Abstract
Replication of the 3.2-kb hepatitis B virus (HBV) genome is driven by the covalently closed circular (ccc) DNA in the nucleus, from which four classes of co-terminal RNAs are transcribed. Genome replication requires just the 3.5-kb pregenomic RNA, which is terminally redundant. Cloning the full-length HBV genome into a vector disrupts its continuity, thus preventing genome replication at the step of pregenomic RNA transcription. This can be overcome by converting the monomeric construct into a tandem dimer, yet the need to ligate two molecules of the HBV genome with vector DNA makes it inefficient and even unsuccessful. To overcome this problem we partially digested the monomeric construct with the unique restriction enzyme used for cloning, and dephosphorylated the linearized monomer before its ligation with another copy of the HBV genome. Alternatively, the monomer was linearized at another unique restriction site inside the HBV genome, followed by its dephosphorylation and ligation with another copy of the HBV genome linearized at the same site. These approaches of two-way molecular ligation greatly improved the efficiency of dimer formation with about 50% of the bacterial colonies screened harboring tandem dimers.
Keywords: hepatitis B virus, covalently closed circular DNA, tandem dimer, pregenomic RNA, replication
1. Introduction
Hepatitis B virus (HBV) infection leads to diverse outcomes ranging from asymptomatic carrier status to fulminant hepatitis with high mortality rate. HBV isolates worldwide can be grouped into at least eight genotypes with over 8% sequence divergence. In addition, mutations accumulate at the immune clearance phase of chronic infection. Increasing evidence suggests that genetic variability in the HBV genome contributes to pathogenesis. For example, genotype C infection is associated with prolonged hepatitis B e antigen (HBeAg) positive phase of infection and increased lifelong risk for hepatocellular carcinoma (HCC) than genotype B infection. The G1896A precore mutation abolishing HBeAg expression has been linked to fulminant hepatitis, while core promoter mutations are implicated in both fulminant hepatitis and HCC development (Tong et al., 2013). Functional characterization of HBV genetic variants through transfection experiments could shed light on their molecular mechanisms of pathogenesis. In this regard, the covalently closed circular (ccc) DNA inside the nucleus of infected hepatocytes is the template for transcription of viral RNAs, the sources of viral protein translation, genome replication, and virion secretion. Transcribed from the 3.2-kb cccDNA template are the 2.4-, 2.1-, and 0.7-kb RNAs of subgenomic length, as well as the terminally redundant, 3.5-kb RNA. A subset of the 3.5-kb RNA, or pregenomic (pg) RNA, serves as the mRNA for the translation of core protein and DNA polymerase (P protein). Furthermore, it is co-packaged with P protein inside core particles assembled from core protein, where it is converted into partially double stranded DNA through a series of enzymatic reactions catalyzed by the P protein. Consequently, HBV genome replication is driven by pg RNA.
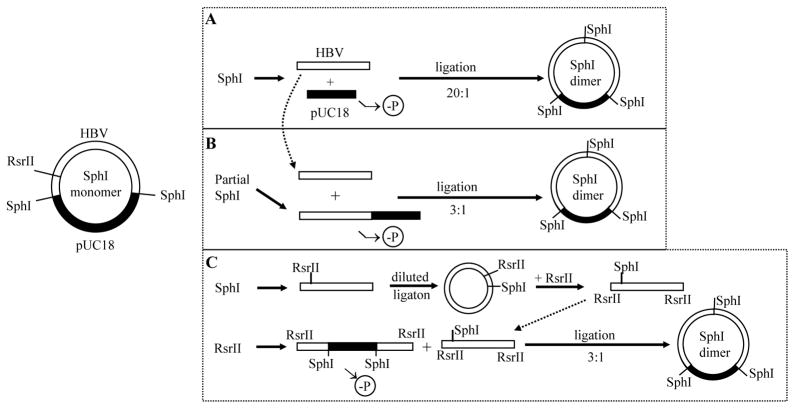
Functional characterization of clinical HBV isolates requires cloning of the intact HBV genome. This can be achieved using a single cutting restriction site on the viral genome, such as SphI. However, linking the HBV genome to a vector disrupts continuity of cccDNA, thus preventing transcription of the terminally redundant pg RNA. Therefore, a monomeric construct is incapable of genome replication (Qin et al., 2011A). One way to overcome this problem is to convert a monomer into a tandem dimer (Sureau et al., 1986; Sells et al., 1987). To achieve this the 3.2-kb HBV DNA is released from the recombinant plasmid by SphI digestion followed by its ligation with SphI cut, dephosphorylated vector DNA at high (10–20:1) molar ratio (Fig. 1, approach A). However, most progeny colonies harbor monomeric HBV construct, which prompted us to develop bacterial colony hybridization with an oligonucleotide probe spanning the tail-to-head junction (the SphI site) of the tandem dimer (Tong et al., 1991). Still, sometimes no dimer could be identified. In the present study, we modified our cloning strategy such that dimer formation required a two-way rather than three-way molecular ligation (Fig. 1, approaches B & C).
Fig. 1.
Strategies to convert an SphI monomer into an SphI dimer. pUC18 vector and HBV DNA are shown in black and white, respectively. In the established method (approach A), SphI-cut HBV DNA was ligated with SphI-cut, dephosphorylated pUC18 vector at high molar ratio. The efficiency of dimer formation is low due to the need for a three-way molecular ligation. In approaches B and C, ligation occurs between the 5.9-kb monomeric HBV DNA and pUC18 vector, through either the SphI site (approach B) or RsrII site (approach C). Such a two-way molecular ligation greatly enhances the efficiency of dimer formation.
2. Materials and methods
2.1. Materials
Nanodrop 2000c Spectrometer from ThermoFisher was used to measure DNA concentration. Tanon 1600 Gel Imaging System from Tanon Science & Technology Co., Ltd. was used to take pictures of ethidium bromide stained DNA gel, and to cut out gel slices containing desired DNA bands. Restriction enzymes (HindIII, NcoI, SphI, RsrII, ScaI), alkaline phosphatase (10 U/μl), and T4 DNA ligase (400 U/μl) were from New England Biolabs. DNA gel extraction kit and PCR cleanup kit were from Axygen. Competent E. coli cells were from Tiangen or self-made. DNA miniprep kit was from Omega. The HBV DNA constructs studied here originated from those previously described (Li et al 2007; Qin et al. 2011B). They are tandem dimers of genotype B, C, D, or G cloned to the SphI site of pUC18 vector (SphI dimers). Another clone of HBV genotype C was cloned to the BamHI site of the same vector (BamHI monomer) (Zhou et al., unpublished).
2.2. Methods
2.2.1. Linearized HBV monomer to serve as vector for dimer construction
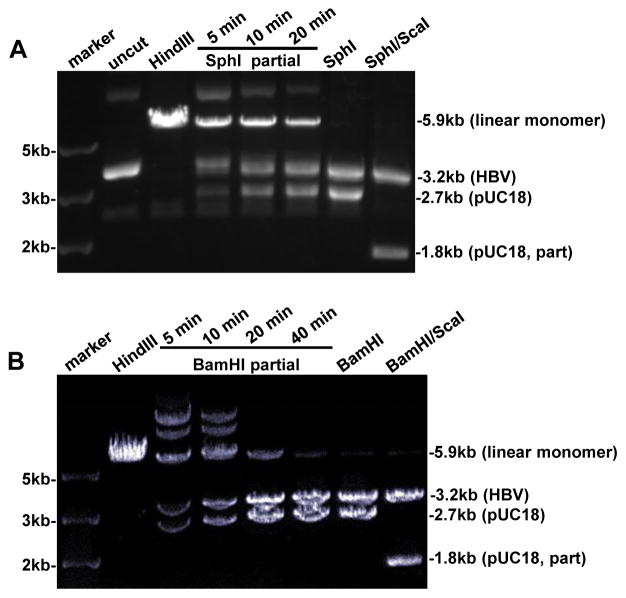
Aliquots of the SphI monomer (1.5–3 μg) were subject to 5–40 min of digestion at 37°C with 2–4 U of SphI, respectively, followed by a further incubation at 65°C for 20 min to inactivate the enzyme (Fig. 2A). The DNA samples were separated in 0.8% agarose gel and the 5.9-kb band corresponding to linearized monomer was cut out from the gel and purified by the gel extraction kit. DNA was eluted in 25–30 μl water and further treated at 37°C for 1 hr with 10 U of alkaline phosphatase to prevent self-ligation. DNA was purified by PCR cleanup kit, eluted in elution buffer, and DNA concentration was determined by Nanodrop 2000c Spectrometer. A similar procedure of BamHI partial digestion was used to obtain the linearized BamHI monomer (Fig. 2B). To linearize the SphI monomer at the RsrII site, 1.5 μg DNA was digested at 37°C for 2 hrs with 10 U of RsrII, followed by further treatment with 10 U of alkaline phosphatase at 37°C for 1 hr. The 5.9-kb linear HBV monomer was gel purified.
Fig. 2.
Preparation of the vector and insert DNA for approach B. Monomeric HBV DNA cloned via the SphI site (panel A) or BamHI site (panel B) was double digested respectively with SphI/ScaI and BamHI/ScaI, and the 3.2-kb HBV DNA was gel purified to serve as the insert for ligation. In addition, the 5.9-kb DNA was partially digested with SphI (panel A) or BamHI (panel B) to linearize the recombinant molecule, which will serve as the vector for ligation. HindIII digested monomer DNA served as the 5.9-kb size marker.
2.2.2. 3.2-kb linear HBV DNA to serve as insert for dimer construction
The SphI monomer (2 μg) was digested at 37°C for > 2 hrs with 15 U each of SphI and ScaI to generate 3.2-kb, 1.8-kb, and 0.9-kb DNA fragments (Fig. 2A). The digests were separated in 1% agarose gel and the 3.2-kb band was cut out. It was purified by the DNA gel extraction kit and eluted in 30 μl of elution buffer. Similarly, the BamHI monomer was doubly digested with BamHI and ScaI followed by gel purification of the 3.2-kb HBV DNA (Fig. 2B). To obtain 3.2-kb HBV DNA with RsrII ends, the purified linear 3.2-kb HBV DNA with SphI ends was ligated at 16°C overnight with 2 μl of T4 DNA ligase in a volume of 60 μl. The ligation product was purified by the PCR cleanup kit, eluted in water, and further digested with 10 U of RsrII. The digest was separated in 1% agarose gel and the 3.2-kb linear DNA was cut out. DNA was extracted from the gel slice (see Fig. 1C for diagram).
2.2.3. DNA ligation and bacterial transformation
DNA ligation was performed at 16°C overnight, usually in 10–20 μl volume using 0.8–1.2 μl of T4 DNA ligase, 15–40 ng of the 5.9-kb monomer DNA, and 3–5 times molar ratio of the 3.2-kb HBV DNA. The three pairs of DNA ligation were: 5.9-kb HBV monomer linearized at the SphI site with SphI cut 3.2-kb HBV DNA; 5.9-kb HBV monomer linearized at the BamHI site with BamHI cut 3.2-kb HBV DNA; and 5.9-kb HBV monomer linearized at the RsrII site with RsrII cut 3.2-kb HBV DNA (Fig. 1). In addition, the 5.9-kb HBV monomers were ligated alone to serve as negative controls. A fraction of the ligation product (5–10 μl) was incubated with 100 μl of competent E. coli on ice for 30 min, followed by heat shock at 42°C for 1 min and chilling on ice for 5 min. LB medium (500 μl) was added and samples were shaken at 37°C for 30 min. Aliquots of 50 μl and 200 μl were spread out onto LB/ampicillin (100 μg/ml) plates, followed by incubation at 37°C overnight.
2.2.4. Plasmid DNA preparation and identification of tandem dimers
Bacterial colonies were picked up from LB/ampicillin plates and inoculated into 5 ml of LB medium supplemented with 100 μg/ml ampicillin. After shaking at 37°C overnight, plasmid DNA was extracted from 3 ml of culture. Gel electrophoresis of uncut plasmid DNA or DNA cut with a set of restriction enzymes was used to identify tandem dimer constructs, as detailed in the Results section (Figs. 3–5).
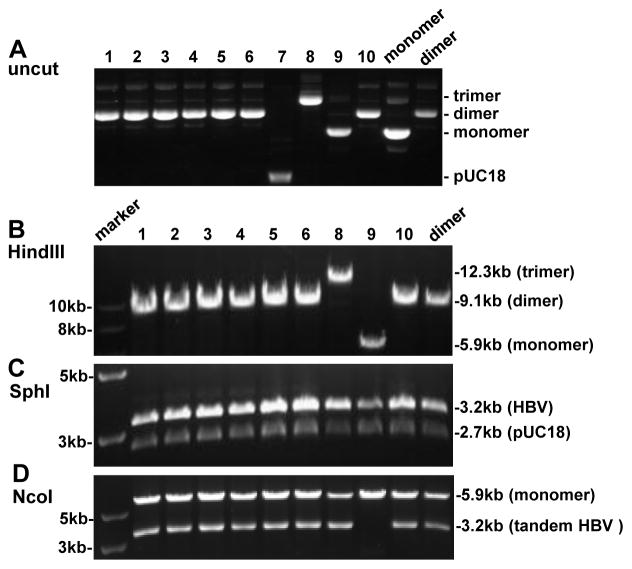
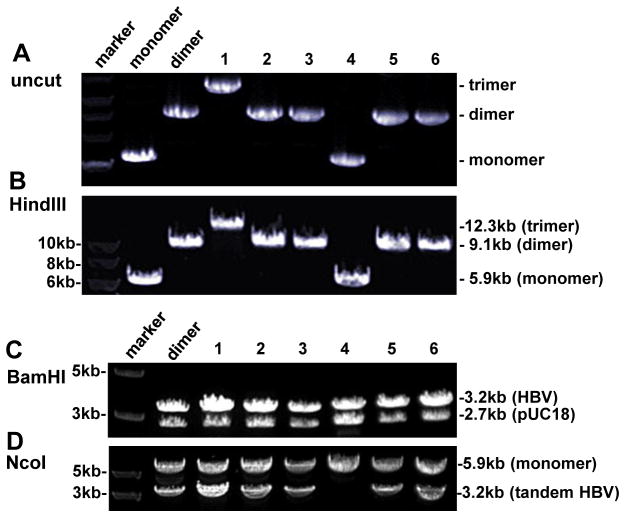
Fig. 3.
Screening for SphI dimers generated by SphI partial digestion of SphI monomer. (A) Uncut DNA from 10 clones with uncut dimer and monomer serving as controls. Clone 7 is probably pUC18 DNA. (B) HindIII digestion of 9 clones with HindIII cut dimer serving as control. The expected size is 9.1 kb for a dimer and 5.9 kb for a monomer. (C) SphI digestion, which will convert both a dimer and a monomer into 3.2-kb and 2.7-kb bands. (D) NcoI digestion. A 3.2-kb band will be released only when a dimer (or trimer) is tandem instead of tail-to-tail or head-to-head.
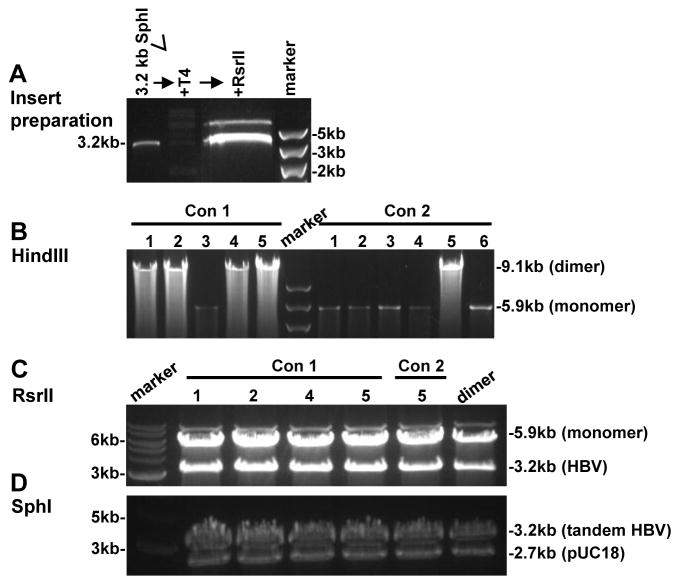
Fig. 5.
Conversion of an SphI monomer into SphI dimer via the RsrII restriction site. (A) Insert preparation. The SphI monomer was digested with SphI and the 3.2-kb genome was gel purified. Diluted HBV DNA was treated with T4 DNA ligase to promote intramolecular ligation. Finally, the ligated DNA was digested with RsrII and the 3.2-kb DNA band was gel purified. (B) HindIII digestion to distinguish dimers from monomers. (C) Digestion of the dimer clones by RsrII to confirm that the two copies in a dimer were ligated via the RsrII site. (C) SphI digestion of the dimer clones to verify whether the two copies are tandem.
3. Results
3.1. Converting SphI monomers into tandem dimers through SphI partial digestion
To increase the efficiency of dimer formation, we ligated HBV DNA to the HBV-pUC18 recombinant plasmid, rather than pUC18 vector. Consequently, a dimer is formed by joining two (3.2-kb + 5.9-kb) rather than three (3.2-kb + 3.2-kb + 2.7-kb) molecules (Fig. 1, compare approaches B & C with approach A). HBV-pUC18 monomer cloned via the SphI site was linearized by SphI partial digestion. As some circular monomers remain after partial digestion, which would contribute to colony formation during the transformation step, we ran the digested DNA in 0.8% agarose gel and cut out the 5.9-kb band (Fig. 2A). The linearized molecule was further treated with alkaline phosphatase to prevent self-ligation. With such measures, we obtained at least 3 times more bacterial colonies from the plate of 5.9-kb + 3.2-kb DNA ligation than the plate of 5.9-kb DNA self-ligation. Gel electrophoresis of uncut miniprep plasmid DNA from randomly picked colonies suggested that some were dimers (Fig. 3A), which was confirmed by digestion with HindIII, an enzyme with a single cleavage site on pUC18 but not the HBV genome (Fig. 3B). SphI would convert both a dimer and a monomer into 3.2-kb and 2.7-kb bands, but the 3.2-kb band will be more intense for a dimer (Fig. 3C). Whether a dimer is tandem (tail-to-head) or not (tail-to-tail or head-to-head) was verified by digestion with NcoI, which cleaves at position 1372 in the HBV genome adjacent to the SphI site (position 1238). Only when the dimer is tandem will NcoI release a 3.2-kb fragment (Fig. 3D).
So far we have converted 42 HBV DNA constructs from SphI monomer into SphI dimer through SphI partial digestion. Out of a total of 162 bacterial colonies screened, 84 (51.9%) were dimers, all of which were tandem (Table 1). The remaining clones harbored monomers (37.0%), pUC18 (9.3%), or trimers (1.9%).
Table 1.
Efficiency of monomer-to-dimer conversion by different methods
| Starting construct | Method of conversion | No. of constructs made | No. of clones examined | trimer* | dimer* | monomer | pUC18 |
|---|---|---|---|---|---|---|---|
| SphI monomer | SphI partial digestion | 42 | 162 | 3 (1.9%) | 84 (51.8%) | 60 (37.0%) | 15 (9.3%) |
| SphI monomer | RsrII digestion | 4 | 17 | 0 | 11 (64.7%) | 6 (35.3%) | 0 |
| BamHI monomer | BamHI partial digestion | 1 | 6 | 1 (16.7%) | 4 (67%) | 1 (16.7%) | 0 |
all were found to be tandem.
3.2. Converting a BamHI monomer into tandem dimer through BamHI partial digestion
A few Korean genotype C genomes harbored two SphI sites, rendering this enzyme unsuitable for cloning of the intact HBV genome. The unique BamHI site (at position 1400) was used instead to clone such HBV genomes into the pUC18 vector. To convert the BamHI monomer into a dimer, it was partially digested with BamHI followed by gel purification of the 5.9-kb DNA (Fig. 2B). The linearized monomer was treated with alkaline phosphatase, and ligated to the 3.2-kb HBV genome generated by complete BamHI/ScaI double digestion (Fig. 2B). Analysis of six randomly picked up colonies identified one monomer, four tandem dimers, and one tandem trimer (Fig. 4 & Table 1).
Fig. 4.
Screening for BamHI dimers generated by BamHI partial digestion of a BamHI monomer. (A) Uncut DNA from 6 clones with uncut dimer and monomer serving as controls. All the clones are dimers except for clone 1 (probably a trimer) and clone 4 (monomer). (B) HindIII digestion of the 6 clones. HindIII cut monomer and dimer served as controls. (C) BamHI digestion, which will generate 3.2-kb and 2.7-kb bands for a monomer, dimer, or trimer. However, the ratio between the two bands differs. (D) NcoI digestion. A 3.2-kb band will be generated from a dimer (or trimer) only when it is tandem.
3.3. Converting SphI monomers into tandem dimers through RsrII digestion
A drawback of the approach of SphI (or BamHI) partial digestion is the need to try out different conditions (enzyme concentration or duration of digestion) to get enough linearized monomer DNA. An alternative approach, which does not require partial digestion, is to use another restriction site unique on the HBV genome but absent in pUC18 (Fig. 1, approach C). In this regard, RsrII cleaves at position 1572 in the HBV genome. The SphI monomer was linearized by digestion with RsrII to completion, followed by treatment with alkaline phosphatase. To obtain the 3.2-kb HBV DNA linearized at the RsrII site, 3 μg of the SphI monomer was digested with SphI and the 3.2-kb HBV DNA was gel purified. It was treated with T4 DNA ligase to circularize the genome, then digested with RsrII and the 3.2-kb DNA was cut out from the gel. Ligation of the 3.2-kb HBV genome with RsrII ends with the 5.9-kb HBV monomer should lead to dimer formation (Fig. 1, approach C). A typical result is shown in Fig. 5. HindIII digestion revealed 4 out of 5 clones to be dimers for one HBV construct (construct 1), in contrast to 1 out of 6 clones for the other HBV construct (construct 2) (Fig. 5A). The remaining clones were monomers. The fact that all these 5 dimer clones generated 3.2-kb and 2.7-kb bands by both SphI digestion and RsrII digestion confirmed them as tandem dimers (Fig. 5B & 5C). So far we have used RsrII digestion to convert SphI monomers of 4 HBV DNA constructs into tandem dimers. Of the 17 clones analyzed, 11 were tandem dimers (Table 1).
4. Discussion
Functional characterization of chimeric HBV DNA constructs or site-directed mutants requires transfection experiments in human hepatoma cell lines such as HepG2 and Huh7. However, molecular cloning disrupts continuity in the HBV genome, which not only interrupts HBV coding sequence, but also prevents the transcription of terminally redundant pg RNA. Therefore, such a monomeric construct is unable to initiate genome replication despite ability to express some of the viral proteins (Qin et al., 2011A). Several approaches have been taken to ensure transcription of pg RNA and consequently replication of HBV DNA. The 3.2-kb HBV genome can be released from the cloning vector by restriction enzyme digestion, gel purified, and re-circularized in vitro (Qin et al., 2011A). The drawback of this approach is the low DNA yield and need for extra steps of DNA manipulation prior to each transfection experiment. Another ingenious method is to use a specialized cloning vector which can be removed inside transfected cells through Cre/loxP-mediated DNA recombination, thus generating authentic cccDNA-like molecules inside cells (Qi et al., 2014). Alternatively, 1.1 copies of terminally redundant HBV DNA that covers the entire length of pg RNA can be inserted to an expression vector downstream of the strong CMV promoter to drive extremely high level of pg RNA transcription and genome replication (Junker et al., 1987). Such a property is useful for the screen of antiviral agents and generation of virus particles for infection experiments, but rather undesirable when comparing biological properties of HBV genetic variants. The minimum HBV genome for efficient replication under the endogenous regulatory elements is 1.3mer with the core promoter (which drives transcription of the 3.5-kb RNAs) located at the 5′ end (Guidotti et al., 1995; Hussain et al. 2009; Zhang et al., 2015). Longer versions such as 1.5mer (Garcia et al., 2009) and tandem dimer also work. Earlier investigators used tandem dimers to establish in vitro HBV DNA replication system (Sureau et al., 1986; Sells et al., 1987).
We have extensively used EcoRI dimers to characterize the functional significance of core promoter mutations in genotype A (Parekh et al., 2003). In addition, SphI dimers were used to compare the biological properties between genotypes B and C (Qin et al., 2011B), and to characterize the functional consequence of a 36-nucleotide insertion in genotype G (Li et al., 2007). To mutate a particular site in the HBV genome, we used a unique restriction site upstream of the mutation site and another unique site downstream of the mutation site to exchange a restriction fragment with PCR-derived mutant sequence. Such a fragment exchange would inevitably convert a tandem dimer into a monomer, which needs to be re-converted to tandem dimer for transfection experiments. Our traditional approach was to purify the 3.2-kb HBV DNA followed by its ligation with the pUC18 vector DNA at high insert-to-vector ratio, but still majority of the clones obtained are monomers (Tong et al., 1991). About 10–20 clones should be screened to identify a tandem dimer, and in some cases no dimer could be obtained. Such a failure might be attributable to poor quality of miniprep DNA (presence of inhibitors of ligation), or low concentration of the T4 DNA ligase. In the present study, we developed alternative methods to generate a tandem dimer through two-way molecular ligation (Fig. 1). In the approach of partial SphI digestion, the critical issue is to get enough linearized HBV monomer by titrating the concentration of SphI and/or duration of digestion. In the approach of ligation via the RsrII site, two rounds of gel purification are needed to obtain the RsrII-cut 3.2-kb HBV DNA. Thus not getting enough insert DNA was the main issue. From our experience, the approach of SphI partial digestion (approach B) gave more consistent results than the approach of RsrII digestion (approach C). So far most of our mutants were converted to SphI dimer by approach B, and the success rate was more than 50%. It is striking that all the dimers (and trimers) we obtained are tandem rather than head-to-tail or tail-to-head. With the approach of RsrII digestion, the success rate ranged from 100% (3/3 for two mutants) to less than 20% (1/6 for one mutant).
Two-way molecular ligation is much more efficient than three-way molecular ligation in converting a monomeric construct into a tandem dimer. The improved dimer formation rate should facilitate the functional characterization of HBV genetic variants through fragment exchange and site-directed mutagenesis. Certainly, due to the unique structural features of virion-associated relaxed circular DNA, our method does not significantly improve the efficiency for construction of replication competent HBV genomes directly from clinical samples.
Highlights.
HBV DNA replication can be initiated from cloned tandem dimers but not monomers.
Novel methods for converting a monomer into a tandem dimer were developed.
Both methods involve the ligation of two rather than three molecules.
About half of the bacterial colonies thus generated contained tandem dimers.
Acknowledgments
This work was supported by a grant from National Science Foundation of China (81371822), as well as NIH grants AI103648, AI107618, AI113394, and AI116639.
Abbreviations
- cccDNA
covalently closed circular DNA
- HBeAg
hepatitis B e antigen
- HBsAg
hepatitis B surface antigen
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- pg RNA
pregenomic RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Garcia T, Li J, Sureau C, Ito K, Qin Y, Wands J, Tong S. Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion. J Virol. 2009;83:11152–11165. doi: 10.1128/JVI.00905-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti L, Matzke B, Schaller H, Chisari F. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain Z, Jung HS, Ryu D, Ryu W. Genetic dissection of naturally ocurring basal core promoter mutations of hepatitis B virus reveals a silent phenotype in the overlapping X gene. J Gen Virol. 2009;90:2272–2281. doi: 10.1099/vir.0.010421-0. [DOI] [PubMed] [Google Scholar]
- Junker M, Galle P, Schaller H. Expression and replication of the hepatitis B virus genome under foreign promoter control. Nucleic Acids Res. 1987;15:10117–10132. doi: 10.1093/nar/15.24.10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Zoulim F, Pichoud C, Kwei K, Villet S, Wands J, Li J, Tong S. Critical role of the 36-nucleotide insertion in hepatitis B virus genotype G on core protein expression, genome replication, and virion secretion. J Virol. 2007;81:9202–9215. doi: 10.1128/JVI.00390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parekh S, Zoulim F, Ahn SH, Tsai A, Li JS, Kawai S, Khan N, Trepo C, Wands JR, Tong S. Genome replication, virion secretion, and e antigen expression of naturally occurring hepatitis B virus core promotermutants. J Virol. 2003;77:6601–6612. doi: 10.1128/JVI.77.12.6601-6612.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z, Li G, Hu H, Yang C, Zhang X, Leng Q, Xie Y, Yu D, Zhang X, Gao Y, Lan K, Deng Q. Recombinant covalently closed circular hepatitis B virus DNA induces prolonged viral persistence in immunocompetent mice. J Virol. 2014;88:8045–8056. doi: 10.1128/JVI.01024-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Zhang J, Garcia T, Ito K, Gutelius D, Li J, Wands J, Tong S. Improved method for rapid and efficient determination of genome replication and protein expression of clinical hepatitis B virus isolates. J Clin Microbiol. 2011A;49:1226–1233. doi: 10.1128/JCM.02340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Tang X, Garcia T, Hussain M, Zhang J, Lok A, Wands J, Li J, Tong S. Hepatitis B virus genotype C isolates with wild-type core promoter sequence replicate less efficiently than genotype B isolates but possess higher virion secretion capacity. J Virol. 2011B;85:10167–10177. doi: 10.1128/JVI.00819-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sells MA, Chen M, Acs G. Production of hepatitis B virus particles in HepG2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci USA. 1987;84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sureau C, Romet-Lemmone J, Mullins J, Essex M. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell. 1986;47:34–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- Tong S, Li JS, Vitvitski L, Benjelloun S, Trepo C. Rapid screening for bacterial colonies harbouring tandem hepatitis B virus sequences by an oligonucleotide probe. J Virol Methods. 1991;32:109–114. doi: 10.1016/0166-0934(91)90190-b. [DOI] [PubMed] [Google Scholar]
- Tong S, Li J, Wands J, Wen Y. Hepatitis B virus genetic variants: Biological properties and clinical implications. Emerging Microbes & Infections. 2013;2:e10. doi: 10.1038/emi2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Xia J, Sun B, Dai Y, Li X, Schalaak JF, Lu M. In vitro and in vivo replication of a chemically synthesized consensus genome of hepatitis B virus genotype B. J Virol Methods. 2015;213:57–64. doi: 10.1016/j.jviromet.2014.11.007. [DOI] [PubMed] [Google Scholar]