Abstract
The emergence of HIV-1 group M subtype B in North American men who have sex with men (MSM) was a key turning point in the HIV/AIDS pandemic. Phylogenetic studies have suggested cryptic subtype B circulation in the United States (US) throughout the 1970s2,3 and an even older presence in the Caribbean3. However, these timing and geographical inferences, based upon partial HIV-1 genomes that postdate the recognition of AIDS in 1981, remain contentious1,4 and the earliest movements of the virus within the US are unknown. We serologically screened >2000 1970s serum samples and developed a highly sensitive new approach for recovering viral RNA from degraded archival samples. Here, we report eight coding-complete genomes from US serum samples from 1978–79 – eight of the nine oldest HIV-1 group M genomes to date. This early, full-genome ‘snapshot’ reveals the US HIV-1 epidemic exhibited surprisingly extensive genetic diversity in the 1970s but also provides strong evidence of its emergence from a pre-existing Caribbean epidemic. Bayesian phylogenetic analyses estimate the jump to the US at ~1970 and place the ancestral US virus in New York City with 0.99 posterior probability support, strongly suggesting this was the crucial hub of early US HIV/AIDS diversification. Logistic growth coalescent models reveal epidemic doubling times of 0.86 and 1.12 years for the US and Caribbean, respectively, suggesting rapid early expansion in each location1. Comparisons with more recent data reveal many of these insights to be unattainable without archival, full-genome sequences. We also recovered the HIV-1 genome from the individual known as ‘Patient 0’5 and show there is neither biological nor historical evidence he was the primary case in the US or for subtype B as a whole. We discuss the genesis and persistence of this belief in the light of these evolutionary insights.
No comprehensive genomic analysis of the emergence and early spread of HIV-1 in North America – where HIV/AIDS was first recognized – has been possible because the only pre-1980 HIV-1 group M genome currently available (strain Z321B) was sampled in Africa. To fill this gap, we performed serological screening and viral genome sequencing of archived serum samples dating back to 1978–79, originally collected from MSM cohort patients in New York City (NYC) and San Francisco (SF). NYC samples were from volunteers in a prospective study of AIDS established in 19846, 378 of whom had been part of an earlier cohort of 8906 men involved in hepatitis B virus (HBV) studies beginning in 19787, and for which stored sera from 1978 and/or 1979 were available8. Previous work showed that 6.6% of these sera from NYC in 1978–79 were HIV-1 seropositive6; 33 of these positive samples were chosen for attempted HIV-1 sequencing. The sera from SF originated from a study of approximately 6875 patients enrolled in the late 1970s in HBV studies at the San Francisco City Clinic9. We tested 2231 of these samples from 1978 and found 83 (3.7%) to be Western blot-positive for HIV-1 antibodies; of these, 20 were randomly chosen for attempted HIV-1 sequencing.
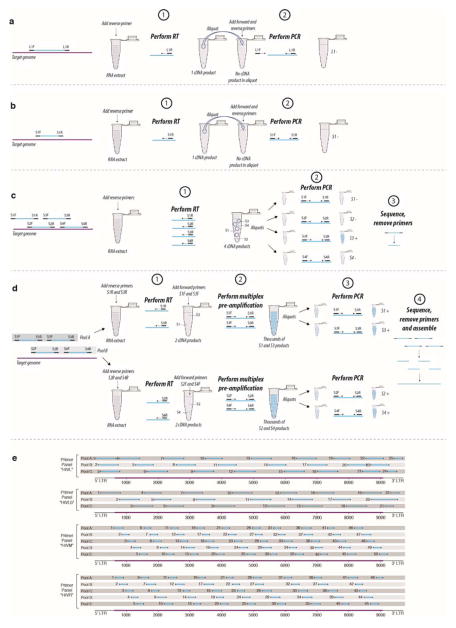
Low template number and degradation arising from long-term storage were major challenges for genomic analysis, as encountered previously with similar samples10: recovered RNA was generally below the limits of quantitation and initial attempts at amplification of reverse transcribed viral RNA failed consistently and indicated viral RNA survived in the 1970s samples only in short fragments. This led us to design an RNA ‘jackhammering’ approach to greatly increase both the ability to detect viral RNA-positive samples and to recover complete genomic HIV-1 sequences from them. Briefly, we use large panels of primers to amplify many short fragments in separate pools, such that amplicons overlap between but not within each pool (ED Fig. 1, Supplementary Table S1). Each pool’s amplicon set fills gaps between those of complementary pools, with the entire panel providing complete genomic coverage. A preliminary, multiplex amplification step, moreover, greatly concentrates target RNA prior to final amplification and sequencing.
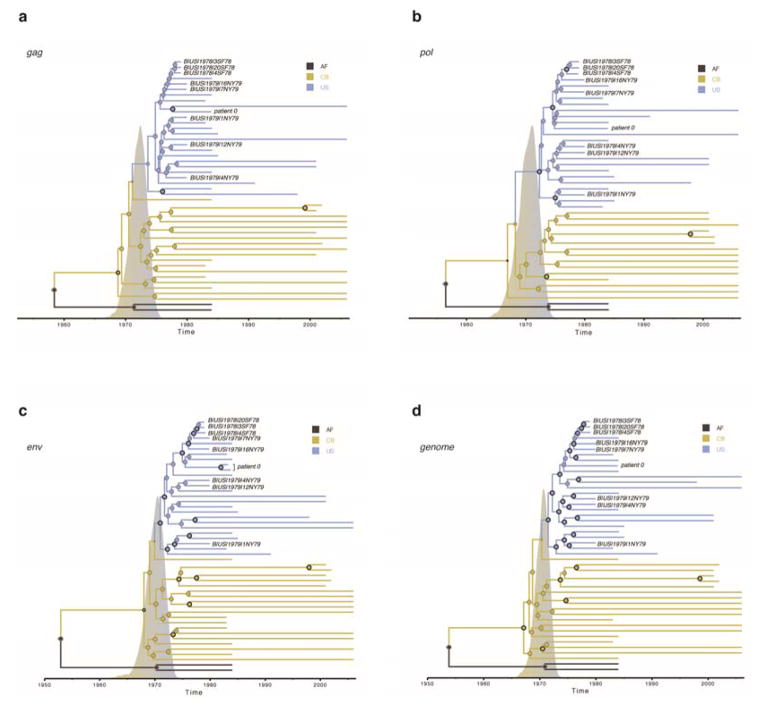
Three samples from SF and five from NYC provided sufficient data to assemble coding-complete sequences. Bayesian phylogenetic analyses of these HIV-1 genomes (Fig. 1, ED Fig. 2) showed that although they are the oldest sampled outside Africa they do not fall on the deepest branches even within subtype B. Instead, the 1970s genomes and the US epidemic as a whole are phylogenetically nested within the more genetically diverse, older subtype B epidemic in Caribbean countries. Separate analyses of gag, pol and env sequences also place the US sequences in a strongly supported monophyletic clade nested within the paraphyletic Caribbean subtype B sequences from Haiti, Dominican Republic, Jamaica, Trinidad and Tobago, and Haitian immigrants in the US (ED Figs. 3 and 4). Molecular clock phylogeographic analysis of the complete genome data supports a subtype B ancestor in the Caribbean (posterior probability > 0.99) dating to 1967 [95% credibility interval 1963–1970] (ED Table 1). This provides genome-wide evidence that the epidemic moved from the Caribbean to the US rather than from the US to the Caribbean3.
Figure 1. Maximum clade credibility (MCC) tree summary of the Bayesian spatio-temporal reconstruction based on complete HIV-1 genome data.
The tips of the tree correspond to year of sampling while branch and node colours reflect the sampling location for the tip branches and the inferred location for the internal branches (AF, Africa; CA, California; CB, Caribbean; GA, Georgia; NY, New York). Tip labels are provided for the newly obtained archival HIV-1 genomes. Diameters of internal node circles reflect posterior location probability values. Thick outer circles represent internal nodes with posterior probability support > 0.95. We also depict the posterior probability density for the time of the introduction event from the Caribbean into the US on the time scale of the tree. A fully annotated tree for this data set (‘full genome 38’, which includes only sequences sampled early in the US epidemic) is shown in ED Fig. 2b; ‘full genome 46’ which includes all available complete genomes basal to the “pandemic clade”3 of subtype B, plus a similar number and date range of US pandemic clade sequences, is shown in ED Fig. 2a. Separate analyses of gag, pol, env, and the coding-complete genomes (including also sequences sampled later in the US epidemic) provide consistent results (ED Figs. 3 and 4).
Location transition estimates recover a relatively precise date (1971 [1969–73], ED Table 1) for the HIV-1 jump from the Caribbean, very shortly before the US most recent common ancestor (MRCA). This narrow timing is aided by the basal relationship of a very close relative from the Caribbean (sequence “H6,” from an individual who entered the US from Haiti in 1981)3 (ED Fig. 2). The probability density of the date of introduction to the US overlaps with the deep branching structure in Caribbean diversity (Fig. 1, ED Fig. 3), indicating that the US clade emerged from the Caribbean epidemic during its early growth phase. We estimated a relatively fast logistic growth rate of 0.62 [0.26,0.99] yr−1 within the Caribbean population (Fig. 3). That of the US population is even higher, 0.81 [0.65,0.98] yr−1, in line with a precipitous spread among existing high-risk sexual networks. These mean growth rate estimates correspond to doubling times of 1.12 years and 0.86 years for the Caribbean and the US, respectively; both the more rapid and longer growth in the US appear to have contributed a higher number of ‘effective infections’ (Fig. 3), with the US overtaking the Caribbean by ~1977 despite the later HIV-1 emergence in the US.
Figure 3. Demographic reconstruction based on the nested coalescent model.
The colour scheme is consistent with that of the phylogeographic analyses in Figs. 1 and 2: the constant-logistic population size estimates (the ‘effective number of infections’, Ne, multiplied by the mean viral generation time, τ) through time are depicted in a black-yellow color range (following the African and Caribbean locations in the phylogeographic analyses) while the logistic population size estimates for the nested US clade are shown in blue (as for the US/NY location in the phylogeographic analyses).
Molecular clock analyses of larger numbers of env sequences revealed similar time of the most recent common ancestor (TMRCA) estimates for the key nodes (Fig. 2, ED Table 1, ED Fig. 5, ED Fig. 6). Interestingly, our modest snapshot of 1970s sequences from NYC and SF (Fig. 2, ED Fig. 5b) encompasses the full diversity exhibited by HIV-1 sequences from later years (i.e. it shares the same MRCA as larger sequence sets sampled in later years): all post-1985 sequences US sequences are nested within the early diversity captured by the limited number of 1970s sequences we recovered (ED Fig. 6)
Figure 2. The early patterns of HIV-1 subtype B spread in the Americas.
The map summarizes the main patterns of spread inferred from the molecular clock phylogeographic analyses. The map inset shows the initial introduction of the subtype B lineage into the Caribbean from Africa. From there, the virus first spreads to NY and subsequently to different locations in the United States. The tree depicts the US clade, plus the most closely related basal HT strain, as inferred from the ‘env 74’ analysis (ED Fig. 5b). Tips of the clade correspond to the year of sampling. Tip branch colours reflect the actual sampling locations as indicated on the map; interior branches depict phylogenetically inferred locations using the same colour scheme. Diameters of internal node circles reflect posterior location probability values. Thick outer circles indicate internal nodes with posterior probability support > 0.95. Thickness of the arrows reflects number of transitions inferred from this tree cluster. Mean dates and 95% credible intervals in yellow and blue represent the date estimates for the MRCA in the Caribbean and the US, respectively, based on the env 74 analysis. Date next to arrow between these locations represents the estimated timing of the corresponding jump. Patient 0 and the earliest sequences from San Francisco (1978) and New York City (1979) are labeled. Maps made with Natural Earth.
A phylogeographic reconstruction including only those US sequences sampled from known locations between 1978–1984 (Fig. 1) demonstrates that the NYC epidemic was already relatively mature and genetically diverse by 1979, tracing back to an MRCA estimated at 1972 [1970–74], and there is strong support the US subtype B ancestor circulated in NYC (posterior probability = 0.99). Indeed, the extensive genetic diversity in the US (and in NYC in particular) in 1978–1979 can only be explained by several years of circulation of the virus prior to 1978–79.
Using sequences sampled from NYC, North Carolina and California relatively late in the epidemic (comparable to the 1978–84 East coast, West coast and Southern sampling), we still infer a US ancestor in NYC, but with only modest support that prevents drawing firm conclusions (pp = 0.67, ED. Fig. 6b, ED Table 1). As a generality, early samples close to the deep branching structure are essential to confidently reconstruct the initial spatio-temporal expansion dynamics in exponentially growing populations.
Compared to NYC, the SF epidemic in 1978 appears to have been established more recently (Figs. 1 and 2, ED Figs. 2b and 5b). It is striking that all three independently-detected complete HIV-1 genomes we found are so closely related; moreover, they form a cluster with three partial env sequences sampled in SF during the same period10 (ED Fig. 5b). This suggests the bulk of the HIV-1 infections in SF in 1978 traced back to a single introduction from NYC in ~1976 (consistent with the lower HIV-1 seroprevalence in the SF cohort).
The sampled sequences thus reveal a series of key founder events in the genesis of subtype B (e.g. Fig. 2, ED Table 1), with the epidemic spreading from the African HIV-1 group M epicenter to the Caribbean by ~1967, from the Caribbean to NYC by ~1971, and from NYC to SF by ~1976 – quickly followed by extensive geographical mixing in the US and beyond.
Reports of one cluster of homosexual men with AIDS linked through sexual contact were important in suggesting the sexual transmission route of an infectious agent before the identification of HIV-15,11. Beginning in California, CDC investigators eventually connected 40 men in ten American cities to this sexual network. Investigators placed one man with Kaposi’s sarcoma (KS) near the center of a sociogram representing this cluster and identified him as ‘Patient 0’ – a ‘non-Californian AIDS patient’ and a possible ‘carrier’ of an infectious agent (ED Fig. 7). Before publication, Patient ‘O’ was the abbreviation used to indicate that this patient with KS resided ‘Out[side]-of-California.’ As investigators numbered the cluster cases by date of symptom onset, the letter ‘O’ was misinterpreted as the number ‘0,’ and the non-Californian AIDS patient entered the literature with that title12. Although the cluster study’s authors repeatedly maintained that Patient 0 was likely not the ‘source’ of AIDS for the cluster or the wider US epidemic, many people have subsequently employed the term ‘patient zero’ to denote an original or primary case, and many still believe the story today13. We therefore recovered the complete HIV-1 genome of Patient 0 and examined it against the backdrop provided by the 1970s sequences.
Though he was labeled as the cluster study’s ‘Index patient’, Patient 0 was neither the first AIDS case to come to CDC researchers’ attention, nor the first to display symptoms. In general, the CDC numbered cases in the order that the reports reached the agency from different cities and employed the terms ‘cases’ and ‘patients’ interchangeably. Patient 0, until he was linked to the cluster and took on his new name, was Case (or Patient) 057. The cluster study’s LA 6 was the CDC’s Case 032, and several cases in the New York section of the cluster5 (ED Fig. 7) were also reported before Patient 0 (and thus brought to investigators’ attention first): NY 3 was Case 001, NY 2 was Case 002, NY 6 was Case 010, and NY 5 was Case 05314.
The information available to CDC investigators to establish symptom onset dates was often fragmentary and thus resisted uniform categorization. Sometimes onset was determined on the basis of lymphadenopathy, other times by the appearance or diagnosis of Kaposi’s sarcoma or Pneumocystis carinii pneumonia. Investigators were unable to link to the cluster several NYC-based cases that had much earlier dates of symptom onset. For example, Case 154 was a middle-aged European man whose reported onset date for KS was January 1975, and Case 153, when he was diagnosed with KS in September 1981, recalled having swollen glands as early as June 197715. Yet even within the cluster, Case 057’s symptoms (lymphadenopathy in December 1979, and a KS lesion diagnosed in May 19805) appeared considerably later than those of several other cases. LA 1 (Case 335) developed a lesion in February 197816, while NY 1 (Case 152) experienced the onset of KS in December 1978, NY 2 (Case 002) in May 1979, and NY 3 (Case 001) in August 197914.
In his book And the Band Played On, Randy Shilts identified ‘Patient Zero’ by name as a highly sexually active French-Canadian flight attendant17. Unlike the initial reports of the cluster, media coverage of Shilts’s book strongly insinuated that this individual was the source of the North American epidemic and an exemplar of dangerous disease transmission18 – ideas which found a global audience (Supplementary Discussion). However, we find that the HIV-1 genome from this individual appears typical of US strains of the time and is not basal to the US diversity, let alone to the deeper Caribbean subtype B diversity, in a manner that might be suggestive of a special role (Figs. 1 and 2). In short, there is no evidence that Patient 0 was the first person infected by this lineage of HIV-1.
In addition to donating plasma for analysis, Patient 0 provided investigators with the names of nearly 10% of his sexual partners over several years5, while many other cluster patients were unable to share more than a handful of names16. This strongly suggests ascertainment bias contributed to his central role in the cluster study and its diagrammatic representation. Later research would also call into question the cluster study’s estimated average latency period of 10.5 months between sexual contact and symptom onset, with a revised average incubation period approaching 10 years for MSM. In retrospect, the study’s sociogram (ED. Fig. 7) almost certainly depicted the sexual contacts of these men years after they had contracted HIV-119 (Supplemental Discussion). Other East coast HIV-1 sequences fall much closer to the main early-California clade we identify than does that of Patient 0 (Fig. 2). Thus, while he did link AIDS cases in New York and Los Angeles through sexual contact, our results refute the widespread misinterpretation that he also infected them with HIV-1.
Much like historical reconstructions, phylogenetic inferences are often generated from data collected long after the critical events occurred. Our work highlights the importance of complete viral genomes from early archival specimens, carefully contextualized through historical analysis, without which this detailed picture of these early landmarks in the HIV/AIDS pandemic would not have been possible.
Methods
HIV-1 serological screening of serum samples from San Francisco from 1978
We tested 2231 samples collected from the cohort of gay and bisexual men in San Francisco in 19789 and detected 83 WB-positives (3.7% prevalence). Samples were first screened by GS HIV-1/HIV-2 Plus O EIA (Bio-Rad Laboratories, Redmond WA) and reactive samples were further tested by WB Genetic Systems HIV-1 Western Blot (Bio-Rad Laboratories, Redmond WA).
HIV-1 nucleic acid amplification
A total of 33 samples of frozen serum previously identified as positive for antibody to HIV-16–8 were assayed from New York City; a total of 20 frozen serum samples from San Francisco9, identified as part of the present study as positive for antibody to HIV-1, were assayed. The New York City samples were from 1978 and 1979 though no complete genomic sequences from 1978 were developed. The San Francisco samples were all from 1978. RNA recovered from samples from both NY and SF was generally undetectable when assaying 5ul aliquots in a Qubit 2.0 flourometer using the Qubit RNA HS reagents (detection limit, 250pg/ul).
Additionally, a sample of PMBC and a sample of serum were both assayed; these had been collected from a single individual in 1983 (Patient 0), and the samples were stored at CDC Atlanta. Other than Patient 0, now deceased, the data recorded were unlinked to individual identifiers and the work was approved by the Human Subjects Protection Program at the University of Arizona.
Four panels of degenerate primers (Supplementary Table S1, ED Figure 1) were designed using a suite of North American subtype B sequences. We aimed to design primers able to amplify both conserved regions and predictably variable sites. Primers within each panel were designed to generate sequence from the 5′ end of gag to the 3′ end of nef and were designed to amplify overlapping fragments. Two panels “HIVL” (N=25) and “HIVLb” (N=22) were designed to amplify fragments of ~500–650 bases in length. Two other panels “HIVM” (N=50) and “HIVR” (N=46) were designed to amplify fragments of ~200–320 bases in length.
Nucleic acids from 100ul aliquots of serum (or PMBCs in the case of Patient 0) were isolated using the QIAamp Viral RNA Mini Kit (Qiagen, Gaithersburg, MD) with 5mg added carrier RNA. Serum samples were then treated with DNase I (Invitrogen, Life Technologies, Carlsbad, CA) prior to reverse transcription. PMBC nucleic acids were left untreated.
Proviral DNA from Patient 0’s PMBCs was amplified with all four primer panels and from multiple separate isolations. Amplification was achieved using Invitrogen Platinum Taq DNA polymerase High Fidelity (Life Technologies, Carlsbad, CA) and run for 55 cycles at an annealing temperature of 52°C. Additionally, attempts were made to amplify longer fragments using PCR SuperMix High Fidelity (Life Technologies, Carlsbad, CA) and forward and reverse primers matched from the HIVLb primer panel for long fragment length followed by nesting with primers for slightly shorter fragment length. A single fragment of slightly more than 7000 bases was generated after multiple attempts with multiple primer combinations and cloned using the Invitrogen TOPO XL PCR Cloning Kit (Life Technologies, Carlsbad, CA). Fragments of individual clones were then amplified using HIVLb forward and reverse primers matched to give approximately 1000-base overlapping fragments and then sequenced.
RNA jackhammering
RNA jackhammering of the serum samples proceeded as follows: Aliquots of RNA extract were reverse transcribed using the GoScript Reverse Transcription System (Promega, Madison, WI) using a program of 4 cycles of 50°C for 30′ followed by 55°C for 30′ and an 85°C final incubation. Primers used were pools of reverse primers from widely spaced amplicons (Supplementary Table S1, ED Fig. 1), typically nine or ten primers per pool in a single reaction tube, with the wide spacing abrogating the possibility of incorporation of an internal primer into any given amplicon. RT products were then briefly amplified in multiplex reactions in the pool-specific tube (denaturation for 3′ at 94°C followed by 30 cycles of 94°C for 30”, 52°C for 30”, 68°C for 30”, and a final extension of 68°C for 5′) with matching forward primer pools (a “preliminary amplification” step). Sequences were then amplified from individual aliquots taken from the pool-specific tubes, via single primer pairs (denaturation for 3′ at 94°C followed by 40 cycles of 94°C for 30”, 52°C for 30”, 68°C for 30”, and a final extension of 68°C for 5′). Two separate isolates were amplified from each sample in this manner, with a minimum of one amplification with each primer panel per isolate. Five out of the 33 (15%) of the NY sera assayed yielded complete HIV-1 genomic data as did 3 out of the 20 (15%) SF sera, suggesting that levels of viral RNA preservation were very similar in each collection.
In ED Fig. 1 we schematically illustrate the RNA jackhammering approach and its advantages over standard RT-PCR procedures for degraded, low input samples. For a conventional RT-PCR approach with a fairly long amplification product we would perform RT and obtain one potentially amplifiable cDNA product. We would then aliquot ~10% of the RT product for amplification in a PCR reaction with forward and reverse primers. Even if the single cDNA product made it into the PCR reaction, the desired amplification product would be too long, and a PCR amplicon would therefore not be obtained. For RT-PCR with a shorter amplification product, more appropriately sized given the damaged RNA in the sample, there is still a 90% chance that it would be deemed a negative sample since most aliquots will not contain the rare cDNA product. Using multiple primer sets will increase the chance of a PCR-positive result, but most PCR reactions remain negative because most aliquots lack target cDNA. Even with a 10 primer-pair pool and 10 final PCR reactions, there may be no amplified product. The RNA jackhammering approach targets large panels of appropriately short amplicons, uses discrete pools of non-overlapping primers pairs for RT, and includes a crucial multiplex pre-amplification step to ensure that each aliquot contains ample template molecules for the final PCR amplifications (a separate reaction for each primer pair in the entire panel).
Sequencing was performed at the University of Arizona Genetics Core using an ABI 3730XL. The Patient 0 sample contained considerable heterogeneity (mixed bases) both in proviral assembly and in viral RNA amplifications. Heterogeneity in the NY and SF samples (all sequences derived from viral RNA) was low. In all cases consensus sequences were used in the phylogenetic analyses. Primer sequences were computationally removed from all sequence data prior to assembling genomic consensus sequences, which yielded coding-complete genomic data with exception of a few small gaps and the 3′ end of the nef gene (Supplementary Table S2).
Validation of the jackhammering approach
To validate this approach we obtained seed stock samples from the NIH AIDS Reagent program of subtype B viruses from the US (US657) and Haiti (HT599) and applied a jackhammering approach with independent runs of both the HIVM and HIVR primer panels (ED Fig. 8).
For US657 we recovered, in total, from both runs combined, 8194nt of high quality data. HIVM and HIVR are independent runs with completely different primer sets, yet where the data overlapped, they were >99.9% similar. Moreover, the few heterogeneities did not line up with heterogeneous primers but fell in regions between primers, demonstrating that differences could not be attributed to the incorporation of primers into the recovered sequences. This was expected both because the wide spacing of amplicons within a single pool of primer pairs prevents incorporation of primers within amplified products and because all primer sequences from final amplification products were computationally removed from the sequences prior to assembly of genomic sequences. There are 3354 bases in the published US657 sequence. Our data covered about 90% of the 3354 bases of previously published US657 sequence (GenBank accession number U04908) and all of our individual amplicons in the region of overlap had US657 as the highest BLAST hit and were >99% similar to the published sequence.
For HT599 the HIVM and HIVR primer panels developed 8545nt of data, 99.6% of the target. HIVM-derived sequence was >99.9% similar to HIVR-derived sequence. We recovered 100% of the overlap with the previously published HT599 sequence (2881nt, GenBank accession number U08447) with 99.5% similarity.
To evaluate discrepancies between the jackhammering-recovered sequences and both US657 and HT599, we compared consensus sequences of combined HIVM and HIVR data with the respective published sequences by adding them to our complete genome alignment and reconstructing a maximum likelihood tree (ED Fig. 8a). As expected, the independently generated sequences from each virus cluster very closely and only have short tips from their common ancestors, resulting from a very small number of substitutions in their overlapping regions. In a regression analysis (ED Fig. 8b), our sequences (with a target symbol) are associated with somewhat smaller residuals then the published sequences (with a circle), indicating our data are likely to be more accurate and, importantly, cannot contain primer remnants as this would result in much larger residuals.
Sequence data
To construct the data sets for the analyses in Fig. 1 and ED Figs. 2–4 we searched the Los Alamos National Laboratories (LANL) HIV database (http://hiv.lanl.gov/) for all available genome-length HIV-1 sequences from Caribbean countries, which had previously been shown to exhibit diverse subtype B lineages that fall basal to a monophyletic “pandemic” clade of subtype that accounts for most US and other non-Caribbean subtype infections3. These included sequences sampled in Haiti, Dominican Republic, Jamaica, and from Haitians who had recently immigrated to the US from Haiti (“H3” and “H5” from 1982, and “H6” and “H7” from 1983, “RF_HAT” from 1983)3. For sequences H3, H5, H6 and H7 pol sequences were not available, but partial gag and full length env sequences were available. For the full genome analyses the pol gene was treated as missing data. We then added a similar number of genomes from the US from a similar time period (1982–2005), plus one each from France and the U.K., as well as outgroup sequences of subtype D from the Democratic Republic of the Congo (D.R.C.). We called this the “full genome 46” data set because it contained 46 genomes. The gag, pol, and env data sets depicted in ED Fig. 3 were each derived from the respective sub-genomic region of this same set of taxa. The subset of “full genome 46” that contained only those US sequences sampled from 1978–84 we called “full genome 38”.
For the env analyses in Fig. 2 and ED Fig. 5 the alignment from ref. 3 was used, with the addition of the sequences generated for the present study, additional Caribbean subtype B sequences from 2000–2005, and four early subtype B partial env sequences from San Francisco10. This alignment we called “env 105”. The subset that contained only those US sequences sampled from 1978–84 we called “env 74”.
For ED Fig. 6 we added to “env 105” a comparable number – relative to those sampled from 1978–1984 from known locations (NY, CA, GA, PA, NJ) (ED Fig. 4b) – of randomly sampled sequences from 1997–2007 from NY, SF, and North Carolina (NC) (the closest available site with sufficient numbers to stand in for the Georgia ones from the 1978–84 sample). We called this alignment “env 133”.
In all cases sequences were manually aligned using Se-Al (http://tree.bio.ed.ac.uk/software/seal/). All sequence alignments, input files, tree files and primer sequences are available at the Dryad Digital Repository (doi:10.5061/dryad.7mv7v).
Recombination analysis and maximum likelihood tree reconstruction
Maximum likelihood (ML) phylogenies were reconstructed using RAXML under on a general time-reversible model of substitution with gamma distributed rate variation among sites18. Bootstrap support values were calculated using 1000 pseudo-replicates. To detect the presence of recombination, we first performed the Phi test19 on every data set (ED Table 1). When the null hypothesis of absence of recombination was rejected (P < 0.05), we subsequently analyzed the data set using RDP420 and produced new alignments in which the minor recombinant regions were deleted from putative recombinants. Re-analyses of these ‘recombination-free’ data sets using the Phi test confirmed the absence of detectable recombination signal (P > 0.05, ED Table 1).
Bayesian phylogenetic inference
Time-measured phylogeographic histories were reconstructed using a Bayesian phylogenetic inference approach implemented in BEASTv1.8.221. Our full probabilistic model combined sequence substitution over an unknown phylogeny calibrated in time units using a molecular clock process with dated tips22, a coalescent tree prior and a discrete diffusion process among discrete location states23. For the sequence substitution process, we used the same model as for the ML reconstructions. We accommodated rate variation among lineages using a lognormal distribution in an uncorrelated relaxed molecular clock model24 and integrated out each sampling date over an uncertainty interval of one year. Visual inspections of root-to-tip divergence as a function of sampling time using TempEst25 indicated strong temporal signal with no clear outlier sequences (ED Fig. 9).
For most analyses, we flexibly modeled changes in effective population size through time by specifying a Bayesian skygrid non-parametric tree prior with a grid of 50 years and yearly effective population size parameters26. (The notion of ‘effective population size’, or ‘effective infections’ in epidemiological applications, comes from population genetics, and is typically lower than the full (i.e. census) population size, reflecting for example variance in reproductive success among individuals – transmissions to new hosts in this context). To estimate viral population growth rates in both the Caribbean and US population, we fitted a ‘nested’ coalescent model to the data set with the largest taxon sampling (env 133). This model fits a constant-logistic demographic function27 to the genealogy excluding the US clade. The initial constant phase was included in the model to accommodate the deep branching between the subtype B sequences and the African subtype D outgroup sequences. Nested within this model, a separate logistic growth model was fitted to the US clade in the genealogy.
The process of discrete diffusion among locations was modeled using a general non-reversible substitution model28. In our analyses including the African subtype D outgroup lineages, we set the root state frequency to one for the African state and zero for all other possible discrete states. We obtained estimates of the transitions among locations (Markov jumps) using a stochastic mapping implementation capable of inferring the complete Markov jump history29,30. We approximate the posterior distribution for our full probabilistic model using Markov chain Monte Carlo (MCMC) sampling. We use BEAGLE in conjunction with BEAST to improve the computational performance of our analyses31. MCMC chains were run for 50,000,000 generations, sampling every 5,000 generations. We diagnosed the runs by examining trace plots and effective samples sizes, and summarized continuous parameters (mean and 95% highest posterior density [HPD] intervals) using Tracer (http://tree.bio.ed.ac.uk/software/tracer/) after discarding a 10% burn-in. Trees were summarized as maximum clade credibility trees using TreeAnnotator and visualized in FigTree (http://tree.bio.ed.ac.uk/software/figtree/).
In two specific phylogeographic analyses we assess i) to what extent sequences sampled early in the US epidemic characterize the subtype B diversity in the US clade (ED Fig. 6a) and ii) to what extent the location state at the origin of the US clade can be estimated using sequences sampled later in the epidemic from three different US states (ED Fig. 6b). For this purpose, we first reconstructed time-measured phylogenies for the env 133 data set using the substitution model, molecular clock model and coalescent model described above and subsequently reconstructed ancestral locations on the inferred posterior distribution of trees.
For ED Fig. 6a we classified US sequences as ‘early’ or ‘late’ depending on whether they were sampled before or after (and including) 1985. For ED Fig. 6b, we first pruned the necessary US sequences from the posterior distributions in order to retain only ‘late’ sequences from NY, NC and CA (matching the sampling from NY, GA and CA in Fig. 2 and ED Fig. 5b). In this case, the support for a NYC ancestral state is likely upheld by the presence of two basal NYC representatives, but location estimates in a star-like tree structure with long tip branches will be critically dependent on how well the diversity of any location is represented in the contemporaneous sampling, as recently noted32.
Comparison of phylogeographic estimates before and after deleting minor recombinant regions from putative recombinants (ED Table 1) indicated highly consistent results.
Extended Data
Extended Data Figure 1. Jackhammering schematic and primer panels and pools.
a through d: detection and amplification of target RNA molecules in old, degraded, low-titre samples. For the purposes of illustration, consider a tube with 1013 RNA molecules, but (because of the low RNA quality) only one molecule that is (i) capable of being primed by the given reverse primer(s) and (ii) long enough to form a 200bp product. a, conventional RT-PCR with a long amplification product, oversized for a sample with RNA less than ~200 bases in length. b, RT-PCR with a shorter amplification product. c, use of multiple primer pairs to increase the chance of a at least one PCR-positive result. d, the jackhammering approach, which overcomes the problems encountered in a through c by (i) targeting an extensive panel of short amplicons appropriately sized to the level of RNA survival in the sample, (ii) conducting RT with pools of primer pairs that amplify discrete, non-overlapping genomic regions, and (iii) employing a multiplex pre-amplification step, in the tube with the RT product, to generate sufficient DNA to ensure that each aliquot from it contains numerous template molecules for final PCR amplification. In this schematic, we show just two primer pairs per pool, but we used pools of ten pairs with our largest primer panels (shown in panel e, HXB2 numbering along HIV-1 genome). With a 10 primer-pair pool, and 10 final reactions, one can reliably recover 10 bands for sequencing. Five such pools (one entire panel of 50 pairs), allows for complete HIV-1 genome recovery even in heavily degraded samples.
Extended Data Figure 2. Maximum clade credibility (MCC) tree summaries of Bayesian spatio-temporal reconstructions based on complete HIV-1 genome data (a: “full genome 46”, b: “full genome 38”).
The tips of the trees correspond to the year of sampling while the branch (and node) colours reflect location: the sampling location for the tip branches and the inferred location for the internal branches (AF, Africa; CB, Caribbean; US, the United States; CA, California, GA, Georgia; NY, New York). The diameters of the internal node circles reflect posterior location probability values. Thick outer circles represent internal nodes with posterior probability support > 0.95. Grey bars indicate the 95% credibility intervals for the internal node ages. The tree in b) represents the fully annotated version of the tree in Fig. 1 in the main manuscript.
Extended Data Figure 3. Maximum clade credibility (MCC) tree summaries of Bayesian spatio-temporal reconstructions based on different genome region data sets.
MCC trees for the same strains are shown for a) gag, b) pol, c) env and d) the complete genome. The tips of the trees correspond to the year of sampling while the branch (and node) colours reflect location: the sampling location for the tip branches and the inferred location for the internal branches (AF, Africa; CB, Caribbean; US, the United States). Tip labels are provided for the newly obtained archival HIV-1 genomes. The diameters of the internal node circles reflect posterior location probability values. Thick outer circles represent internal nodes with posterior probability support > 0.95. We also depict the posterior probability densities for the time of the introduction event from the Caribbean into the U.S on the time scale of the trees.
Extended Data Figure 4. Maximum likelihood phylogenies for the different genome region data sets.
We analyzed the same data sets as in ED Fig. 3. The diameters of the internal node circles reflect bootstrap support values. We manually colored the branches in a similar way as for the Bayesian phylogeographic reconstructions.
Extended Data Figure 5. Maximum clade credibility (MCC) tree summaries of Bayesian spatio-temporal reconstructions based on different env data sets (a: “env 105”, b: “env 74”).
The tips of the trees correspond to the year of sampling while the branch (and node) colours reflect location: the sampling location for the tip branches and the inferred location for the internal branches (AF, Africa; CB, Caribbean; US, the United States, CA, California; GA, Georgia; NJ, New Jersey, NY, New York; PA, Pennsylvania). The diameters of the internal node circles reflect posterior location probability values. Thick outer circles represent internal nodes with posterior probability support > 0.95. We also depict the posterior probability density for the time of the introduction event from the Caribbean into the U.S on the time scales of the trees. The three partial env sequences from SF in 197810 are highlighted with bullets.
Extended Data Figure 6. Maximum clade credibility (MCC) tree summaries of Bayesian spatio-temporal reconstruction comparing early and late strains (a: “env 133”, b: only “late” sequences from “env 133”).
In a), we classified US sequences as ‘early’ or ‘late’ depending on whether they were sampled before or after (and including) 1985. In b), the analysis was conducted on an empirical tree distribution of “env 133” from which we pruned early US sequences (in grey), but we still annotate the reconstruction on the complete phylogenies for reference. The tips of the tree correspond to the year of sampling while the branch (and node) colours reflect location: the sampling location for the tip branches and the inferred location for the internal branches (AF, Africa; CB, Caribbean; US early, the United States sampled < 1985; US late, the United States sampled in or after 1985; CA, California; GA, Georgia; NC, North Carolina, NY, New York). The diameters of the internal node circles reflect posterior location probability values. Thick outer circles represent internal nodes with posterior probability support > 0.95.
Extended Data Figure 7. A cluster of 40 early AIDS patients linked through sexual contact.
(Reprinted from Figure 1 of reference 5 with permission from Elsevier).
Extended Data Figure 8. Jackhammering validation with reference viruses.
a, The consensus sequences for primer panels HIVM and HIVR (‘RMcon’ suffix) were included, with previously published sequences for a US (US657) virus and a Haitian (HT599) virus, in a maximum likelihood tree. The two clusters of paired sequences are highlighted by coloured boxes. b, Plot of the root-to-tip genetic distance against sampling time for the tree in a). The colors for the data points are consistent with those used for sampling locations in the phylogenies (the two African outgroup tips are not shown for clarity). The data points with black circles represent the published sequences while the data points with a target symbol represent the newly obtained sequences.
Extended Data Figure 9. Plots of the root-to-tip genetic distance against sampling time for different genome region data sets (gag, pol, env and complete genome).
We used TempEst25 to obtain exploratory regressions based on the maximum likelihood trees (ED Fig. 4). Each data point represents a tip; colors are consistent with those used for sampling locations in the phylogenies. The US data points with black circles represent the new genomes dating back to 1978–1979. The data point with the target symbol represents the Patient 0 genome. In each plot, we provide the R2 for the regression and the slope, reflecting the evolutionary rate (in substitutions per site per year).
ED Table 1.
Molecular clock, phylogeographic and recombination estimates for the different data sets
| Data set | TMRCA (subtype B & D) | TMRCA (subtype B) | Location probability (subtype B) | Jump time (CB to US) | TMRCA (US subtype B) | Location probability (US subtype B) | Evolutionary rate | Rate, coefficient of variation | Phi test p-value |
|---|---|---|---|---|---|---|---|---|---|
| “full genome 46”, ED Fig. 2 & ED Fig. 3 | 1953 (1946, 1961) | 1967 (1963, 1970) | CB: > 0.99 | 1970 (1968, 1973) | 1972 (1969, 1973) | US: > 0.99 | 0.0027 (0.0024, 0.0030) | 0.25 (0.20, 0.31) | 0.99 |
| “full genome 38”, Fig. 1 & ED Fig. 2 | 1955 (1946, 1962) | 1967 (1963, 1970) | CB: 0.99 | 1971 (1968. 1973) | 1972 (1970, 1974) | NY: > 0.99 | 0.0024 (0.0021, 0.0027) | 0.26 (0.20, 0.32) | 0.99 |
| “gag”, ED Fig. 3 | 1958 (1950, 1964) | 1969 (1964, 1972) | CB: > 0.99 | 1972 (1969, 1974) | 1974 (1971, 1975) | US: > 0.99 | 0.0023 (0.0020, 0.0026) | 0.23 (0.14, 0.33) | 0.77 |
| “pot”, ED Fig. 3 | 1956 (1947, 1965) | 1967 (1961, 1972) | CB: 0.92 | 1970 (1966, 1973) | 1973 (1969, 1974) | US: > 0.99 | 0.0015 (0.0013, 0.0017) | 0.29 (0.20, 0.37) | 0.21 |
| “env”, ED Fig. 3 | 1953 (1943, 1962) | 1968 (1964, 1972) | CB: > 0.99 | 1970 (1966, 1973) | 1971 (1968, 1974) | US: 0.99 | 0.0037 (0.0032, 0.0043) | 0.25 (0.16, 0.34) | <0.01 |
| “env, recomb. free”* | 1952 (1940, 1961) | 1968 (1964, 1972) | CB: 0.99 | 1970 (1966, 1973) | 1971 (1967, 1973) | US: 0.99 | 0.0039 (0.0031, 0.0047) | 0.26 (0.18, 0.35) | 0.59 |
| “env 105”, ED Fig. 5 | 1954 (1947, 1961) | 1968 (1964, 1971) | CB: > 0.99 | 1970 (1968, 1972) | 1971 (1969, 1973) | US: > 0.99 | 0.0047 (0.0042,0.0052) | 0.23 (0.18,0.28) | 0.01 |
| “env 105, recomb. free”* | 1955 (1947, 1961) | 1968 (1974, 1970) | CB: > 0.99 | 1970 (1968.1972) | 1971 (1969, 1972) | US: > 0.99 | 0.0047 (0.0041,0.0053) | 0.23 (0.18, 0.28) | 0.26 |
| “env 74”, ED Fig. 5 | 1957 (1948, 1963) | 1969 (1963, 1971) | CB: > 0.99 | 1971 (1969, 1973) | 1972 (1969, 1974) | NY: 0.97 | 0.0044 (0.0038, 0.0050) | 0.28 (0.21, 0.36) | <0.01 |
| “env 74, recomb. free”* | 1957 (1948, 1964) | 1969 (1964, 1972) | CB: 0.99 | 1971 (1968, 1973) | 1972 (1970, 1974) | NY: 0.97 | 0.0046 (0.0038, 0.0054) | 0.31 (0.23, 0.39) | 0.91 |
| “env 133”, ED Fig. 6† | 1952 (1944, 1958) | 1966 (1963, 1969) | CB: 0.99 | 1969 (1966, 1971) | 1969 (1967, 1971) | NY: 0.67 | 0.0045 (0.0041, 0.0048) | 0.20 (0.16,0.23) | 0.76 |
The recombination free (“recomb. free”) data sets were obtained by deleting the minor recombinant regions from the putative recombinants identified using RDP4.
the empirical trees from the “env 133” analysis were used for two different ancestral reconstructions (ED Fig. 6); here we list the location estimates for the analysis that considered different US states for the late samples (ED Fig. 6b).
Supplementary Material
Acknowledgments
We thank Cladd Stevens and Dollene Hemmerlein for facilitating access to archival sera; Guan-Zhu Han, Adam Bjork, William Switzer, Vickie Sullivan, Ryan Ruboyianes and Patrick Sprinkle for technical assistance; Thomas Spira and Michele Owen for geographical data on some published sequences; and the NIH AIDS Reagent program for providing reference virus samples US657 and HT599. William W. Darrow led the initial 1982 cluster investigation and provided R.A.M. access to his copies of archival CDC documents. This work was supported by NIH/NIAID R01AI084691 and the David and Lucile Packard Foundation (M.W.); the Wellcome Trust (080651), the University of Oxford’s Clarendon Fund, the Economic and Social Research Council (PTA-026-27-2838), and a J. Armand Bombardier Internationalist Fellowship (R.A.M.); the Research Fund KU Leuven (Onderzoeksfonds KU Leuven, Program Financing no. PF/10/018) and the ‘Fonds voor Wetenschappelijk Onderzoek Vlaanderen’ (FWO) (G066215N) (P.L); and NSF DMS 1264153, NIH R01 HG006139 and NIH R01 AI107034 (M.A.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions:
M.W., H.W.J., P.L. and R.A.M. conceived the study. T.D.W and M.W. designed the RNA jackhammering method. T.D.W. generated the sequences. B.A.K provided serum samples from New York City. W.H and T.G. acquired specimens and provided serological data. D.E.T. provided conceptual input. M.W., M.A.S. and P.L. prepared the data sets and performed the phylogenetic analyses. R.A.M. performed the historical analyses. M.W., H.W.J., P.L. and R.A.M. wrote the paper. All authors discussed the results and commented on the manuscript. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The HIV-1 sequences reported here have been deposited in GenBank under accession numbers KJ704787-KJ704797.
Competing Financial Interests Statement
The authors declare no competing financial interests. A patent, “Methods and systems for RNA or DNA detection and sequencing” (U.S. patent application 62/325,320), has been filed with the U.S. Patent Office. It will be used to facilitate the nonexclusive licensing of this methodology.
References
- 1.Holmes EC. When HIV spread afar. Proc Natl Acad Sci USA. 2007;104:18351–18352. doi: 10.1073/pnas.0709179104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korber BT, et al. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;9:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- 3.Gilbert MT, et al. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci USA. 2007;104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pape JW, et al. The epidemiology of AIDS in Haiti refutes the claims of Gilbert et al. Proc Natl Acad Sci USA. 2008;105:E13. doi: 10.1073/pnas.0711141105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auerbach DM, Darrow WW, Jaffe HW, Curran JW. Cluster of cases of the acquired immune deficiency syndrome: patients linked by sexual contact. Am J Med. 1984;76:487–492. doi: 10.1016/0002-9343(84)90668-5. [DOI] [PubMed] [Google Scholar]
- 6.Stevens CE, et al. Human T-cell lymphotropic virus type III infection in a cohort of homosexual men in New York City. JAMA. 1986;255:2167–2172. [PubMed] [Google Scholar]
- 7.Szmuness A, et al. A controlled clinical trial of the efficacy of the hepatitis B vaccine (Heptavax B): A final report. Hepatology. 1981;1:377–385. doi: 10.1002/hep.1840010502. [DOI] [PubMed] [Google Scholar]
- 8.Koblin BA, et al. Mortality trends in a cohort of homosexual men in New York City, 1978–1988. Am J Epidemiology. 1992;136:646–656. doi: 10.1093/oxfordjournals.aje.a116544. [DOI] [PubMed] [Google Scholar]
- 9.Jaffe HW, et al. The acquired immunodeficiency syndrome in a cohort of homosexual men: a six-year follow-up study. Ann Intern Med. 1985;103:210–214. doi: 10.7326/0003-4819-103-2-210. [DOI] [PubMed] [Google Scholar]
- 10.Foley B, Pan H, Buchbinder S, Delwart EL. Apparent founder effect during the early years of the San Francisco HIV type 1 epidemic (1978–1979) AIDS Res Hum Retrov. 2000;16:1463–1469. doi: 10.1089/088922200750005985. [DOI] [PubMed] [Google Scholar]
- 11.Task Force on Kaposi’s Sarcoma and Opportunistic Infections, CDC. A cluster of Kaposi’s sarcoma and Pneumocystis carinii pneumonia among homosexual male residents of Los Angeles Orange Counties California. Morb Mort Wkly Rep. 1982;31:305–307. [PubMed] [Google Scholar]
- 12.McKay RA. Doctoral thesis. Univ. of Oxford; 2011. Imagining ‘Patient Zero’: Sexuality, Blame, and the Origins of the North American AIDS Epidemic. [Google Scholar]
- 13.Harden VA. AIDS at 30: A History. Potomac Books; Washington, D.C: 2012. pp. 159–184. [Google Scholar]
- 14.Darrow WW. Trip report to New York City, July 12–16 and August 3–6, 1982. CDC Task Force on AIDS, internal communication. Sep 3, 1982.
- 15.Darrow WW. Time-space clustering of KS cases in the City of New York: evidence for horizontal transmission of some mysterious microbe. CDC Task Force on Kaposi’s Sarcoma and Opportunistic Infections, internal communication. Mar 3, 1982.
- 16.Darrow WW, Auerbach DM. Los Angeles cluster: background. CDC Task Force on Kaposi’s Sarcoma and Opportunistic Infections, internal communication. May 12, 1982.
- 17.Shilts R. And the Band Played On: Politics, People, and the AIDS Epidemic. St. Martin’s Press; New York: 1987. [Google Scholar]
- 18.McKay RA. ‘Patient Zero’: the absence of a patient’s view of the early North American AIDS epidemic. Bull Hist Med. 2014;88:161–194. doi: 10.1353/bhm.2014.0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss AR. In response to: AIDS without end. N Y Rev Books. 1988 Dec 8;35(60) [Google Scholar]
- 20.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bruen TC, Philippe H, Bryant D. A simple and robust statistical test for detecting the presence of recombination. Genetics. 2006;172:2665–2681. doi: 10.1534/genetics.105.048975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution. 2016;1:vev003. doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology and Evolution. 2012;29:1969. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rambaut A. Estimating the rate of molecular evolution: incorporating non-contemporaneous sequences into maximum likelihood phylogenies. Bioinformatics. 2000;16:395. doi: 10.1093/bioinformatics/16.4.395. [DOI] [PubMed] [Google Scholar]
- 25.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLoS Computational Biology. 2009;5:e1000520. doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rambaut A, Lam TT, de Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst. Virus Evolution. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gill MS, et al. Improving Bayesian population dynamics inference: a coalescent-based model for multiple loci. Molecular Biology and Evolution. 2013;30:713. doi: 10.1093/molbev/mss265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faria NR, et al. HIV epidemiology. The early spread and epidemic ignition of HIV-1 in human populations. Science. 2014;346:56. doi: 10.1126/science.1256739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edwards CJ, et al. Ancient hybridization and an Irish origin for the modern polar bear matriline. Current Biology. 2011;21:1251. doi: 10.1016/j.cub.2011.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minin VN, Suchard MA. Counting labeled transitions in continuous-time Markov models of evolution. Journal of Mathematical Biology. 2007;56:391. doi: 10.1007/s00285-007-0120-8. [DOI] [PubMed] [Google Scholar]
- 32.Lemey P, et al. Unifying Viral Genetics and Human Transportation Data to Predict the Global Transmission Dynamics of Human Influenza H3N2. PLoS Pathogens. 2014;10:e1003932. doi: 10.1371/journal.ppat.1003932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suchard MA, Rambaut A. Many-core algorithms for statistical phylogenetics. Bioinformatics. 2009;25:1370. doi: 10.1093/bioinformatics/btp244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Graf T, et al. Contribution of Epidemiological Predictors in Unraveling the Phylogeographic History of HIV-1 Subtype C in Brazil. J Virol. 2015;89:12341–12348. doi: 10.1128/JVI.01681-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.