3 | 2.6 |
| I7 | 30 | AA | F | Cad | 15 days | Acute pyelonephritis |
Negative | 0; 0 | Negative at time of biopsy |
n/a | 1 month after biopsy; Klebsiella oxytoca, Citrobacter koseri <50 000 |
Antibiotics | DGF | DGF 8.4 |
8.1 | Lost | Lost |
| I8 | 46 | C | M | Cad | 8 days | Acute pyelonephritis |
Trace focal |
0; 0 | Negative at time of biopsy |
n/a | 2 months after biopsy; Klebsiella, Enterobacter ESBL <1000 |
Antibiotics, followed by corticoster- oids |
DGF | DGF 10.9 |
5.7 | 8.2 | 2.1 |
| I9 | 51 | C | F | LRD | 2 months | Acute pyelonephritis |
Negative | 0; 0 | Negative at time of biopsy |
Mixed flora (contamination) |
n/a | Antibiotics | 1.9 | 4.6 | 2.6 | 2.7 | Expired |
| I10 | 37 | C | M | Cad | 15 days | Acute pyelonephritis, |
Negative | 77; 33 | Negative at time of biopsy |
n/a | n/a | Antibiotics, followed by corticoster- oids |
DGF | DGF 5.3 |
9.3 | 1.1 | 2.4 |
| I11 | 62 | AA | M | Cad | 20 days | Acute pyelonephritis |
Weak focal |
0; 0 | Negative at time of biopsy |
n/a | N/A | Antibiotics | 8.8 | 6.3 | 4.3 | 1.8 | 2 |
| I12 | 66 | C | M | Cad | 13 days | Acute pyelonephritis |
Negative | 0; 0 | Negative at time of biopsy |
n/a | N/A | Steroids | 2.8 | 3.9 | 2.1 | 2.1 | 1.6 |
LRD, living-related donor; Cad, cadaveric donor; PRA, panel-reactive HLA antibodies; S. cr., serum creatinine in mg/dL; DGF, delayed graft function (dialysis within first week post-transplant); Ph, Phillipino; MRSE, methicillin-resistant Staphylococcus epidermidis.
Among the 15 biopsies with diagnosis of APN (Table 2), eight had concomitant positive urine cultures (I1, I2, I3, I5, I6, I13, I14, and I15). The remaining 7 of 15 biopsies (I4, I7, I8, I9, I10, I11, and I12) had negative urine culture results despite the presence of histologic features of APN on the biopsy and supportive urinalysis findings. Such cases pose a diagnostic dilemma for a pathologist. We included them in this study to see whether their gene expression pattern resembled that of infection or rejection. As described in our previous study,6 “concomitant urine cultures” was defined as within 10 days of the date of biopsy (preceding or following biopsy). This is an arbitrary cutoff, used for the purpose of this study. The urinalysis findings in all these patients are shown in Table 3.
TABLE 3.
Urinalysis results for the transplant patients with acute pyelonephritis (n=15) and patients with acute rejection (n=8)
| Biopsy | Concomitant urine culture result |
Urinalysis results | ||||
|---|---|---|---|---|---|---|
| Bacteria | WBCs/hpf | RBCs/hpf | Leukocyte esterase | Nitrites | ||
| I1 | Positive | Present | 20–29 | Rare | Large | Negative |
| I2 | Positive | Present | 30–49 | Rare | Moderate | Negative |
| I3 | Positive | Not available | Not available | Not available | Not available | Not available |
| I5 | Positive | Absent | 5–9 | 30–49 | Small | Negative |
| I6 | Positive | Present | 10–14 | Absent | Small | Negative |
| I13 | Positive | Present | 30–49 | Absent | Trace | Negative |
| I14 | Positive | Present | >50 | 1–2 | Moderate | Positive |
| I15 | Positive | Absent | >50 | 5–9 | Moderate | Negative |
| I4 | Negative | Present | >50 | 3–4 | Large | Negative |
| I7 | Negative | Present | 10–14 | 15–19 | Small | Negative |
| I8 | Negative | Present | 10–14 | 1–2 | Small | Negative |
| I9 | Negative | Present | 5–9 | Absent | Negative | Negative |
| I10 | Negative | Absent | 20–29 | 20–29 | Large | Negative |
| I11 | Negative | Absent | Rare | 20–29 | Negative | Negative |
| I12 | Negative | Absent | 10–14 | 15–19 | Small | Negative |
| R1 | Negative | Present | 3–4 | 1–2 | Negative | Negative |
| R2 | Positive | Absent | 3–4 | 1–2 | Moderate | Negative |
| R3 | Negative | Absent | 1–2 | Absent | Negative | Negative |
| R4 | Negative | Present | 20–29 | 5–9 | Moderate | Negative |
| R5 | Positive | Present | 20–29 | Rare | Large | Negative |
| R6 | Negative | Absent | 5–9 | >50 | Large | Negative |
| R7 | Negative | Absent | 5–9 | 5–9 | Trace | Negative |
| R8 | Negative | Absent | Not available | Not available | Not available | Not available |
All renal allograft recipients at our institution receive trimethoprim/sulfamethoxazole (Bactrim) prophylaxis along with nystatin and ganciclovir. Patients with biopsy diagnosis of APN received additional antimicrobial treatment to treat the episode of pyelonephritis. Ciprofloxacin is the drug most frequently used, but additional antibiotics are given depending on antibiotic sensitivity results. Induction immunosuppression consisted of high-dose solumedrol or prednisone taper over the first 5–8 days along with four to five doses of antithymocyte globulin (ATG).
2.2 | Biopsy samples for validation of Nanostring results by PCR
The 33 biopsies described in Tables 1, 2 and additional 22 samples (shown in Table 4) were subsequently tested by quantitative PCR for validation of the gene expression results obtained by Nanostring. These additional 22 samples also included biopsies with APN, AR, NP and living donor baseline biopsies as normal controls.
TABLE 4.
Additional 22 biopsy samples for validation of results
| Biopsy | Age (years) |
Race | Sex | Type of transplant |
Duration between transplant and biopsy |
Biopsy diagnosis |
PRA Class I; Class II |
Duration between biopsy and urine culture result |
Urine culture result |
Colony count (CFU)/mL |
Baseline S. Cr. before biopsy |
S. cr. at time of biopsy |
S. cr. 1 month post- biopsy |
S. cr. 1 year post-biopsy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Baselines | ||||||||||||||
| B8 | 29 | AA | F | LRD | N/A | Unremarkable renal cortex |
N/A | I day before | Negative | N/A | N/A | N/A | N/A | N/A |
| B9 | 64 | C | M | LUD | N/A | Unremarkable renal cortex |
N/A | No cultures | N/A | N/A | N/A | N/A | N/A | N/A |
| B10 | 66 | C | F | LRD | N/A | Unremarkable renal cortex |
N/A | No cultures | N/A | N/A | N/A | N/A | N/A | N/A |
| AR | ||||||||||||||
| R9 | 53 | AA | M | Cad | 2 years 5 months |
ACR 1B; C4d neg |
0; 0 | 1 day before, 2 days after |
Negative | N/A | 2.2 | 3 | 2.3 | Not available |
| R10 | 47 | C | M | LRD | 15 days | ACR III; C4d neg |
0; 0 | 6 days before | Negative | N/A | 1.4 | 3.6 | 1.6 | Not available |
| R11 | 43 | C | M | Cad | 17 days | ACR 1B; C4d neg |
0; 0 | 3 days before, same day |
Negative | N/A | 3.5 | 7.5 | 1.8 | 1.9 |
| R12 | 43 | C | M | Cad | 7 months | ACR 1B; C4d neg |
0; 0 | Same day | Negative | N/A | 1.6 | 2.6 | 2.6 | 7 |
| R13 | 48 | C | F | LUD | 11 months | ACR 1B; C4d neg |
0; 3 | 1 day before, 2 days after |
Negative | N/A | 2.7 | 3 | 2.6 | 3 |
| R14 | 27 | AA | M | LRD | 1 year, 1 month |
ACR 1A; C4d neg |
0; 0 | 4 days before | Negative | N/A | 2.5 | 3.2 | 3.2 | 2.8 |
| APN with positive urine cultures at time of biopsy | ||||||||||||||
| I16 | 52 | C | F | LUD | 2 years | Acute pyelonephritis |
0; 0 | 1 day before | E. coli | >100 000 | 1.4 | 2.4 | 4.3 | 1.7 |
| I17 | 32 | C | F | Cad | 15 days | Acute pyelonephritis |
19; 0 | 5 days before | MRSE | 50 000 | 1.1 | 3.6 | 3.6 | 1.0 |
| I18 | 41 | AA | M | Cad | 1 month 17 days |
Acute pyelonephritis |
40; 0 | 2 days before |
Klebsiella pneumonia |
50 000 | 5.5 (DGF) | 7.3 | 8.2 | 9.5 (failed) |
| I19 | 50 | C | F | Cad | 2 months 10 days |
Acute pyelonephritis |
0; 0 | 9 days before |
Serratia marcescens |
100 000 | 2.2 | 11 | 3.1 | 2.6 |
| APN with negative urine cultures at time of biopsy | ||||||||||||||
| I20 | 31 | Hispanic | F | LRD | 7 days | Acute pyelonephritis |
0; 50 | 1 day before, 2 days after |
Negative | N/A | 1.2 | 1.7 | 4.7 | 1.1 |
| I21 | 39 | C | M | Cad | 7 weeks | ATN, Acute pyelonephritis |
0; 0 | 13 days before | Negative | N/A | 2.3 | 4.9 | 2.5 | 2.2 |
| I22 | 57 | C | F | LUD | 10 days | Acute pyelonephritis |
0; 0 | 2 days after | Negative | N/A | 5.2 | 7.77 | 7.3 | 2.12 |
| I23 | 67 | AA | M | Cad | 4 months 3 weeks |
Acute pyelonephritis |
0; 0 | 10 days before | Negative | N/A | 1.8 | 11 | Not available |
Failed |
| I24 | 31 | C | M | Cad | 4 months | Acute pyelonephritis |
0; 0 | 1 day after | Negative | N/A | 1.5 | 3.1 | 2.1 | 1.7 |
| I25 | 58 | C | M | Cad | 2 years | Acute pyelonephritis |
0; 24 | 4 days after | Negative | N/A | 2.2 | 2.6 | 2.5 | Not available |
| Native kidney pyelonephritis (NP) | ||||||||||||||
| NP5 | 48 | C | F | Nephrectomy | N/A | Acute pyelonephritis |
N/A | Day of surgery | Enterococcus | >10 000 | 0.6 | 1.1 | 0.9 | 0.6 |
| NP6 | 61 | AA | F | Nephrectomy | N/A | Acute pyelonephritis |
N/A | 1 month before surgery |
Enterobacter | >100 000 | 1 | 1.2 | Not available |
1 |
| NP7 | 46 | C | F | Nephrectomy | N/A | Acute and chronic pyelonephritis |
N/A | 8 days before surgery |
Negative | N/A | 0.7 | 0.7 | Not available |
0.8 |
S. cr., serum creatinine; MRSE, methicillin-resistant Staphylococcus epidermidis; DGF, delayed graft function.
2.2.1 | RNA isolation
Formalin-fixed paraffin-embedded (FFPE) tissue sections from the allograft and native kidney biopsies were deparaffinized using xylene, and total RNA was prepared using the RNeasy FFPE kit (QIAGEN, Valencia, CA, USA). Total RNA quality and quantity were assessed by Nanodrop spectrophotometry (ThermoFisher Scientific, Waltham, MA, USA).
2.2.2 | mRNA NanoString assay
The digital multiplexed NanoString nCounter Gx Human Immunology Kit (NanoString Technologies Seattle, WA) with a code set of 513 genes was used. Major classes of cytokines, their receptors, chemokine ligands and receptors, interferons and their receptors, the TNF receptor superfamily, and the KIR family genes are represented. Details are provided in supplemental data Table S1. It allows 12 samples per cartridge. Therefore, 12 of 33 samples were tested on this kit (B2, B3, B4, I1, I2, I3, I6, R1, R2, R3, R4, R5). However, during the course of this study, NanoString updated this kit to nCounter Human Immunology V2 panel which retained the original genes on the panel but had an expanded code set of 581 genes. Therefore, the remaining 21 samples were tested on the newer expanded V2 kit. Comparison list of genes on each panel is provided in Supplemental data (Table S2) Three of the 12 initial samples were rerun on the V2 kit, and we confirmed reproducibility of the results (Supplemental data Table S3).
2.2.3 | Normalization and statistical analysis
Normalization and data analysis were performed separately for the 12 samples that used the GX Immunology kit and the 24 samples that used the updated V2 Immunology kit. Technical normalization was performed using positive controls to adjust counts for each gene target in the assay. Data after technical normalization were log2-transformed first. To reduce the false-positive rate, genes of over 80% samples with an expression level 2 SD below the mean expression of the negative controls were excluded from further analysis. Quartile normalization was used for normalization across samples. Overall 510 genes from GX kit and 572 genes from the V2 kit were analyzed for differential expression, respectively. A linear model was used to compare the gene expression between APN, NP, AR, and baseline. To improve the estimates of variability and statistical tests for differential expression, variance smoothing methods were employed.12 P values were adjusted by controlling the mean number of false positives at 5 (out of ~500 genes) (i.e., α=0.01).13
2.2.4 | Construction of heat maps and principle component analysis (PCA)
Heat maps were created to show median-centered expression of selected genes and clustering pattern among the biopsies, using Cluster 3.0 and JavaTreeView software algorithms. Using the normalized mRNAs expression values, PCA plots were constructed to show overall clustering pattern of the groups.
2.2.5 | Quantitative PCR validation
Total RNA from the FFPE samples were extracted as described previously4 and reversed-transcribed into cDNA using SuperScript VILO cDNA synthesis kit (Invitrogen, Carlsbad, CA, USA) (Supplemental Methods).
2.2.6 | Immunohistochemistry
Three biopsies with APN (I1, I15, and I7) and three biopsies with acute rejection (from our biopsy archives) were stained for CXCL10 and CXCL2. Formalin-fixed paraffin-embedded (FFPE) tissue sections were immunostained using a three-step avidin–biotin complex peroxidase system (Vectastain Elite ABC Vector Laboratories, Burlingame, CA, USA).
2.2.7 | Ingenuity pathway analysis
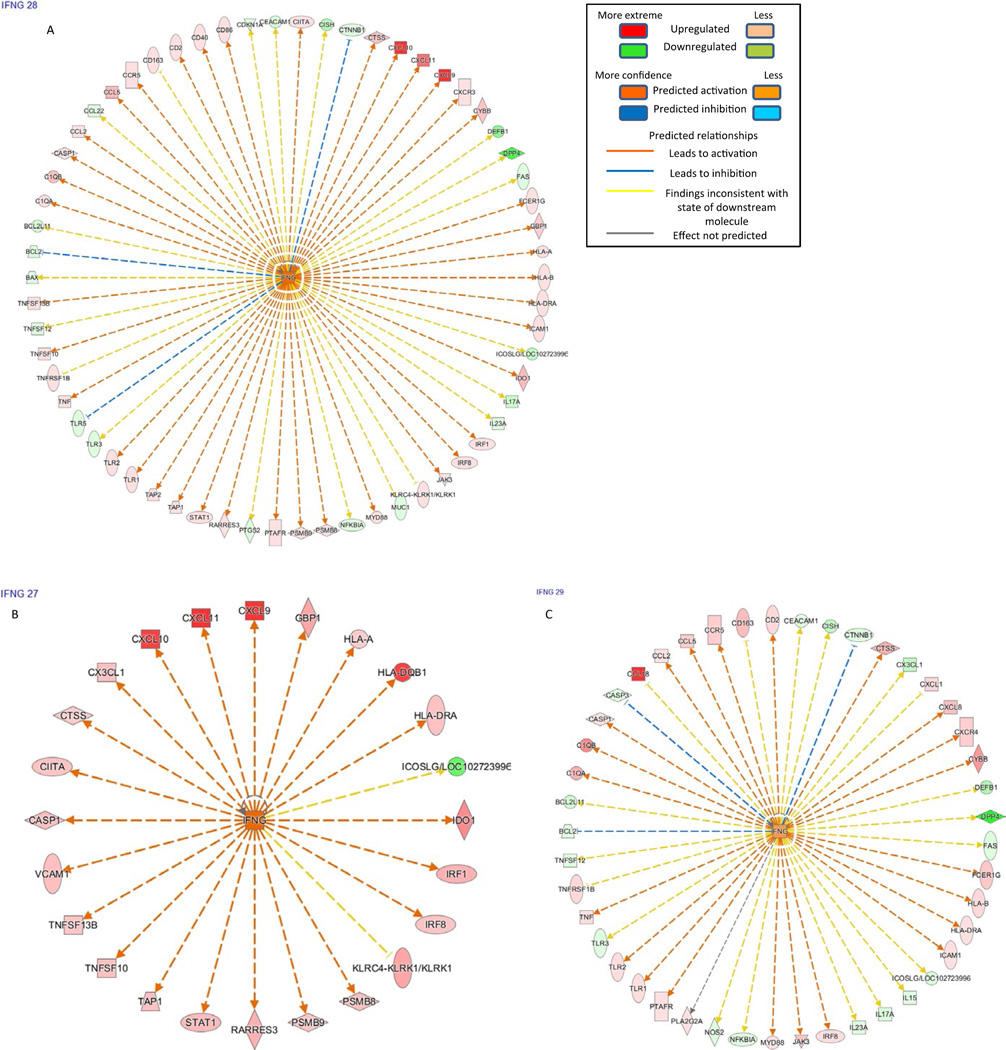
Data were analyzed through the use of QIAGEN’s Ingenuity® Pathway Analysis (IPA®, QIAGEN, Redwood City, CA, USA, www.qiagen.com/ingenuity).13 Top expressed genes which met both criteria of at least 1.5-fold change and P-value .05 were used for IPA. We identified upstream regulators that are predicted to be activated or inhibited based on activation z-score. The z-score algorithm is designed to produce either a prediction of activation or inhibition (or no prediction) as well as to reduce the chance that random data will generate significant predictions. The software does this using information in the Ingenuity® Knowledge Base to explain observed changes in expression in the various groups in the dataset. We then ran a comparison analysis of the upstream regulatory molecules for the disease groups (AR, APN, and NP). Significant molecules are sorted by score −log(P-value) from the Fisher’s exact test across all observations. The predicted activation (or inhibition) state of these molecules is determined by z-score algorithm calculated by IPA. Orange color is used to indicate activation and blue color for inhibition.
3 | RESULTS
Clinicopathologic findings of the patients are shown in Tables 1 and 2. Urinalysis findings are shown in Table 3. The additional 22 cases used for validation are shown in Table 4.
3.1 | Differential gene expression by NanoString multiplexed assay
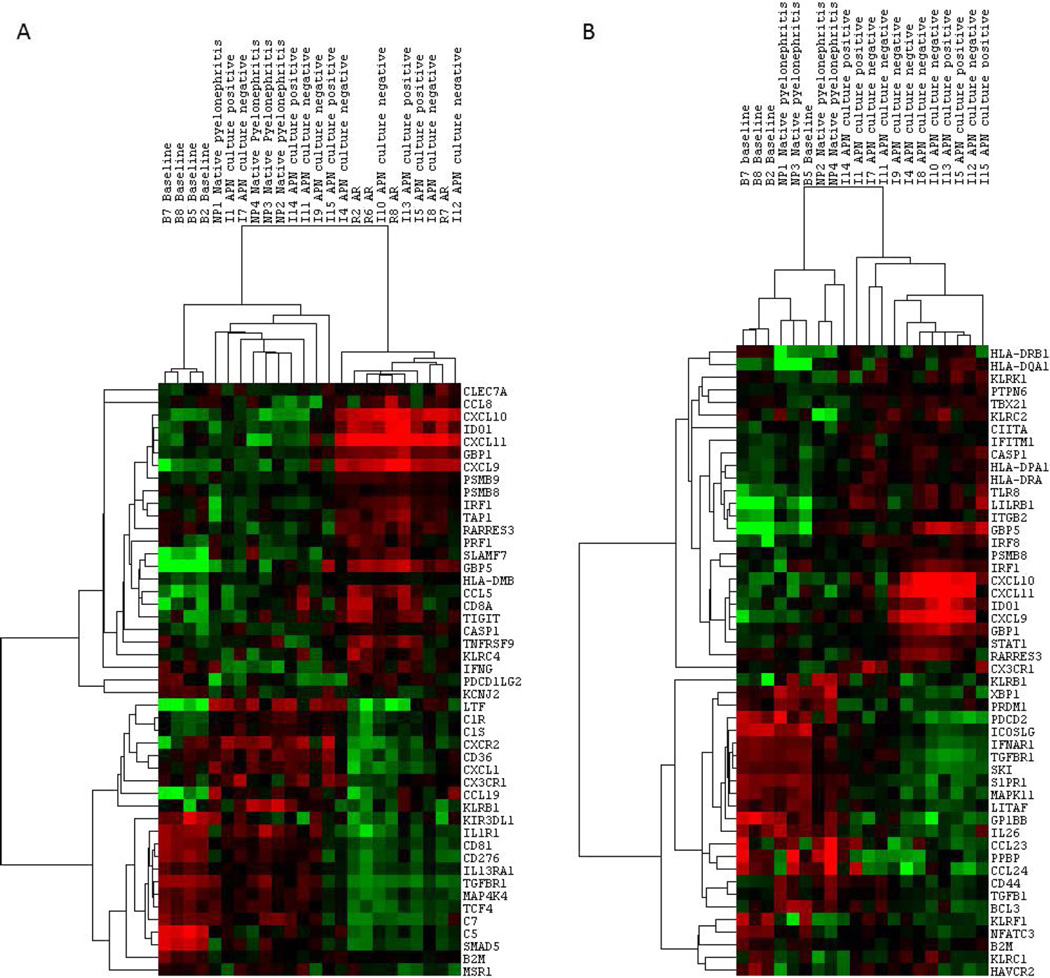
Top differentially expressed genes between APN, AR, and NP are shown in Table 5 (P<.05). We found CXCL9, CXCL10, CXCL11, and IDO1 to be the most highly upregulated genes in AR (up to 40-, 39- and 25-fold, respectively, above normal) and showing largest fold difference compared to APN and NP. Other genes such as SLAMF7, GBP5, IRF1, TIGIT, Granzyme B, CCL5, and CCL19 show only mild upregulation in AR. The only gene showing statistically significant upregulation in APN and NP as compared to AR was LTF (lactoferrin). CXCL1 was higher in APN and NP as well but did not show statistical significance. These comparisons are shown in the form of heat maps in Fig. 1A. Top differentially expressed genes between APN and NP are shown in Fig. 1B. The genes showing largest fold difference between allograft APN and NP were CXCL9, CXCL10, CXCL11, IDO1, HLA-DRB1 and HLA-DQA1, HLA-DQB1, GBP5, IRF1. These genes are expressed higher in allograft APN (albeit not as much as in AR) compared to NP. Other genes showing higher expression in APN are those involved in T-cell costimulation including KLRK1, KLRC2, LILRB1. CXCL1 shows statistically significant upregulation in NP over normal (2.8-fold, P=.008) but not in APN.
TABLE 5.
Differential gene expression between AR and APN, P<.05
| log2 average |
AR vs baseline |
APN vs baseline |
NP vs baseline |
APN vs AR |
APN vs NP |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | AR | APN | NP | Baseline | Fold change | P-value | Fold change | P-value | Fold change | P-value | Fold change | P-value | Fold change | P-value |
| IDO1 | 10.29 | 8.25 | 6.51 | 6.69 | 12.19 | 0 | 2.95 | .0147 | −1.13 | .809 | −4.17 | 8.00×10−04 | 3.33 | .0033 |
| CXCL10 | 11.54 | 9.12 | 6.67 | 6.18 | 41.19 | 0 | 7.69 | .002 | 1.41 | .6314 | −5.26 | .004 | 5.48 | .0036 |
| CXCL11 | 10.36 | 7.92 | 5.31 | 5.68 | 25.47 | 1.00×10−04 | 4.7 | .0178 | −1.29 | .7227 | −5.56 | .0048 | 6.08 | .0029 |
| CXCL9 | 12.65 | 10.57 | 8.02 | 7.36 | 39.11 | 0 | 9.24 | .0014 | 1.59 | .5369 | −4.17 | .0153 | −1.15 | .5191 |
| LTF | 8.92 | 11.51 | 12.43 | 7.16 | 3.38 | .1391 | 20.38 | 2.00×10−04 | 33.33 | 1.00×10−04 | 6.03 | .0064 | 1.11 | .5803 |
| CD36 | 6.57 | 7.48 | 7.67 | 6.99 | −1.33 | .2933 | 1.4 | .1586 | 1.61 | .0975 | 1.88 | .0055 | −1.89 | .3006 |
| C1R | 10.2 | 11 | 10.85 | 8.94 | 2.4 | .0014 | 4.19 | 0 | 3.85 | 0 | 1.75 | .0057 | 1.3 | .1666 |
| CLEC7A | 8.29 | 7.51 | 7.13 | 6.65 | 3.11 | 1.00×10−04 | 1.81 | .0083 | 1.39 | .1891 | −1.72 | .0076 | 2.55 | .0076 |
| GBP1 | 11.38 | 10.03 | 8.68 | 8.21 | 8.98 | 0 | 3.52 | .0018 | 1.39 | .4571 | −2.56 | .0077 | 1.1 | .7216 |
| SLAMF7 | 9.14 | 8.06 | 7.92 | 5.67 | 11.06 | 0 | 5.22 | 0 | 4.76 | 2.00×10−04 | −2.13 | .0089 | 1.71 | .0225 |
| PSMB9 | 10.6 | 9.69 | 8.91 | 8.14 | 5.49 | 0 | 2.93 | 2.00×10−04 | 1.72 | .0787 | −1.89 | .0091 | 1.17 | .4029 |
| C1S | 9.51 | 10.22 | 10 | 8.36 | 2.21 | .0028 | 3.63 | 0 | 3.12 | 1.00×10−04 | 1.64 | .0112 | 1.54 | .1751 |
| CCL5 | 9.67 | 8.48 | 7.85 | 6 | 12.7 | 0 | 5.55 | 0 | 3.57 | .0047 | −2.27 | .0135 | 3.06 | .0066 |
| GBP5 | 9.75 | 8.32 | 6.71 | 4.56 | 36.65 | 0 | 13.59 | 0 | 4.35 | .0063 | −2.7 | .0144 | −1.18 | .2904 |
| CD81 | 11.17 | 11.73 | 11.96 | 12.62 | −2.7 | 0 | −1.85 | .001 | −1.58 | .0274 | 1.47 | .0152 | 5.84 | .0039 |
| CX3CR1 | 7.13 | 7.91 | 6.94 | 6.59 | 1.45 | .1845 | 2.48 | 6.00×10−04 | 1.27 | .3858 | 1.71 | .0159 | 1.95 | .0035 |
| KIR3DL1 | 4.32 | 5.02 | 4.96 | 5.78 | −2.78 | 5.00×10−04 | −1.69 | .0198 | −1.77 | .0324 | 1.63 | .0165 | 1.04 | .8332 |
| PSMB8 | 10.7 | 10.07 | 9.14 | 9.22 | 2.8 | 1.00×10−04 | 1.81 | .0051 | −1.05 | .8243 | −1.54 | .0174 | 1.9 | .001 |
| CXCR2 | 5.28 | 6.26 | 6.45 | 5.74 | −1.37 | .3711 | 1.43 | .24 | 1.64 | .1748 | 1.97 | .0175 | −1.15 | .6197 |
| CASP1 | 8.7 | 8.14 | 7.22 | 6.52 | 4.51 | 0 | 3.08 | 0 | 1.61 | .0261 | −1.47 | .0199 | 1.9 | 3.00×10−04 |
| PRF1 | 8.24 | 7.51 | 7.13 | 6.26 | 3.96 | 0 | 2.38 | .001 | 1.82 | .0375 | −1.67 | .0217 | 1.3 | .2212 |
| KLRC4 | 6.95 | 6.2 | 5.46 | 5.48 | 2.76 | .0015 | 1.64 | .051 | −1.02 | .9583 | −1.69 | .0232 | 1.66 | .0263 |
| TNFRSF9 | 6.06 | 5.28 | 5.23 | 4.57 | 2.81 | .0018 | 1.64 | .0587 | 1.59 | .1329 | −1.72 | .0236 | 1.04 | .8734 |
| PDCD1LG2 | 6.75 | 5.92 | 5.73 | 6 | 1.69 | .1131 | −1.05 | .8438 | −1.21 | .5608 | −1.79 | .0246 | 1.14 | .5822 |
| KCNJ2 | 7.5 | 6.93 | 6.53 | 7.07 | 1.35 | .1846 | −1.1 | .6085 | −1.45 | .1021 | −1.49 | .0252 | 1.32 | .1083 |
| C5 | 5.95 | 6.37 | 6 | 7.87 | −3.85 | 0 | −2.86 | 0 | −3.67 | 0 | 1.34 | .0268 | 1.29 | .0488 |
| IRF1 | 9.72 | 8.78 | 7.63 | 7.98 | 3.35 | .0031 | 1.74 | .086 | −1.27 | .5215 | −1.92 | .0273 | 2.22 | .0086 |
| IL-1R1 | 6.5 | 7.32 | 7.79 | 8.05 | −2.94 | .003 | −1.67 | .0768 | −1.2 | .5861 | 1.76 | .03 | −1.39 | .1938 |
| IL-13RA1 | 9.12 | 9.54 | 9.53 | 9.63 | −1.43 | .044 | −1.06 | .648 | −1.08 | .6635 | 1.34 | .0308 | 1.01 | .9483 |
| CCL8 | 7.22 | 6.3 | 6.16 | 5.97 | 2.38 | .028 | 1.26 | .4743 | 1.14 | .7215 | −1.89 | .0317 | 1.1 | .7398 |
| TGFBR1 | 6.86 | 7.43 | 8.33 | 8.65 | −3.45 | 0 | −2.33 | 2.00×10−04 | −1.25 | .3502 | 1.49 | .0327 | −1.85 | .0015 |
| HLA-DMB | 10.22 | 9.76 | 9.37 | 8.13 | 4.27 | 0 | 3.1 | 0 | 2.38 | 1.00×10−04 | −1.37 | .033 | 1.31 | .0707 |
| CD8A | 8.2 | 7.19 | 6.63 | 5.61 | 6.01 | 2.00×10−04 | 3 | .004 | 2.04 | .0948 | −2 | .0338 | 1.47 | .2216 |
| TAP1 | 8.32 | 7.5 | 6.63 | 6.75 | 2.98 | .0032 | 1.69 | .0748 | −1.09 | .8078 | −1.75 | .0342 | 1.83 | .0241 |
| C7 | 9.29 | 10.1 | 11.04 | 10.63 | −2.56 | .0095 | −1.45 | .1969 | 1.32 | .4052 | 1.75 | .0345 | −1.92 | .0155 |
| KLRB1 | 5.99 | 6.72 | 7.86 | 5.58 | 1.33 | .3436 | 2.2 | .0044 | 4.76 | 0 | 1.65 | .0352 | −2.22 | .0017 |
| RARRES3 | 10.19 | 9.27 | 8.09 | 8.62 | 2.96 | .0088 | 1.56 | .1792 | −1.45 | .3427 | −1.89 | .0359 | 2.26 | .0093 |
| TCF4 | 7.89 | 8.34 | 8.84 | 8.51 | −1.54 | .0299 | −1.12 | .4587 | 1.27 | .2264 | 1.36 | .0367 | −1.41 | .0203 |
| B2M | 14.81 | 14.27 | 13.59 | 13.96 | 1.79 | .0153 | 1.24 | .2733 | −1.29 | .2636 | −1.45 | .0377 | 1.6 | .0107 |
| MSR1 | 7.78 | 7.21 | 7.43 | 7.23 | 1.47 | .1275 | −1.02 | .9411 | 1.15 | .5658 | −1.49 | .0399 | −1.18 | .4015 |
| SMAD5 | 6.85 | 7.32 | 7.03 | 8.74 | −3.7 | 0 | −2.7 | 0 | −3.5 | 0 | 1.38 | .0404 | 1.22 | .2043 |
| TIGIT | 6.88 | 6.12 | 5.7 | 4.79 | 4.26 | 2.00×10−04 | 2.52 | .0025 | 1.89 | .062 | −1.69 | .0414 | 1.33 | .2496 |
| MAP4K4 | 8.3 | 8.77 | 9.01 | 9.07 | −1.69 | .0133 | −1.23 | .2242 | −1.04 | .84 | 1.38 | .0428 | −1.19 | .2718 |
| CD276 | 6.89 | 7.36 | 7.42 | 8.14 | −2.38 | 4.00×10−04 | −1.72 | .0055 | −1.65 | .026 | 1.39 | .0485 | −1.04 | .7903 |
| CCL19 | 9.06 | 10.07 | 10.32 | 6.61 | 5.46 | 9.00×10−04 | 11 | 0 | 12.5 | 0 | 2.01 | .0488 | −1.19 | .618 |
| IFNG | 7.22 | 6.45 | 5.92 | 6.28 | 1.93 | .0685 | 1.12 | .6922 | −1.28 | .4813 | −1.72 | .0489 | 1.44 | .176 |
| CXCL1 | 8.87 | 9.7 | 10.4 | 8.86 | 1 | .9904 | 1.79 | .075 | 2.86 | .0084 | 1.78 | .0499 | −1.61 | .0986 |
Figure 1.
(A) Unsupervised hierarchical cluster analysis showing median-centered expression of 47 differentially expressed genes between AR and APN by NanoString assay (P<.05). Each column represents a biopsy sample. Each row represents individual mRNA. The color in each cell reflects the relative level of expression of the corresponding mRNA. Increasing intensities of red mean higher expression, and increasing intensity of green means lower expression. All four groups (baselines, NP, APN, and AR biopsies are shown) run on the nCounter Human Immunology V2 panel are shown. The degree of relatedness is represented by the dendrogram at the top of the panel. There is good intragroup clustering among the baseline biopsies and the NP biopsies, but the APN biopsies show a spectrum. (B) Hierarchical cluster analysis showing differential gene expression between allograft APN and NP (P<.01). CXCL9, CXCL10, CXCL11, IDO1 do show differential expression between APN and NP. Their expression in NP biopsies is much lower than that seen in APN biopsies. Thus, chemokine pattern in allograft APN does not exactly resemble native kidney pyelonephritis. APN, acute pyelonephritis; AR, acute rejection; CXCL, CXC chemokine ligand; NP, native kidney pyelonephritis
We performed the same analysis after separating out the culture negative cases from the APN group (Table 6). We did not find any genes showing statistically significant differences between culture-positive and culture-negative APN biopsies. The culture-negative APN biopsies also showed similar differences from AR.
TABLE 6.
Separated out culture-positive and culture-negative patients from the APN group and compared for the top differentially expressed genes. No significant differences are seen between culture positive and culture negative APN biopsies.
| APN culture positive vs AR |
APN culture negative vs AR |
APN culture positive vs APN culture negative |
||||
|---|---|---|---|---|---|---|
| Gene | Fold change | P-value | Fold change | P-value | Fold change | P-value |
| IDO1 | −3.57 | .0074 | −4.55 | .001 | −1.28 | .5177 |
| CXCL10 | −5.88 | .0086 | −5 | .012 | 1.23 | .7128 |
| CXCL11 | −6.25 | .0092 | −5 | .0145 | 1.27 | .6723 |
| CXCL9 | −5 | .0199 | −3.7 | .0429 | 1.39 | .5718 |
| LTF | 7.09 | .0115 | 5.37 | .0191 | −1.32 | .6581 |
| CXCL1 | 2.03 | .0417 | 1.62 | .128 | −1.25 | .443 |
| CLEC7A | −2 | .0031 | −1.54 | .0396 | 1.31 | .1607 |
| CD36 | 2.08 | .0061 | 1.75 | .0214 | −1.19 | .4236 |
| PSMB9 | −2.13 | .0068 | −1.69 | .0375 | 1.26 | .307 |
| C1R | 1.85 | .0092 | 1.67 | .0182 | −1.11 | .5949 |
| CXCR2 | 2.39 | .0093 | 1.72 | .0703 | −1.39 | .2339 |
| IL-8 | 4.03 | .0164 | 1.85 | .2342 | −2.17 | .1106 |
| MSR1 | −1.72 | .0165 | −1.35 | .1467 | 1.28 | .188 |
| TNFAIP6 | 1.94 | .0169 | −1.1 | .685 | −2.13 | .0025 |
| RARRES3 | −2.27 | .0216 | −1.67 | .1166 | 1.37 | .2915 |
| KLRB1 | 1.9 | .0219 | 1.49 | .1143 | −1.27 | .3008 |
| IRAK3 | 1.58 | .0235 | 1.18 | .3487 | −1.33 | .0923 |
| GBP1 | −2.44 | .0283 | −2.63 | .0128 | −1.08 | .8382 |
| CASP1 | −1.52 | .0309 | −1.43 | .0463 | 1.06 | .7072 |
| KLRC4 | −1.79 | .0313 | −1.61 | .056 | 1.11 | .6408 |
| C1S | 1.62 | .0333 | 1.65 | .0195 | 1.02 | .9256 |
| CD81 | 1.48 | .0347 | 1.46 | .0289 | −1.01 | .9389 |
| HLA-DMB | −1.43 | .0412 | −1.33 | .0758 | 1.07 | .6312 |
| HLA-DPB1 | −1.64 | .0413 | −1.12 | .5848 | 1.45 | .0724 |
| KIR_Activating_ Subgroup_2 |
1.71 | .0418 | 1.08 | .7577 | −1.59 | .0437 |
| CX3CR1 | 1.69 | .0426 | 1.72 | .0267 | 1.02 | .94 |
| PSMB8 | −1.54 | .0429 | −1.56 | .0305 | 1 | .9899 |
| TNFRSF10C | 1.58 | .0452 | 1.26 | .2589 | −1.25 | .2475 |
| IL-1A | −1.61 | .0483 | −1.19 | .4248 | 1.35 | .142 |
| HLA-DMA | −1.41 | .0486 | −1.19 | .2809 | 1.19 | .2409 |
| CR1 | 1.74 | .0488 | 1.49 | .1224 | −1.16 | .5118 |
| CD86 | −1.45 | .0507 | −1.18 | .3468 | 1.23 | .1953 |
| CCL8 | −2 | .0508 | −1.82 | .0633 | 1.08 | .7879 |
The genes validated by PCR are highlighted in bold.
3.2 | Validation by PCR
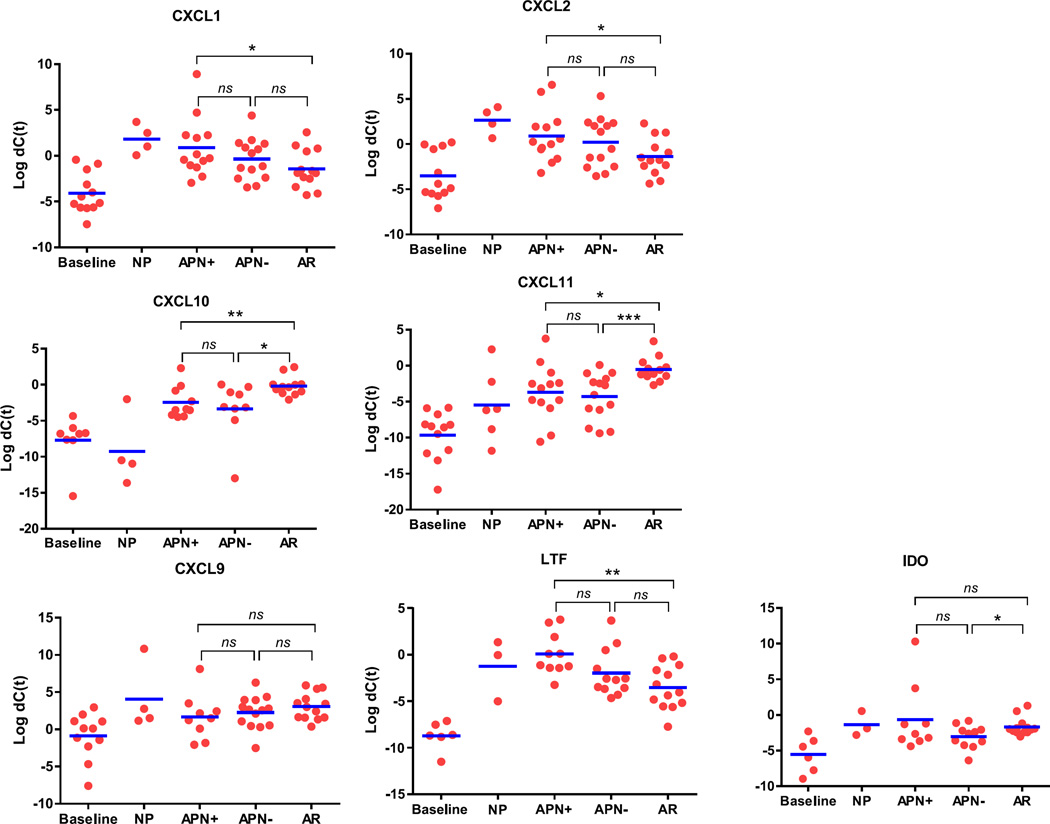
Eight of the top differentially expressed genes were validated using quantitative PCR. This was performed on all of the 33 biopsy samples as well as the 22 additional biopsies used for overall validation of the results (Fig. 2). APN culture-positive and culture-negative samples are shown separately. CXCL10 and CXCL11 are consistently higher in AR as compared to APN (both culture positive and culture negative) and NP. CXCL1, CXCL2, and LTF were statistically higher in APN (culture positive) compared to AR with similar trend in NP. Culture negative APN biopsies, however, did not show statistical difference in CXCL1, CXCL2, and LTF expression as compared to AR. Expression of IDO1, however, showed statistical difference only between culture-negative APN and AR. CXCL9 did not show statistical differences as seen on NanoString. Similar to NanoString results, culture-positive and culture-negative APN did not show statistically significant differences for any of these genes. The P-values for these comparisons are shown in Table 7.
Figure 2.
Validation of eight selected genes by quantitative real-time PCR. Each dot represents one biopsy. Gene expression is represented as ΔCt after normalizing to the housekeeping gene GAPDH. Validation was performed on all 33 samples analyzed by NanoString as well as 22 additional biopsies. P values were calculated using a Student’s t test and are shown in Table 7. ns, not significant, *P<.05, **P<.01, ***P<.001. Three samples of NP did not show amplification. Trends are similar to those seen by NanoString. CXCL9 and IDO-1, however, did not show statistical significance as seen on NanoString. APN, acute pyelonephritis; AR, acute rejection; CXCL, CXC chemokine ligand; NP, native kidney pyelonephritis
TABLE 7.
P values for the dot plot shown in Figure 2
| CXCL1 | CXCL2 | CXCL10 | CXCL11 | CXCL9 | LTF | IDO | |
|---|---|---|---|---|---|---|---|
| APN culture+ vs AR | 0.0395 | 0.0288 | 0.0075 | 0.0119 | 0.1919 | 0.0012 | 0.4686 |
| APN culture− vs AR | 0.2088 | 0.1061 | 0.0172 | 0.0009 | 0.2967 | 0.1174 | 0.025 |
| APN culture+ vs APN culture− | 0.2563 | 0.5251 | 0.5454 | 0.6675 | 0.6051 | 0.0607 | 0.1317 |
| APN culture+ vs NP | 0.5849 | 0.2657 | 0.0184 | 0.4077 | 0.2844 | 0.4385 | 0.8027 |
| NP vs AR | 0.0127 | 0.0027 | 0.0002 | 0.0041 | 0.514 | 0.1694 | 0.7522 |
APN, acute pyelonephritis; AR, acute rejection; NP, native pyelonephritis. The orange shaded boxes highlight the statistically significant P -values. P < 0.05.
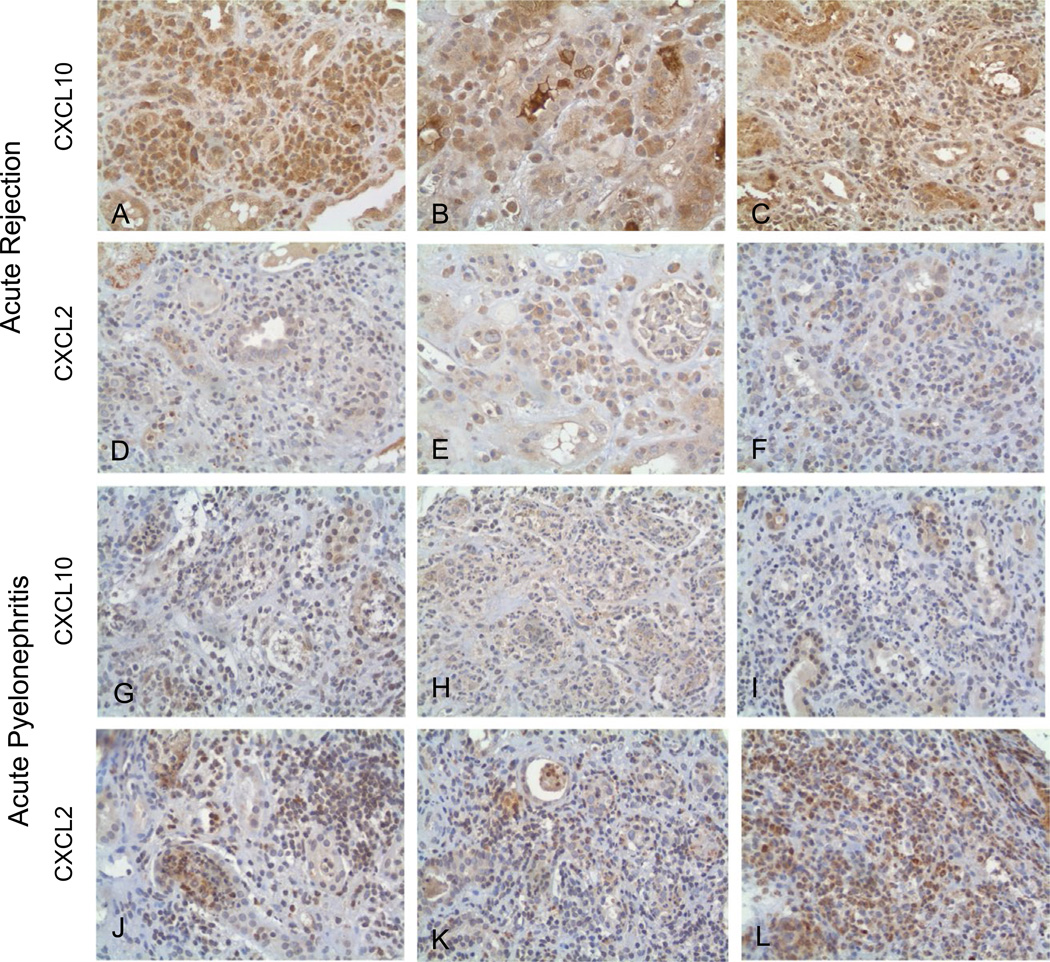
3.3 | Immunohistochemical staining
The three biopsies with AR showed diffuse staining for CXCL10 (Fig. 3A–C) but no staining for CXCL2 (Fig. 3D–F). In the three biopsies with APN, there was weak to no staining for CXCL10 (Fig. 3G–I) but strong staining for CXCL2 (Fig. 3J–L).
Figure 3.
Immunoperoxidase staining of FFPE tissue with rabbit anti-human polyclonal antibody to CXCL10 and CXCL2 (400×). (A–C) Three cases of acute rejection stained for CXCL10. Diffuse staining of interstitial inflammatory cells including lymphocytes and plasma cells is seen for CXCL10. Constitutive expression is seen in distal tubular epithelial cells. (D–F) The same cases of AR stained for CXCL2 show scant to negative staining. (G–I) Three cases of acute pyelonephritis (I1, I15, and I7) stained for CXCL10 show scant to negative staining. (J–L) The same three cases of APN (I1, I15, and I7) are stained for CXCL2. Prominent staining in the inflammatory cells is seen. APN, acute pyelonephritis; AR, acute rejection; CXCL, CXC chemokine ligand
3.4 | Principle component analysis using NanoString data
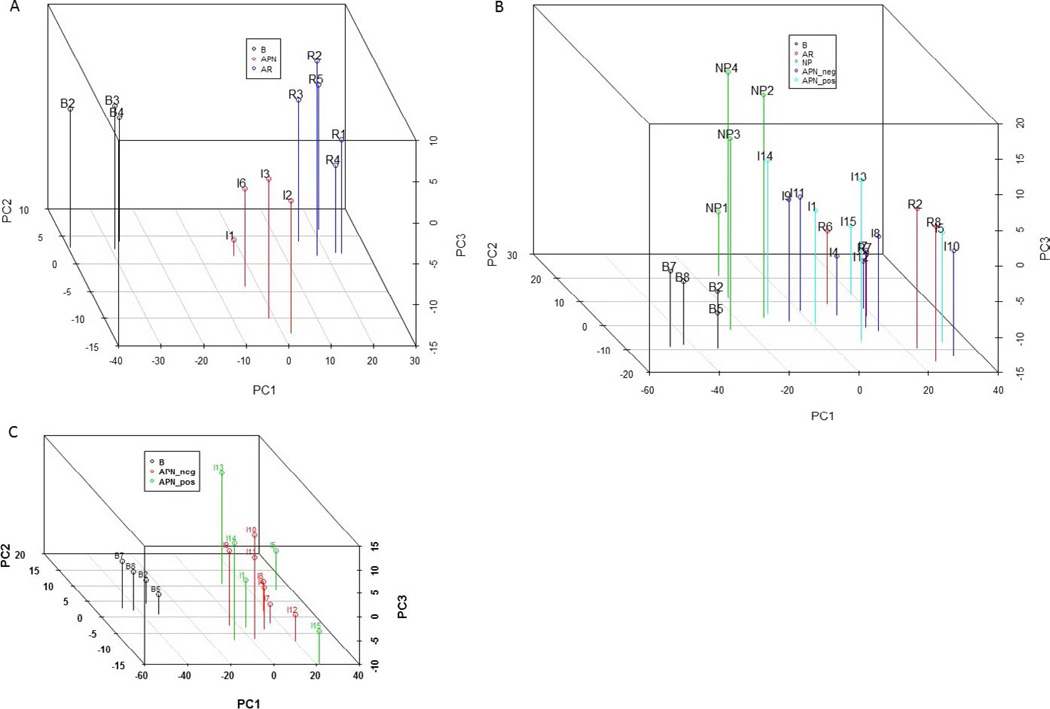
Figure 4A shows the PCA plot of 12 samples run on the GX Immunology panel. These include 3 normal baselines, 4 culture-positive APN, and 5 AR. All four of the APN samples (I1, I2, I3, and I6) had positive urine culture results, and these patients recovered graft function with antibiotic treatment. These four APN biopsies did cluster separately from AR. Figure 4B shows PCA plot of the remaining 21 samples with all the groups (baselines, APN culture positive and negative, NP and AR). The normal baseline biopsies clustered together and the NP biopsies clustered together, distinct from allograft AR and APN. The AR and APN samples, however, showed overlap. The APN biopsies with positive and negative urine cultures also clearly showed overlapping gene expression as shown in Fig. 4C. No separate clustering was seen.
Figure 4.
Principle component analysis (PCA) of gene expression data from NanoString. This includes all genes after filtering for the low expressors. (A) Shows the 12 samples analyzed on NanoString nCounter Gx Human Immunology Kit (NanoString Technologies). The samples are labeled in the figure. The three groups—3 normal kidney baseline biopsies, 4 biopsies with allograft APN and 5 biopsies with AR clustered separately. (B) Shows remaining 21 samples (samples B2, R2, and I1 were rerun) analyzed on nCounter Human Immunology V2 panel. The normal baseline biopsies and the four samples of NP clustered separately. The biopsies with allograft APN show a spectrum and overlap with AR, irrespective of urine culture results. (C) APN culture-positive and culture-negative biopsies do not cluster separately. APN, acute pyelonephritis; AR, acute rejection; NP, native kidney pyelonephritis
3.5 | Ingenuity Pathway Analysis—upstream regulators in acute rejection, allograft pyelonephritis, and native kidney pyelonephritis
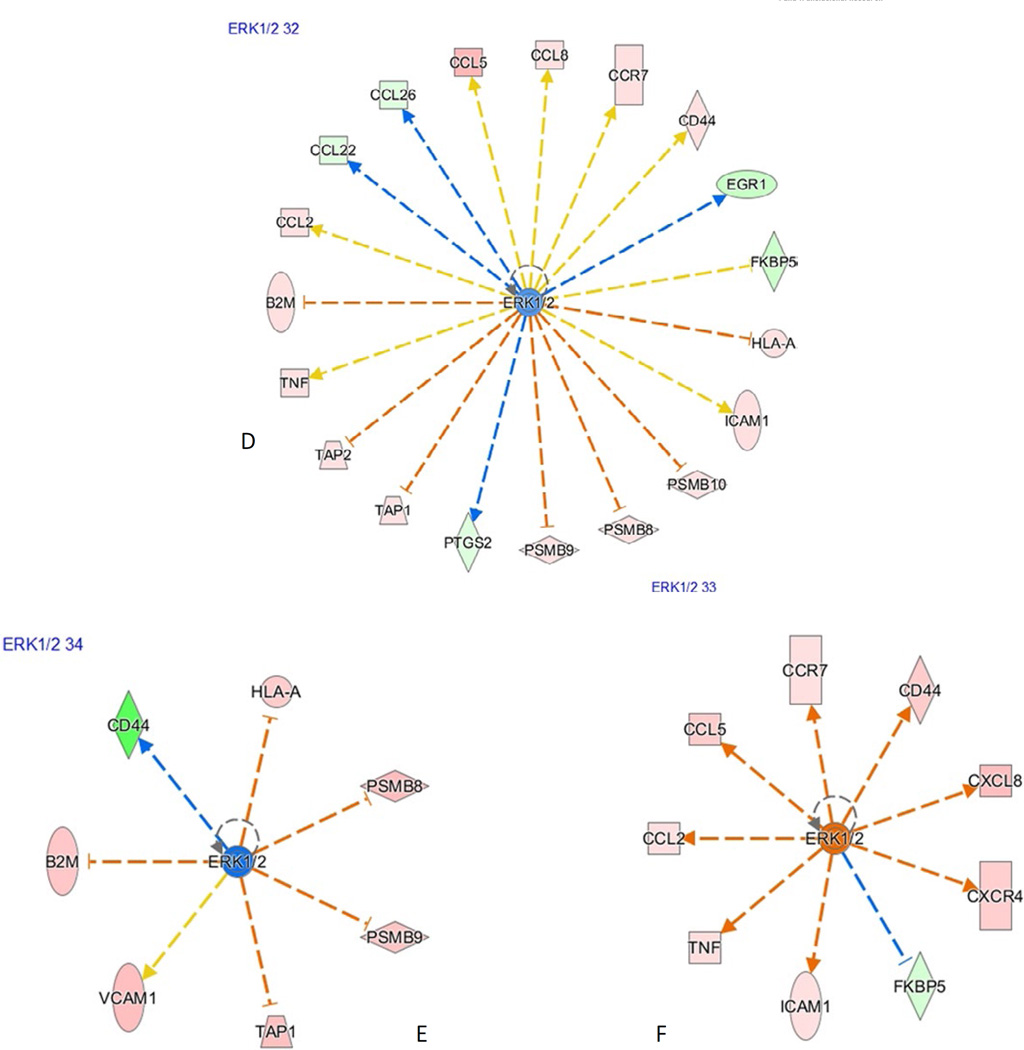
Comparison of the major upstream regulators (with z-score greater than 1) between the disease groups is shown in Table 8. B-cell receptor (BCR) complex and the MAPK subfamilies—ERK1/2, MAPK1, appear to be active in native kidney pyelonephritis, in contrast to AR and allograft APN (culture-positive and culture-negative subgroups). IFN-γ is the major upstream regulatory molecule in AR along with IFN-α, IL-12 complex, IL-18, IL-27. APN shows activation of similar pathways as in AR and shows subtle differences from NP (Fig. 5A–F).
TABLE 8.
Upstream regulatory molecules in AR, APN culture-positive, APN culture-negative, native pyelonephritis using Ingenuity pathway analysis, showing z-scores. © 2000–2015 QIAGEN. All rights reserved
| Upstream regulators |
AR | APN culture positive |
APN culture negative |
NP |
|---|---|---|---|---|
| IL-27 | 4.658 | 3.8935 | 4.3333 | 3.503 |
| EBI3 | 3.8312 | 3.4218 | 3.6996 | 2.7894 |
| IFNG | 3.2101 | 2.6045 | 2.9437 | 0.9913 |
| MAPK1 | −3.3264 | −2.3757 | −2.8673 | −0.9045 |
| IRF4 | −3.05796 | −2.7196 | −3.3023 | 0.2182 |
| Interferon- alpha |
3.08814 | 1.6787 | 2.9861 | 0.3428 |
| IL-12 (complex) | 2.9496 | 1.708 | 2.7634 | 1.6843 |
| IFNL1 | 2.7997 | 2.1783 | 2.6186 | 0.2773 |
| ERK1/2 | −1.0566 | 0.6602 | 0.4849 | 2.9153 |
| IL-18 | 2.4275 | 1.8676 | 2.6921 | 1.0687 |
| TNF | 2.4727 | 1.7964 | 2.0825 | 0.5429 |
| STAT1 | 1.66119 | 0.4527 | 1.6666 | 0.239 |
| P38 MAPK | 1.3962 | 1.5366 | 2.1335 | 0.7475 |
| NFkB (complex) | 1.35419 | 0.5369 | 0.5166 | 0.0143 |
| BCR (complex) | 0.63907 | 1.0397 | 1.3655 | 2.1801 |
Orange—predicted activation; blue—predicted inhibition; white—no prediction can be made and darker colors indicate higher z-score.
Figure 5.
Ingenuity pathway analysis results. The color code key is shown in top right corner. Downstream effectors of IFNG in AR (A), APN (B), and NP (C) are shown. IFNG and its major effectors CXCL9, CXCL10, CXCL11 are upregulated in AR and APN, but not in NP. Downstream effectors of ERK1/2 in AR (D), APN (E), and NP (F) are shown. ERK1/2 and most of the downstream effectors appear to be upregulated NP but not in AR and APN. In AR and APN, the effectors that are supposed to be activated by ERK1/2 are downregulated. The effectors that are supposed to be blocked by ERK1/2 are upregulated because of inhibition of ERK1/2 itself. APN, acute pyelonephritis; AR, acute rejection; CXCL, CXC chemokine ligand; NP, native kidney pyelonephritis
4 | DISCUSSION
Although biopsy examination remains the gold standard to diagnose allograft rejection, extensive efforts are in progress to develop less-invasive tools using blood14–21 and urine22–27 biomarkers. Interferon-gamma-controlled chemokines (CXCL9, CXCL10) and cytotoxic T lymphocyte granule contents (granzyme B, perforin) have been shown to be highly expressed in acute rejection. CXCL10 has been shown to be a candidate urinary biomarker for acute rejection.20–23,28–30 CXCL9, CXCL10, and CXCL11 are CXCR3 ligands and potent T-cell chemoattractants and are universally induced during cell-mediated immune responses.31,32 However, no such markers have been identified specifically for graft pyelonephritis, which is a common differential diagnostic consideration in the early post-transplant period. Granzyme B mRNA levels in the urine were shown to be statistically different between UTI and AR.25 However, gene expression in pyelonephritis (infection of the renal parenchyma) is likely to differ from that in lower urinary tract infections. In fact, our results did not show significant difference in expression of Granzyme B in the kidney between AR and APN. In some of the previous studies measuring chemokine levels in the urine of AR patients, the positive predictive values have been relatively low.33 In the study by Hricik et al.,33 using 2760 urinary supernatants, the PPV of CXCL9 to predict AR was 67.6%. Eliminating infectious etiologies improved the PPV of CXCL9 to 73.3% for AR. Also, the mean CXCL10 protein levels were found to be similar in patients with AR and infection. Therefore, it appears that there is likely to be significant overlap in gene expression between AR and APN which has not been addressed before. That is the focus of our study.
We confirm that CXCR3 ligands CXCL9, CXCL10, CXCL11 show high expression in AR with large fold differences as compared to APN and NP. Our results support previous study results.22–25 Additionally, we found significantly elevated IDO1 in AR (not been reported in previous studies). IDO1 is a metabolic enzyme which catalyzes the rate limiting step of tryptophan catabolism in plasmacytoid dendritic cells.34,35 Other T-cell-associated genes (TIGIT, SLAM7, IRF1, GNZB, GBP1, and GBP5),36–38 show only small fold difference between AR and APN and therefore may not be helpful as differentiating biomarkers. Only few genes on this NanoString panel show higher expression in APN as compared to AR. We identified Lactoferrin as one of them. CXCL1 and CXCL2 also showed higher expression in APN and NP by PCR. CXCL1 and CXCL2 are known to be neutrophil chemo-attractants.39,40 The fold differences, however, are small, and larger studies are needed to test their feasibility as potential biomarkers for graft pyelonephritis.
However, what we find interesting is that, although CXCL10 and CXCL11 expression is significantly elevated in AR compared to APN and NP, these chemokine genes also show significant difference in expression between graft APN and NP (native kidney pyelonephritis). They are found to be significantly higher in graft APN as compared to NP (also confirmed by PCR). Also, CXCL9, IDO-1, Class II HLA show higher expression in APN as compared to NP as seen on NanoString assay. Therefore, there are subtle differences in renal allograft pyelonephritis and native kidney pyelonephritis. This is better understood by studying the overall gene expression profiles of these groups with the help of principle component analysis (Fig. 4) and Ingenuity pathway analysis. It showed similarities between graft APN and AR and differences between graft pyelonephritis (APN) and native kidney pyelonephritis (NP).
The T-cell-dominant upstream regulatory molecules (IFN-γ, IFN-α, IL-18, IL-12)37 are predicted to be activated in both AR and APN (culture positive and culture negative), but not NP. Conversely, B-cell receptor (BCR) complex and the MAPK subfamily—ERK1/2, are predicted to be active in native kidney pyelonephritis, but in allograft APN and AR, there is predicted inhibition of the ERK1/2 and MAPK1. With the p38 MAPK pathway, we found mild activation in graft APN but not in native kidney pyelonephritis. The four major MAPK subfamilies (JNKs, ERK1/2, p38MAPKs, and MAPK-1) are reported to have divergent roles in antimicrobial immune response.41–44 Phagocytosis of bacteria activates ERK1/2 in human neutrophils and limits bacterial replication, but p38 activation was shown to have opposite effects.45–48 Therefore, ineffective bacterial phagocytosis in APN resulting in lack of response to antibiotics in some cases may be speculated. Whether it is an effect of the immunosuppressive treatment is not known but is certainly a possibility. Also there is no predicted activation of TNF and NFκ-B in native kidney pyelonephritis as compared to graft APN and AR, probably suggesting a more controlled inflammatory reaction in NP as compared to AR and APN. Thus, the pathogenesis of allograft APN and native kidney pyelonephritis may not be exactly the same. The PCA plot (Fig. 4B) does show this overlap between APN and AR, but NP biopsies appear to cluster separately. Although two of the AR biopsies (R2 and R5) were associated with positive urine cultures, these were due to lower urinary tract infections, which is common in transplant patients. They are unlikely to interfere with gene expression analysis of the kidney tissue. Also R6 had mixed cellular and humoral rejection, but we have confirmed by statistical methods that the gene expression profiles of these three AR samples were close to or within the range of the other AR samples and by no way were they outliers. They are therefore unlikely to be the cause of the overlap seen between the APN and AR samples. Also, Rabant et al. recently published their findings showing similar interferon-gamma signature in antibody-mediated rejection.49
Based on these results, we propose that in allograft APN (in contrast to native kidney pyelonephritis), a combination of antimicrobial immune response and alloimmune response may be playing a role. Whether they occur simultaneously or one follows the other is difficult to postulate. It may have a temporal relation and may depend on the timing of the biopsy as to which component of the immune response is dominant and that may determine the response to antibiotics. The possibility that infection can trigger a rejection in the graft has been postulated for a long time in older transplant literature.50,51 Our previous study6 showed that 10 of 30 patients with culture-positive APN had graft loss (six of them within one year post-biopsy) despite treatment with antibiotics. Four other patients showed improvement in graft function after adding steroids to their antibiotic treatment.
Toll-like receptors (TLRs), specifically TLR-4, is important in protection against ascending urinary tract bacterial infection.52 TLRs have also been shown to be expressed by renal tubular epithelial cells and collecting duct cells in ascending urinary tract infection.53 Our results did not show significantly elevated levels of TLR transcripts in APN or NP. However, the biopsy is only a snap shot and upregulation of receptor genes can be transitory. We did, however, find very high levels of gene expression for lactoferrin (LTF) in both APN and NP (but not in AR), and this may prove to be a good urinary biomarker to differentiate APN and AR. LTF is an iron-binding glycoprotein in secondary granules of polymorphonuclear leukocytes, found in various body secretions such as saliva, tears and milk. It has antimicrobial activity and interacts with molecules in the TLR4 pathway, such as CD14 and LPS-binding protein in macrophages.54,55
Other causes of allograft inflammation such as allergic interstitial nephritis and polyomavirus nephropathy were not investigated in the present study. Interstitial nephritis is rarely diagnosed in renal allograft especially in the early post-transplant period, and polyomavirus infection can be diagnosed by other modalities such as quantitative serum PCR and immunohistochemistry.
In summary, this is a preliminary exploratory study but it highlights two important issues about renal allograft pyelonephritis. We did find few genes with differential expression between graft APN and AR. These include CXCL10, CXCL11, CXCL1, CXCL2, and LTF. But we have to emphasize that the fold differences between graft APN and AR are small, leaving room for overlap. In fact, the overall gene expression pattern and pathway analysis reveal important similarities in the activated upstream molecules (especially interferon-gamma) between graft APN and AR, but not with native pyelonephritis. The other important issue highlighted in our study is that, a subset of allograft biopsies with histologic features of APN may have negative urine culture results and pose a diagnostic dilemma. These cases did not show any significant differences in gene expression from culture-positive APN biopsies. In fact, they show some overlap with both—APN and AR. From the practical point of view, we recommend that if transplant patients do not show improvement with antibiotics alone despite histologic features of APN, then a trial of steroids may be helpful. This is especially true if urine cultures are negative, and the chemokine profile shows a rejection pattern with high levels of interferon-γ-induced genes CXCL10 and CXCL11 and low lactoferrin. Although biomarkers may offer some hope, extensive validation studies are required before these can be used in clinical practice. This is an exploratory study with small sample numbers and it is possible that the gene expression markers identified may be non-specifically modulated by inflammation. Diagnosis and treatment of APN in renal allografts can be difficult. No single test (neither biopsy nor urine cultures nor biomarkers) can be used as a gold standard for diagnosis of APN in renal allografts or prediction of response to antibiotics. They have to be used in combination.
Supplementary Material
Acknowledgments
We want to acknowledge Hansjuerg Alder in the Comprehensive Cancer Core Facility and Kirsteen Maclean at NanoString Technology Seattle, Washington, for helpful advice in the analysis for the NanoString mRNA data; Lianbo Yu in the Department of Biostatistics for advice on statistical methods.
Footnotes
CONFLICT OF INTEREST
The authors of this manuscript have no conflicts of interest to disclose.
Additional Supporting Information may be found online in the supporting information tab for this article.
REFERENCES
- 1.Takai K, Tollemar J, Wilczek HE, Groth CG. Urinary tract infections following renal transplantation. Clin Transplant. 1998;12:19–23. [PubMed] [Google Scholar]
- 2.Schmaldienst S, Dittrich E, Hörl WL. Urinary tract infections after renal transplantation. Curr Opin Urol. 2002;12:125–130. doi: 10.1097/00042307-200203000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Pelle G, Vimont S, Levy PP, et al. Acute pyelonephritis represents a risk factor impairing long term kidney graft function. Am J Transplant. 2007;7:889–907. doi: 10.1111/j.1600-6143.2006.01700.x. [DOI] [PubMed] [Google Scholar]
- 4.Shin DH, Kim EJ, Lee S, Kim SJ, Oh J. Early-onset graft pyelonephritis is predictive of long term outcome of renal allografts. Tohoku J Exp Med. 2015;236:175–183. doi: 10.1620/tjem.236.175. [DOI] [PubMed] [Google Scholar]
- 5.Rollino C, Beltrame G, Ferro M, Quattrocchio G, Sandrone M, Quarello F. Acute pyelonephritis in adults: a case series of 223 patients. Nephrol Dial Transplant. 2012;27:3488–3493. doi: 10.1093/ndt/gfr810. [DOI] [PubMed] [Google Scholar]
- 6.Oghumu S, Bracewell A, Nori U, et al. Acute pyelonephritis in renal allografts – a new role for microRNAs? Transplantation. 2014;97:559–568. doi: 10.1097/01.TP.0000441322.95539.b3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta G, Shapiro R, Girnita A, et al. Neutrophilic tubulitis as a marker for urinary tract infection in renal allograft biopsies with C4d deposition. Transplantation. 2009;87:1013–1018. doi: 10.1097/TP.0b013e31819ca304. [DOI] [PubMed] [Google Scholar]
- 8.Mohamed N, Aggarwal V, Cole E, John R. Histopathologic detection of rejection in acute allograft pyelonephritis. Transplantation. 2012;94:e46. doi: 10.1097/TP.0b013e318265c4b8. [DOI] [PubMed] [Google Scholar]
- 9.Meehan SM, Nadasdy T. Tubulointerstitial diseases. In: Zhou XJ, Laszik Z, Nadasdy T, D’Agati V, Silva FG, editors. Silva’s Diagnostic Renal Pathology. New York, NY: Cambridge University Press; 2009. pp. 407–435. [Google Scholar]
- 10.Nickeleit V, Mengel M, Colvin R. Renal transplant pathology. In: Jeanette JC, Olson JL, Silva FG, D’Agati VD, editors. Heptinstall’s Pathology of the Kidney. 7th. Philadelphia, PA: Wolters Kluwer; 2015. pp. 1321–1460. [Google Scholar]
- 11.Solez K, Colvin RB, Racusen LC, et al. Banff “05 Meeting Report: differential diagnosis of chronic allograft injury and elimination of chronic allograft nephropathy (CAN) Am J Transplant. 2007;7:518–526. doi: 10.1111/j.1600-6143.2006.01688.x. [DOI] [PubMed] [Google Scholar]
- 12.Smyth GK. Linear models and empirical Bayes methods for assessing expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 13.Gordon A, Glazko G, Qiu X, Yakovlev A. Control of the mean number of false discoveries, Bonferroni and stability of multiple testing. Ann Appl Stat. 2007;1:179–190. [Google Scholar]
- 14.Anglicheau D, Suthanthiran M. Noninvasive prediction of organ graft rejection and outcome using gene expression patterns. Transplantation. 2008;86:192–199. doi: 10.1097/TP.0b013e31817eef7b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vasconcellos LM, Schachter AD, Zheng XX, et al. Cytotoxic lymphocyte gene expression in peripheral blood leukocytes correlates with rejecting renal allografts. Transplantation. 1998;66:562–566. doi: 10.1097/00007890-199809150-00002. [DOI] [PubMed] [Google Scholar]
- 16.Dugre FJ, Gaudreau S, Belles-Isles M, Houde I, Roy R. Cytokine and cytotoxic molecule gene expression determined in peripheral blood mononuclear cells in the diagnosis of acute renal rejection. Transplantation. 2000;70:1074–1080. doi: 10.1097/00007890-200010150-00014. [DOI] [PubMed] [Google Scholar]
- 17.Netto MV, Fonseca BA, Dantas M, Saber LT, Castro MC, Ferraz AS. FAS-ligand and perforin expression during acute cellular rejection episodes after kidney transplantation: comparison between blood and renal aspirates. In: Granzyme B, editor. Transplant Proc. Vol. 34. 2002. pp. 476–478. [DOI] [PubMed] [Google Scholar]
- 18.Sabek O, Dorak MT, Kotb M, Gaber AO, Gaber L. Quantitative detection of T-cell activation markers by real-time PCR in renal transplant rejection and correlation with histopathologic evaluation. Transplantation. 2002;74:701–707. doi: 10.1097/00007890-200209150-00019. [DOI] [PubMed] [Google Scholar]
- 19.Shin GT, Kim SJ, Lee TS, Oh CK, Kim H. Gene expression of perforin by peripheral blood lymphocytes as a marker of acute rejection. Nephron Clin Pract. 2005;100:c63–c70. doi: 10.1159/000085050. [DOI] [PubMed] [Google Scholar]
- 20.Veale JL, Liang LW, Zhang Q, et al. Noninvasive diagnosis of cellular and antibody-mediated rejection by perforin and granzyme B in renal allografts. Hum Immunol. 2006;67:777–786. doi: 10.1016/j.humimm.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Graziotto R, Del Prete D, Rigotti P, et al. Perforin, Granzyme B, and fas ligand for molecular diagnosis of acute renal-allograft rejection: analyses on serial biopsies suggest methodological issues. Transplantation. 2006;81:1125–1132. doi: 10.1097/01.tp.0000208573.16839.67. [DOI] [PubMed] [Google Scholar]
- 22.Galichon P, Hertig A, Rondeau E. Urinary-cell mRNA and acute kidney-transplant rejection. N Eng J Med. 2013;369:1860–1861. doi: 10.1056/NEJMc1310006. [DOI] [PubMed] [Google Scholar]
- 23.Suthanthiran M, Schwartz JE, Ding R, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Eng J Med. 2013;369:20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li B, Hartono C, Ding R, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Eng J Med. 2001;344:947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 25.Dadhania D, Muthukumar T, Ding R, et al. Molecular signatures of urinary cells distinguish acute rejection of renal allografts from urinary tract infection. Transplantation. 2003;75:1752–1754. doi: 10.1097/01.TP.0000063931.08861.56. [DOI] [PubMed] [Google Scholar]
- 26.Yannaraki M, Rebibou JM, Ducloux D, et al. Urinary cytotoxic molecular markers for a noninvasive diagnosis in acute renal transplant rejection. Transpl Int. 2006;19:759–768. doi: 10.1111/j.1432-2277.2006.00351.x. [DOI] [PubMed] [Google Scholar]
- 27.Muthukumar T, Ding R, Dadhania D, et al. Serine proteinase inhibitor-9, an endogenous blocker of granzyme B/perforin lytic pathway, is hyperexpressed during acute rejection of renal allografts. Transplantation. 2003;75:1565–1570. doi: 10.1097/01.TP.0000058230.91518.2F. [DOI] [PubMed] [Google Scholar]
- 28.Lo DJ, Weaver TA, Kleiner DE, et al. Chemokines and their receptors in human renal allotransplantation. Transplantation. 2011;91:70–77. doi: 10.1097/TP.0b013e3181fe12fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romagnani P, Crescioli C. CXCL10: a candidate biomarker in transplantation. Clin Chim Acta. 2012;413:1364–1373. doi: 10.1016/j.cca.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 30.Grau V, Gemsa D, Steiniger B, Garn H. Chemokine expression during acute rejection of rat kidneys. Scand J Immunol. 2000;51:435–440. doi: 10.1046/j.1365-3083.2000.00719.x. [DOI] [PubMed] [Google Scholar]
- 31.Husain S, Resende MR, Rajwans N, et al. Elevated CXCL10 (IP-10) in bronchoalveolar lavage fluid is associated with acute cellular rejection after human lung transplantation. Transplantation. 2014;97:90–97. doi: 10.1097/TP.0b013e3182a6ee0a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller M, Carter S, Hofer MJ, Campbell IL. Review: the chemokine receptor CXCR3 and its ligands CXCL9, CXCL10, and CXCL11 in neuroimmunity – a tale of conflict and conundrum. Neuropathol Appl Neurobiol. 2010;36:368–387. doi: 10.1111/j.1365-2990.2010.01089.x. [DOI] [PubMed] [Google Scholar]
- 33.Hricik DE, Nickerson P, Formica RN, et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant. 2013;13:2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Orabona C, Grohmann U. Indoleamine 2,3-dioxygenase and regulatory function: tryptophan starvation and beyond. Methods Mol Biol. 2011;677:269–280. doi: 10.1007/978-1-60761-869-0_19. [DOI] [PubMed] [Google Scholar]
- 35.Pallotta MT, Orabona C, Volpi C, et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat Immunol. 2011;12:870–878. doi: 10.1038/ni.2077. [DOI] [PubMed] [Google Scholar]
- 36.Choy JC. Granzymes and perforin in solid organ transplant rejection. Cell Death Differ. 2010;17:567–576. doi: 10.1038/cdd.2009.161. [DOI] [PubMed] [Google Scholar]
- 37.Delves PJ, Martin SJ, Burton DR, Roitt IM. Roitt’s Essential Immunology. 11th. Malden, MA: Blackwell Publishing; 2006. The Production of Effectors, Chapter 9. [Google Scholar]
- 38.Haudek-Prinz VJ, Klepeisz P, Slany A, et al. Proteome signatures of inflammatory activated primary human peripheral blood mononuclear cells. J Proteomics. 2012;76:150–162. doi: 10.1016/j.jprot.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.De Filippo K, Henderson RB, Laschinger M, Hogg N. Neutrophil chemokines KC and macrophage-inflammatory protein-2 are newly synthesized by tissue macrophages using distinct TLR signaling pathways. J Immunol. 2008;180:4308–4315. doi: 10.4049/jimmunol.180.6.4308. [DOI] [PubMed] [Google Scholar]
- 40.De Filippo K, Dudeck A, Hasenberg M, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013;121:4930–4937. doi: 10.1182/blood-2013-02-486217. [DOI] [PubMed] [Google Scholar]
- 41.Vassalli G, Milano G, Moccetti T. Role of mitogen-activated protein kinases in myocardial ischemia-reperfusion injury during heart transplantation. J Transplant. 2012;2012:928954. doi: 10.1155/2012/928954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong C, Davis RJ, Flavell RA. MAP kinases in the immune response. Annu Rev Immunol. 2002;20:55–72. doi: 10.1146/annurev.immunol.20.091301.131133. [DOI] [PubMed] [Google Scholar]
- 43.Chen R, Li X, Lu S, et al. Role of extracellular signal-regulated kinases 1 and 2 and p38 mitogen-activated protein kinase pathways in regulating replication of Penicillium marneffei in human macrophages. Microbes Infect. 2014;16:401–408. doi: 10.1016/j.micinf.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 44.Baldassare JJ, Bi Y, Bellone CJ. The role of p38 mitogen-activated protein kinase in IL-1 beta transcription. J Immunol. 1999;162:5367–5373. [PubMed] [Google Scholar]
- 45.Feng GJ, Goodridge HS, Harnett MM, et al. Extracellular signal-related kinase (ERK) and p38 mitogen-activated protein (MAP) kinases differentially regulate the lipopolysaccharide-mediated induction of inducible nitric oxide synthase and IL-12 in macrophages: leishmania phosphoglycans subvert macrophage IL-12 production by targeting ERK MAP kinase. J Immunol. 1999;163:6403–6412. [PubMed] [Google Scholar]
- 46.Hii CS, Stacey K, Moghaddami N, Murray AW, Ferrante A. Role of the extracellular signal-regulated protein kinase cascade in human neutrophil killing of Staphylococcus aureus and Candida albicans and in migration. Infect Immun. 1999;67:1297–1302. doi: 10.1128/iai.67.3.1297-1302.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kiigler S, Schüller S, Goebel W. Involvement of MAP-kinases and phosphatases in uptake and intracellular replication of Listeria monocytogenes in J774 macrophage cells. FEMS Microbiol Lett. 1997;157:131–136. doi: 10.1111/j.1574-6968.1997.tb12763.x. [DOI] [PubMed] [Google Scholar]
- 48.Klug K, Ehlers S, Uhlig S, Reiling N. Mitogen-activated protein kinases p38 and ERK1/2 regulated control of Mycobacterium avium replication in primary murine macrophages is independent of tumor necrosis factor-alpha and interleukin-10. Innate Immun. 2011;17:470–485. doi: 10.1177/1753425910377799. [DOI] [PubMed] [Google Scholar]
- 49.Rabant M, Amrouche L, Lebreton X, et al. Urinary C-X-C motif chemokine 10 independently improves the noninvasive diagnosis of antibody-mediated kidney allograft rejection. J Am Soc Nephrol. 2015;26:2840–2851. doi: 10.1681/ASN.2014080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Audard V, Amor M, Desvaux D, et al. Acute graft pyelonephritis: a potential cause of acute rejection in renal transplant. Transplantation. 2005;80:1128–1130. doi: 10.1097/01.tp.0000174343.05590.9f. [DOI] [PubMed] [Google Scholar]
- 51.Locke JE, Zoachary AA, Warren DS, et al. Proinflammatory events are associated with significant increases in breadth and strength of HLA-specific antibody. Am J Transplant. 2009;9:2136–2139. doi: 10.1111/j.1600-6143.2009.02764.x. [DOI] [PubMed] [Google Scholar]
- 52.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 53.Chassin C, Goujon JM, Darche S, et al. Renal collecting duct epithelial cells react to pyelonephritis-associated Escherichia coli by activating distinct TLR4-dependent and -independent inflammatory pathways. J Immunol. 2006;177:4773–4784. doi: 10.4049/jimmunol.177.7.4773. [DOI] [PubMed] [Google Scholar]
- 54.Curran CS, Demick KP, Mansfield JM. Lactoferrin activates macrophages via TLR4-dependent and -independent signaling pathways. Cell Immunol. 2006;242:23–30. doi: 10.1016/j.cellimm.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 55.Ando K, Hasegawa K, Shindo K, et al. Human lactoferrin activates NF-kappaB through the Toll-like receptor 4 pathway while it interferes with the lipopolysaccharide-stimulated TLR4 signaling. FEBS J. 2010;277:2051–2066. doi: 10.1111/j.1742-4658.2010.07620.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.