Abstract
Chromosome instability contributes to the multistep oncogenesis of cancer cells. Kaposi’s sarcoma (KS), an angiogenic vascular spindle cancer of endothelial cells, displays stage advancement with lesions at early stage being hyperproliferative, whereas lesions at late stage are clonal or multiclonal and can exhibit a neoplastic nature and chromosome instability. Although infection with KS-associated herpesvirus (KSHV) has been associated with the initiation and promotion of KS, the mechanism of KS neoplastic transformation remains unclear. We show that KSHV infection of primary human umbilical vein endothelial cells induces abnormal mitotic spindles and centrosome duplication. As a result, KSHV-infected cells manifest chromosome instability, including chromosomal misalignments and laggings, mitotic bridges, and formation of micronuclei and multinucleation. Our results indicate that KSHV infection could predispose cells to malignant transformation through induction of genomic instability and contributes to the development of KS.
Introduction
Genetic alterations as a result of chromosome instability play a pivotal role in the pathogenesis of a variety of human cancers. Alterations such as chromosome deletions, translocations, inversions, and microsatellite instability often lead to the activation of a proto-oncogene(s) and/or inactivation of a tumor suppressor gene(s), contributing to cellular transformation and subsequent tumorigenesis (1). It is generally believed that environmental clastogens, fragile site induction, or telomere dysfunction could lead to chromosome instability. Besides these factors, viral infection and subsequent integration of viral genetic materials into cellular chromosomes have been implicated in the initiation of structural and numerical chromosome instability (2, 3).
A number of viruses, including DNA and RNA viruses, have been found to induce either random or specific chromosome aberrations (2, 3). For example, adenoviruses type 2 and 7 cause random aberrations, whereas type 17 causes chromosome 17 uncoiling (4). Among her-pesviruses, human cytomegalovirus causes a variety of chromosome alterations, most of which are chromatid breaks and chromosome pulverizations resembling prematurely condensed S-phase chromatin (5, 6). Herpes simplex virus also induces similar chromosome aberrations early in an infection (7).
Kaposi’s sarcoma (KS) is an angiogenic vascular spindle tumor of endothelial cells associated with infection by a γ-2 herpesvirus, KS-associated herpesvirus (KSHV). KSHV is also etiologically associated with several other lymphoproliferative malignancies, including primary effusion lymphoma and multicentric Castleman’s disease (8). Although some early studies have described KS as a cytokine-driven hyperplasia, more recent studies have shown that KS lesions are clonal or multiclonal, thus manifest a neoplastic-transforming nature, and they have various genomic abnormalities, including chromosome instability (9). For example, cells of primary cultures derived from KS biopsies contained a number of chromosome rearrangements and deletions (10–12). Structural chromosome rearrangements were also found in blood lymphocytes and fibroblasts from patients with KS (13). Chromosome instability such as microsatellite instability has also been described in primary effusion lymphoma (14). Besides gross chromosomal abnormalities, genetic alterations have been found in some specific genes or gene loci in KSHV-related malignancies and KSHV-infected primary effusion lymphoma cell lines. Amplification of FGF4 and INT2 oncogenes and mutations in p53 was observed in KS lesions (15, 16), whereas loss of another tumor suppressor gene, p16INK4a, was commonly found in primary effusion lymphoma (17). Together, these studies suggest that genetic alterations, including chromosomal abnormalities, may be involved in the pathogenesis of KS and other KSHV-related malignancies. Nevertheless, it has not been determined whether KSHV-induced genetic alterations contribute to KSHV-induced neoplastic transformation. In this study, we directly assessed whether KSHV induced chromosome instability in a highly efficient KSHV infection model involving primary human umbilical vein endothelial cells (HUVECs). We found that KSHV induced chromosome instability and aberrant mitosis such as chromosome misalignments and laggings, mitotic bridges, micronucleus, multipolar mitotic spindles, and multinucleation. These results provide direct evidence to support an essential role for KSHV in the neoplastic transformation of cells into KS tumors.
Materials and Methods
KSHV Infection of HUVECs
HUVECs obtained from Clonetics (San Diego, CA) were cultured in Endothelial Cell Growth Medium Bullet Kit (Clonetics), containing human epidermal growth factor, human fibroblast growth factor B, vascular endothelial growth factor, ascorbic acid, hydrocortisone, long R3 insulin-like growth factor I, and heparin, as instructed by the manufacturer, and infected with KSHV as described previously (18, 19). The virus stock used for the study infected ~50% of HUVECs.
Detection of Chromosome Instability
Micronuclei were detected by staining cells with the DNA dye 4′,6-diamidino-2-phenylindole (DAPI). Cells grown on chamber slides were fixed with freshly prepared 2% paraformaldehyde for 10 min at room temperature and permeabilized with 1% Triton X-100 in 3% BSA for 20 min at 65°C. The cells were stained with DAPI at 1 µg/ml for 1 min at room temperature and analyzed with a fluorescence microscope. Chromosomal misalignments and laggings, and mitotic bridges were similarly examined after DAPI staining of mitotic cells.
Detection of Mitotic Spindles
Mitotic spindles were detected by staining α-tubulin. Cells, prepared as described previously, were blocked with 3% BSA for 20 min at room temperature. A monoclonal antibody against α-tubulin (Sigma-Aldrich, St. Louis, MO) was incubated with the cells at a 1:500 dilution in 3% BSA for 1 h at room temperature. After washing, the cells were incubated with a rabbit antimouse secondary antibody conjugated with TRITC (DAKO, Glostrup, Denmark) at a 1:40 dilution in 3% BSA for 45 min. Cells then were washed in PBS, counterstained with DAPI, and analyzed using a fluorescence microscope.
Detection of Centrosomes
Centrosomes were detected by staining γ-tubulin with a monoclonal antibody against γ-tubulin (GTU-88, Sigma-Aldrich) at a 1:200 dilution for 45 min at room temperature, washing, and incubating with a TRITC-conjugated rabbit antimouse secondary antibody before preparation for examination, as described previously.
Results
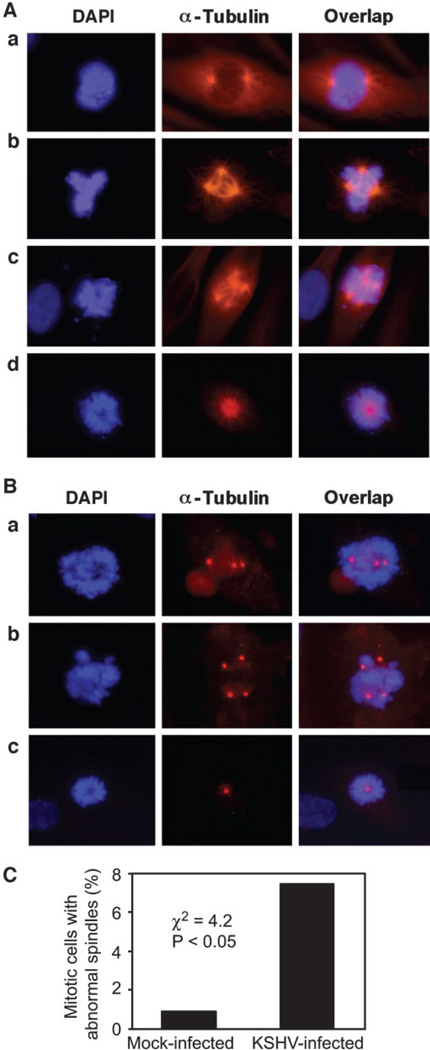
During normal cell mitosis, bipolar metaphase spindles are formed to ensure proper alignment of chromosomes in the central equator plate in metaphase, followed by segregation into two daughter cells in anaphase and telophase (Fig. 1A, a). The disruption of normal mitotic spindles results in chromosome instability (20). To determine whether a KSHV infection induced chromosome instability, we first examined whether KSHV infection disrupted normal mitotic spindles. HUVECs were infected with an infectious recombinant KSHV BAC36 (18). This cell model recapitulates KSHV infection in KS tumor cells (19). KSHV infection induced abnormal spindles, including multipolar and monopolar spindles (Fig. 1A, b–d). Abnormal mitotic spindles were observed in 7.5% of the mitotic cells, a rate that was 8.3-fold higher than that of control mock-infected cells (Fig. 1C; χ2 = 4.2; P < 0.05). There were more infected cells with multipolar spindles than those with monopolar spindles (95% versus 5%).
Figure 1.
Induction of abnormal mitotic spindles and centrosome numbers and chromosomal misalignments during Kaposi’s sarcoma-associated herpesvirus (KSHV) infection of human umbilical vein endothelial cells (HUVECs). A, 4′,6-diamidino-2-phenylindole (DAPI) and α-tubulin staining shows normal mitotic spindles and chromosomal alignments in mock-infected HUVECs (a) and abnormal mitotic spindles and chromosomal misalignments in three different representative fields of KSHV-infected HUVECs (b– d). B, DAPI and γ-tubulin staining shows abnormal centrosome numbers and chromosomal misalignments in three different representative fields of KSHV-infected HUVECs. C, quantification of abnormal mitotic spindles in mock- or KSHV-infected mitotic cells.
Mitotic spindle is formed by the dynamic nucleation of centrosomes, whose number in each cell is under tight control by the cell cycle machinery (21). A defect in centrosome amplification could lead to an abnormal number of mitotic spindle poles. As expected, KSHV infection induced abnormal centrosome duplication. We observed multiple (Fig. 1B, a and b) and single centrosomes (Fig. 1B, c) in KSHV-infected cells with abnormal mitotic spindles.
The disruption of mitotic spindles could result in chromosomal misalignments in metaphase (22). Chromosomal misalignments in KSHV-infected cells with abnormal mitotic spindles (Fig. 1A, b–d) and abnormal centrosome numbers (Fig. 1B) were observed. Misaligned chromosomes were frequently delocalized from the metaphase plate with some connecting to the plate but not aligning with it (misaligned), whereas others were totally separating from the plate (unaligned).
Chromosomal laggings and formation of mitotic bridges in telophase and anaphase commonly are seen in cells with chromosome instability, alterations that could lead to chromosomal deletions and formation of aneuploidy cells (22). KSHV infection induced chromosomal laggings in anaphase and telophase with some connecting to, whereas others were separating from, the main aligned chromosomes (Fig. 2A, b and c). Some KSHV-infected cells with chromosomal lagging had also other mitotic aberrations such as mitotic bridges (Fig. 2A, c). Interestingly, chromosomal misalignments were also observed in KSHV-infected cells with normal mitotic spindles (Fig. 2A, b and c) and normal centrosome numbers (data not shown), indicating that the induction of abnormal mitotic spindles is not the sole mechanism underlining these chromosomal aberrations. Overall, chromosomal laggings were significantly higher in KSHV-infected cells than in control mock-infected cells (14% versus 2.1%; P < 0.05; Fig. 2B).
Fig. 2.
Induction of chromosomal laggings and mitotic bridges during Kaposi’s sarcoma-associated herpesvirus (KSHV) infection of human umbilical vein endothelial cells (HUVECs). A, 4′,6-diamidino-2-phenylindole (DAPI) and α-tubulin staining shows chromosomal laggings (b and c) and mitotic bridges (c–f) in KSHV-infected cells. Some cells have chromosomal laggings and mitotic bridges (c). Mock-infected cells have neither chromosomal laggings nor mitotic bridges (a). B, quantification of chromosomal laggings in mock- or KSHV-infected mitotic cells. C, quantification of mitotic bridges in mock- or KSHV-infected mitotic cells.
The formation of mitotic bridges is another indicator of chromosomal aberrations (22). We observed mitotic bridges in KSHV-infected cells (Fig. 2A, c–f). The majority of the bridges were observed in telophase and anaphase; however, postmitotic bridges between two daughter cells that had finished cytokinesis (Fig. 2A, f) also were observed. The thin connection between the two nuclei of daughter cells was obvious even when the chromosomes were under-going decondensation. The percentage of cells with mitotic bridges was fivefold higher in KSHV-infected cells than in control mock-infected cells (8.4% versus 1.6% χ2 = 3.9; P < 0.05; Fig. 2C). Mitotic bridges in KSHV-infected cells with normal mitotic spindles again were observed, indicating that, similar to chromosomal laggings, induction of abnormal mitotic spindles was not the sole mechanism underlining development of this particular type of chromosomal aberration.
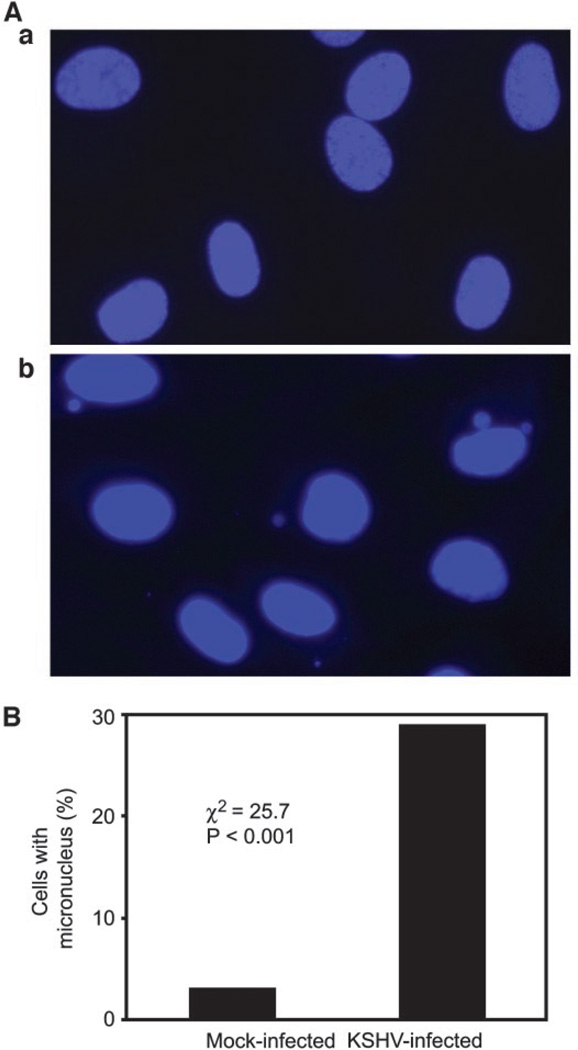
One of the consequences of mitotic chromosomal abnormalities is the loss of whole or partial chromosomes, which can be monitored by measuring the formation of micronuclei (20). KSHV infection of HUVECs induced micronuclei, which either were in the cytoplasm or as buds attached to the nuclei (Fig. 3A, b). We observed micronuclei in as many as 29% of KSHV-infected cells, a rate that was 15-fold greater than that of control mock-infected cells (χ2 = 25.7; P < 0.001; Fig. 3B). Because cells having micronuclei were in greater proportion than cells having mitotic chromosomal abnormalities and/or cells with abnormal mitotic spindles, it appears that the former cells had accumulated micronuclei, or some other mechanism(s) must be involved in the increased formation of micronuclei.
Fig. 3.
Induction of micronuclei during Kaposi’s sarcoma-associated herpesvirus (KSHV) infection of human umbilical vein endothelial cells (HUVECs). A, micronuclei detected by 4′,6-diamidino-2-phenylindole staining of mock-infected HUVECs (a) or HUVECs infected by KSHV (b). B, quantification of micronuclei in mock- or KSHV-infected cells.
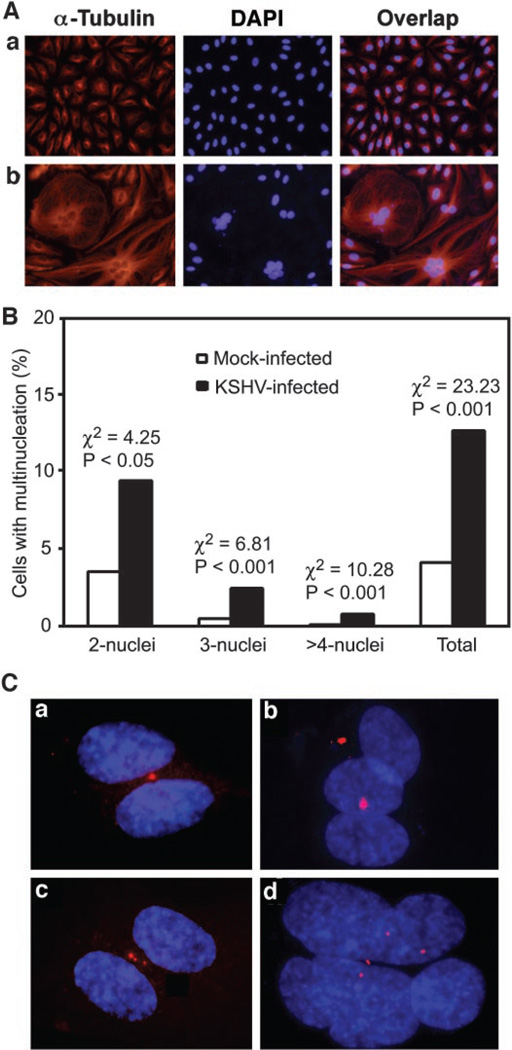
Abnormalities in mitosis could result in aneuploidy and multinucleation. KSHV infection induced multinucleation (Fig. 4A, b). The percentage of cells with multiple nuclei was significantly higher in KSHV-infected cells than that of control mock-infected cells (1.28% versus 0.41% χ2 = 23.23; P < 0.001; Fig. 4B). KSHV-infected cells with abnormal numbers of nuclei often exhibited other atypias, such as enlargement and irregular shape of the nuclei, and enlargement of the entire cells (Fig. 4A, b). Furthermore, cells with multinucleation had abnormal centrosome numbers with either one or multiple centrosomes (Fig. 4C), indicating that abnormal centrosome duplication is associated with KSHV-induced multinucleation.
Fig. 4.
Induction of multinucleation during Kaposi’s sarcoma-associated herpesvirus (KSHV) infection of human umbilical vein endothelial cells (HUVECs). A, 4′,6-diamidino-2-phenylindole (DAPI) and α-tubulin staining shows multinucleation in KSHV-infected cells (b) but not in mock-infected cells (a). B, quantification of multinucleation in mock- or KSHV-infected cells. C, DAPI and γ-tubulin staining shows abnormal centrosome numbers in cells with multinucleation (a–d represent four different fields).
Discussion
Despite the link between KSHV infection and development of KS, direct cellular transformation by KSHV in cell culture has only been rarely observed, suggesting that other cellular genetic alterations are required for the oncogenesis of KS lesions (23). The development of KS resembles a multistep oncogenesis model with early-stage KS lesions often being hyperproliferative and late-stage KS lesions progressing to neoplastic cancers that contain various genomic abnormalities (9). We have shown that KSHV infection of HUVECs induces multiple chromosomal abnormalities, including chromosomal misalignments and laggings, mitotic bridges, micronuclei, and multinucleation, any of which could predispose the cells to malignant transformation.
The fidelity of segregation of genetic materials during mitosis highly depends on tight cell cycle control of centrosome duplication and spindle formation (21). In a normal cell, there is one centromere in each sister chromatid, which is bound by the spindle thread to ensure proper segregation of the two sister chromatids in cytokinesis. If each sister chromatid is associated with more than one centromere, the chromatid would be attached by two or more spindles in the opposite direction. The outcomes would be abnormal chromosome behaviors, such as those that we have observed in this study, which could lead to chromosomal deletions, translocations, inversions, and even losses of whole chromosomes and cell aneuploidy. KSHV infection induces mitotic spindle and centrosome abnormalities, which could be a mechanism underlining some of the observed chromosomal abnormalities.
Activation of mitogen-activated protein kinase has been implicated in uncontrolled centrosome duplication leading to abnormal mitotic spindles and chromosomal abnormalities (24). KSHV infection activates mitogen-activated protein kinase pathways through binding of its glycoprotein B to integrin receptors (25). Various KSHV-encoded proteins can also activate mitogenic pathways (8). Inhibition of tumor suppressor genes, such as p53 (26), BRCA1 (27), BRCA2 (28), or GADD45 (29), could disrupt the mitotic checkpoints that normally ensure the proper attachment of kinetochores to microtubules of the mitotic spindles and alignment of chromosomes at the metaphase plate, thus leading to induction and/or accumulation of abnormal mitotic spindles and centrosomes that could result in chromosome instability (30). Several KSHV genes are known to inhibit tumor suppressor pathways, among which latent nuclear antigen 1 encoded by orf73 is a multifunction major latent antigen and inhibits p53 and pRb tumor suppressor pathways (31, 32). Latent nuclear antigen 1 also tethers KSHV episomes to cellular chromosomes, which could further contribute to chromosomal abnormalities (33). Therefore, initial viral entry and subsequent expression of viral proteins could lead to the induction of chromosome instability.
We have shown previously that KSHV infection of HUVECs induces cellular proliferation, followed by an early crisis phase with many cells dying, an outcome that has been attributed to active viral replication (19). Induction of chromosome instability by KSHV infection could certainly trigger cell cycle checkpoints and contribute to the observed cell death in the crisis phase. Nevertheless, a few of the KSHV-infected cells overcome the crisis, continue to proliferate, and establish a latent viral infection with a life span substantially longer than that of mock-infected cells, which generally undergo senescence after 4–5 weeks of culture (19). These hyperproliferative cells, which are reminiscent of early-stage KS tumor cells, would need to over-come controls at cell cycle checkpoints to survive KSHV-induced chromosome instability. KSHV-encoded genes with inhibitory activities toward tumor suppressor pathways should have important roles in disrupting cell cycle checkpoints, therefore facilitating KSHV-induced oncogenesis (8).
KSHV induction of KS fits a multistep oncogenesis model (Fig. 5). KSHV infection leads to an activation of mitogenic pathways and an inhibition of tumor suppressor pathways via binding to virus receptor(s) and activation of viral oncogenes to result in the induction of chromosome instability. The majority of the cells undergo cell death crisis because of the activation of cell cycle checkpoints. However, several KSHV genes could disrupt the cell cycle checkpoints via the inhibition of tumor suppressor pathways. In addition, a number of KSHV genes can also activate cell survival signals. A few cells consequently survive, continue to proliferate, and accumulate genomic alterations that eventually lead to the development of neoplastic tumors.
Fig. 5.
Multistep oncogenesis model for Kaposi’s sarcoma-associated herpesvirus (KSHV) induction of Kaposi’s sarcoma.
Acknowledgments
We thank Dr. Charles Gauntt for helpful discussions and editing of the manuscript.
Grant support: NIH Grants CA096512 and HL60604, Charlotte Geyer Foundation, and Association for International Cancer Research (S-J. Gao).
References
- 1.Gisselsson D. Chromosome instability in cancer: how, when, and why. Adv Cancer Res. 2003:1–29. doi: 10.1016/s0065-230x(03)87164-6. [DOI] [PubMed] [Google Scholar]
- 2.Lavia P, Mileo AM, Giordano A, Paggi MG. Emerging roles of DNA tumor viruses in cell proliferation: new insights into genomic instability. Oncogene. 2003;22:6508–6516. doi: 10.1038/sj.onc.1206861. [DOI] [PubMed] [Google Scholar]
- 3.Fortunato EA, Spector DH. Viral induction of site-specific chromosome damage. Rev Med Virol. 2003;13:21–37. doi: 10.1002/rmv.368. [DOI] [PubMed] [Google Scholar]
- 4.Steffensen DM, Szabo P, McDougall JK. Adenovirus 12 uncoiler region of human chromosome 1 in relation to the 5s rRNA genes. Exp Cell Res. 1976;100:436–439. doi: 10.1016/0014-4827(76)90176-2. [DOI] [PubMed] [Google Scholar]
- 5.Luleci G, Sakizli M, Gunalp A. Selective chromosome damage caused by human cytomegalovirus. Acta Virol. 1980;24:341–345. [PubMed] [Google Scholar]
- 6.AbuBakar S, Au WW, Legator MS, Albrecht T. Induction of chromosome aberrations and mitotic arrest by cytomegalovirus in human cells. Exp Mol Mutag. 1988;12:409–420. doi: 10.1002/em.2860120409. [DOI] [PubMed] [Google Scholar]
- 7.Peat DS, Stanley MA. Chromosome damage induced by herpes simplex virus type 1 in early infection. J Gen Virol. 1986;67:2273–2277. doi: 10.1099/0022-1317-67-10-2273. [DOI] [PubMed] [Google Scholar]
- 8.Moore PS, Chang Y. Kaposi’s sarcoma-associated herpesvirus immunoevasion and tumorigenesis: two sides of the same coin? Annu Rev Microbiol. 2003;57:609–639. doi: 10.1146/annurev.micro.57.030502.090824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ensoli B, Sgadari C, Barillari G, Sirianni MC, Sturzl M, Monini P. Biology of Kaposi’s sarcoma. Eur J Cancer. 2001;37:1251–1269. doi: 10.1016/s0959-8049(01)00121-6. [DOI] [PubMed] [Google Scholar]
- 10.Popescu NC, Zimonjic DB, Leventon-Kriss SLBJ, Lunardi-Iskandar Y, Gallo RC. Deletion and translocation involving chromosome 3(p14) in two tumorigenic Kaposi’s sarcoma cell lines. J Natl Cancer Inst. 1996;88:450–455. doi: 10.1093/jnci/88.7.450. [DOI] [PubMed] [Google Scholar]
- 11.Bovi PD, Donti E, Knowles DM, et al. Presence of chromosomal abnormalities and lack of AIDS retrovirus DNA sequences in AIDS-associated Kaposi’s sarcoma. Cancer Res. 1986;46:6333–6338. [PubMed] [Google Scholar]
- 12.Casalone R, Albini A, Righi R, Granata P, Toniolo A. Nonrandom chromosome changes in Kaposi sarcoma: cytogenetic and FISH results in a new cell line (KSIMM) and literature review. Cancer Genet Cytogenet. 2001;124:16–19. doi: 10.1016/s0165-4608(00)00241-7. [DOI] [PubMed] [Google Scholar]
- 13.Cerimele D, Contu L, Scappaticci S, Cottoni F. Kaposi’s sarcoma in Sardina: an epidemiologic and genetic investigation. Ann NY Acad Sci. 1984;437:216–227. doi: 10.1111/j.1749-6632.1984.tb37140.x. [DOI] [PubMed] [Google Scholar]
- 14.Gaidano G, Pastore C, Gloghini A, et al. Microsatellite instability in KSHV/HHV-8 positive body-cavity-based lymphoma. Hum Pathol. 1997;28:748–750. doi: 10.1016/s0046-8177(97)90187-8. [DOI] [PubMed] [Google Scholar]
- 15.Kiuru-Kuhlefelt S, Sarlomo-Rikala M, Larramendy ML, et al. FGF4 and INT2 oncogenes are amplified and expressed in Kaposi’s sarcoma. Mod Pathol. 2000;13:433–437. doi: 10.1038/modpathol.3880074. [DOI] [PubMed] [Google Scholar]
- 16.Scinicariello F, Dolan MJ, Nedelcu I, Tyring SK, Hilliard JK. Occurrence of human papillomavirus and p53 gene mutations in Kaposi’s sarcoma. Virology. 1994;203:153–157. doi: 10.1006/viro.1994.1466. [DOI] [PubMed] [Google Scholar]
- 17.Platt G, Carbone A, Mittnacht S. p16INK4a loss and sensitivity in KSHV associated primary effusion lymphoma. Oncogene. 2002;21:1823–1831. doi: 10.1038/sj.onc.1205360. [DOI] [PubMed] [Google Scholar]
- 18.Zhou FC, Zhang YJ, Deng JH, et al. Efficient infection by a recombinant Kaposi’s sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J Virol. 2002;76:6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao SJ, Deng JH, Zhou FC. Productive lytic replication of a recombinant Kaposi’s sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J Virol. 2003;77:9738–9749. doi: 10.1128/JVI.77.18.9738-9749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Parry EM, Parry JM, Corso C, et al. Detection and characterization of mechanisms of action of aneugenic chemicals. Mutagenesis. 2002;17:509–521. doi: 10.1093/mutage/17.6.509. [DOI] [PubMed] [Google Scholar]
- 21.Kramer A, Neben K, Ho AD. Centrosome replication, genomic instability and cancer. Leukemia. 2002;16:767–775. doi: 10.1038/sj.leu.2402454. [DOI] [PubMed] [Google Scholar]
- 22.Wassmann K, Benezra R. Mitotic checkpoints: from yeast to cancer. Curr Opin Genet Dev. 2001;11:83–90. doi: 10.1016/s0959-437x(00)00161-1. [DOI] [PubMed] [Google Scholar]
- 23.Flore O, Rafii S, Ely S, O’Leary JJ, Hyjek EM, Cesarman E. Transformation of primary human endothelial cells by Kaposi’s sarcoma-associated herpesvirus. Nature. 1998;394:588–592. doi: 10.1038/29093. [DOI] [PubMed] [Google Scholar]
- 24.Saavedra HI, Fukasawa K, Conn CW, Stambrook PJ. MAPK mediates RAS-induced chromosome instability. J Biol Chem. 1999;274:38083–38090. doi: 10.1074/jbc.274.53.38083. [DOI] [PubMed] [Google Scholar]
- 25.Akula SM, Pramod NP, Wang FZ, Chandran B. Integrin á3ā1 (CD 49c/29) is a cellular receptor for Kaposi’s sarcoma-associated herpesvirus (KSHV/HHV-8) entry into the target cells. Cell. 2002;108:407–419. doi: 10.1016/s0092-8674(02)00628-1. [DOI] [PubMed] [Google Scholar]
- 26.Carroll PE, Okuda M, Horn HF, et al. Centrosome hyperamplification in human cancer: chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene. 1999;18:1935–1944. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- 27.Xu X, Weaver Z, Linke SP, et al. Centrosome amplification and a defective G2-M cell cycle checkpoint induce genetic instability in BRCA1 exon. Mol Cell. 1999;3:389–395. doi: 10.1016/s1097-2765(00)80466-9. [DOI] [PubMed] [Google Scholar]
- 28.Tutt A, Gabriel A, Bertwistle D, et al. Absence of Brca2 causes genome instability by chromosome breakage and loss associated with centrosome amplification. Curr Biol. 1999;9:1107–1110. doi: 10.1016/s0960-9822(99)80479-5. [DOI] [PubMed] [Google Scholar]
- 29.Hollander MC, Sheikh MS, Bulavin DV, et al. Genomic instability in Gadd45a-deficient mice. Nat Genet. 1999;23:461–466. doi: 10.1038/13802. [DOI] [PubMed] [Google Scholar]
- 30.Fisk HA, Mattison CP, Winey M. Centrosomes and tumour suppressors. Curr Opin Cell Biol. 2002;14:700–705. doi: 10.1016/s0955-0674(02)00385-x. [DOI] [PubMed] [Google Scholar]
- 31.Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402:889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- 32.Radkov SA, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat Med. 2000;6:1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- 33.Ballestas ME, Chatis PA, Kaye KM. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science. 1999;284:641–644. doi: 10.1126/science.284.5414.641. [DOI] [PubMed] [Google Scholar]