Abstract
Hailey-Hailey disease (HHD) is an autosomal dominant trait characterized by erythematous and oozing skin lesions preponderantly involving the body folds. In the present unusual case, however, unilateral segmental areas along the lines of Blaschko showing a rather severe involvement were superimposed on the ordinary symmetrical phenotype. Based on this observation and similar forms of mosaicism as reported in other autosomal dominant skin disorders, we postulated that in such cases, 2 different types of segmental involvement can be distinguished. Accordingly, the linear lesions as noted in the present case would exemplify type 2 segmental HHD. In the heterozygous embryo, loss of heterozygosity occurring at an early developmental stage would have given rise to pronounced linear lesions reflecting homozygosity or hemizygosity for the mutation. By analyzing DNA and RNA derived from blood and skin samples as well as keratinocytes of the index patient with various molecular techniques including RT-PCR, real-time PCR, and microsatellite analysis, we found a consistent loss of the paternal wild-type allele in more severely affected segmental skin regions, confirming this hypothesis for the first time, to our knowledge, at the molecular and cellular level.
Introduction
Hailey-Hailey disease (HHD) (OMIM 169600), also known as familial benign chronic pemphigus, is a rare cutaneous disorder inherited as an autosomal dominant trait (1). It is caused by germline mutations in the ATP2C1 gene (OMIM 604384) on chromosome 3q21–q24, encoding an adenosine triphosphate–powered (ATP-powered) calcium channel pump (2, 3).
The disorder usually becomes manifest in the third or fourth decade of life with erythema, vesicles, and erosions involving the body folds, particularly the groin and axillary regions. Other sites of the body, such as the neck, perianal, and submammary regions, may likewise be affected. The distribution of cutaneous lesions is usually rather symmetrical (1), which is consistent with a general rule that autosomal dominant skin disorders show a generalized and bilateral involvement. However, several recent reports indicate that exceptions to this rule exist. On rare occasions, the disorder may show a band-like or otherwise segmental arrangement reflecting mosaicism (4, 5).
According to a new genetic concept we postulated, 2 different types of segmental manifestation can be distinguished in autosomal dominant skin diseases (Figure 1) (4, 5). Type 1 reflects heterozygosity for a de novo postzygotic mutation occurring at an early stage of embryogenesis. The cutaneous lesions within the affected segments show a degree of severity similar to that of the nonsegmental phenotype caused by a germline mutation. Outside the segmental areas, the skin is both clinically and genetically normal (4, 5).
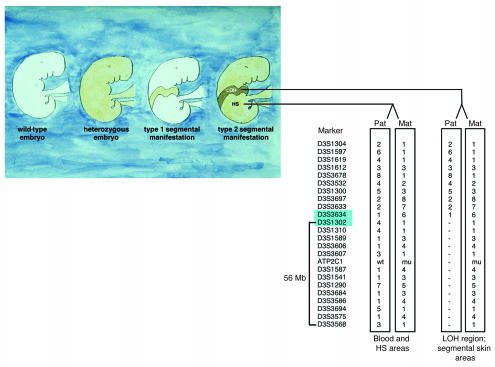
Figure 1.
Origin of 2 types of segmental manifestation in autosomal dominant skin disorders. Left to right: healthy phenotype (2 wild-type alleles); common diffuse manifestation (heterozygous germline mutation); type 1 segmental manifestation, reflecting heterozygosity for a postzygotic somatic mutation; type 2 segmental manifestation, reflecting the result of a germline mutation in combination with somatic LOH. Comparative haplotype analysis in DNA samples obtained from peripheral blood, heterozygous skin (HS) areas, and keratinocytes from segmentally affected skin regions of the index patient revealing consistent loss of one allele distal of marker D3S1302 to marker D3S3568, a chromosomal region of 56 Mb harboring the ATP2C1 gene. Of note, it is the paternal (pat) wild-type allele that is consistently lost in more severely affected segmental skin areas. Loss of the paternal wild-type allele occurred between markers D3S3634 and D3S1302 (markers shaded in blue). Mat, maternal.
By contrast, the type 2 manifestation tends to occur in heterozygous embryos which later develop a nonsegmental, diffuse distribution of skin lesions. A postzygotic mutation occurring at an early developmental stage would result in loss of heterozygosity (LOH) and give rise, in a segmental area, to a homozygous or hemizygous state of the underlying mutation. Clinically, this would be reflected by rather pronounced segmental lesions being superimposed on the ordinary nonsegmental phenotype (4, 5). The concept of type 2 segmental involvement, however, has so far not been proven at the cellular and molecular level.
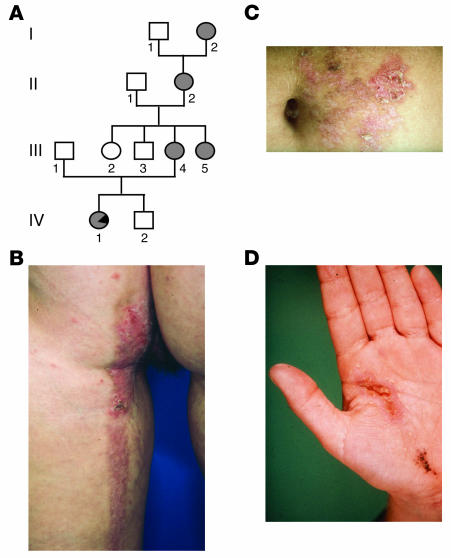
Recently, a case of type 2 segmental HHD in a member of a 4-generation family of German origin was reported (individual IV-1 in Figure 2A) (6). Remarkably, the unilateral linear lesions first appeared at the age of 3 months and persisted into adulthood with frequent exacerbations (Figure 2, B–D) (7), whereas a nonsegmental involvement of body folds was first noted at the age of 24 years (6).
Figure 2.
Pedigree and clinical manifestations. (A) Pedigree of the nuclear family studied with regard to the occurrence of Hailey-Hailey disease. Affected individuals are indicated by filled symbols. Note that in individual IV-1 the segmental type 2 manifestation is indicated by a filled symbol with a black triangle. (B–D) Clinical manifestation in individual IV-1 with more severely affected skin regions showing a unilateral segmental pattern on (B) the back of the left leg and the left buttock, (C) the left abdomen, and (D) the left hand.
Because previous therapeutic attempts, including dermabrasion, had been unsuccessful, 3 linear segments were now excised completely (6). This enabled us to examine skin samples obtained from segmental and nonsegmental areas. In addition to histopathological evaluation of paraffin-embedded tissue, several of the excised skin samples, corresponding to areas of either segmental or nonsegmental involvement, were subjected to cellular and molecular analysis. The epidermis was separated from the dermis, and keratinocyte cultures were initiated as previously described in detail (8).
Using different molecular techniques, including RT-PCR, real-time PCR, heteroduplex analysis, automated sequencing, laser-assisted microdissection, and haplotype analysis, we were able to demonstrate LOH in the segmental area of severely affected skin.
This study provides the first molecular evidence to our knowledge in favor of the recently postulated concept that in autosomal dominant skin disorders, dichotomous forms of segmental manifestation can be distinguished (4, 5). In the present case, postzygotic LOH was found to result in a type 2 segmental manifestation of HHD.
Results
Mutation analysis and verification.
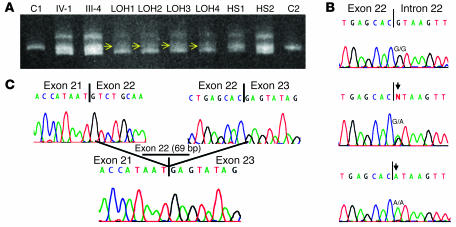
Initial mutation screening of leukocyte-derived DNA by heteroduplex analysis using conformation-sensitive gel electrophoresis (CSGE) revealed a complex heteroduplex formation in the PCR fragment containing exon 22 of the ATP2C1 gene (Figure 3A). This heteroduplex formation was not observed in 300 chromosomes from unrelated Caucasian individuals studied for control purposes, indicating the presence of a mutation (data not shown).
Figure 3.
Mutation analysis studies. (A) Heteroduplex analysis of PCR products containing exon 22 of the ATP2C1 gene performed on DNA samples derived from leukocyte DNA of the index patient (IV-1) and her mother (III-4) and DNA derived from keratinocytes originating from regions with either LOH (LOH1–LOH4) or the diffuse phenotype (HS1 and HS2), as well as DNA from 2 controls (C1 and C2). Note the complex heteroduplex in individuals IV-1 and III-4, indicative of a mutation and the loss of the homoduplex wild-type band (middle band) observed in LOH regions (LOH1–LOH4) when compared to heterozygous skin regions (HS1 and HS2), indicated by yellow arrows. (B) Splice site mutation 2146+1G→A identified in the germline in all affected individuals, consisting of a G-to-A transition, indicated by an arrow (middle panel). Of note, in regions with LOH, we observed only the mutant A signal (arrow, lower panel) and if there was any wild-type G-signal, it was low. (C) Consequence of mutation 2146+1G→A on the cDNA level, leading to skipping of exon 22 (69 bp).
Direct sequencing of newly synthesized PCR fragments containing exon 22 showed a heterozygous guanine-to-adenine substitution at the first base of the consensus splice donor site of exon 22, resulting in a splice site mutation designated 2146+1G→A (Figure 3B, middle lane).
Consequence of mutation 2146+1G→A.
The effect of mutation 2146+1G→A was studied on the cDNA level by RT-PCR. Subcloning of the PCR product and subsequent automated sequencing analysis of 20 clones revealed that the splice site mutation resulted in the skipping of exon 22, which consists of 69 bp (Figure 3C).
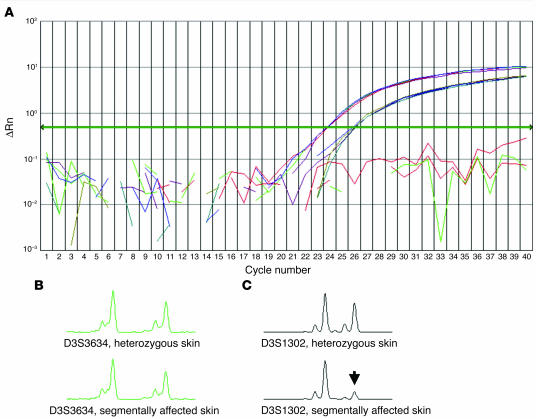
Quantification of wild-type and mutant ATP2C1 cDNA content within heterozygous skin areas and in heterozygous keratinocytes, as determined with quantitative real-time RT-PCR, revealed that the ratio of wild-type to mutant cDNA was approximately 3.8:1 (Figure 4A).
Figure 4.
Genetic consequences in nonsegmental and segmental skin areas. (A) Analysis of the relative amount of wild-type (blue curve) and mutant (red curve) ATP2C1 gene copies in a nonsegmental skin area by allele-specific quantitation using a real-time PCR TaqMan assay. The measured ratio of 3.8 ± 0.2 indicates that the splice site mutation most likely leads to nonsense-mediated mRNA decay. Results shown as a background-adjusted quantification of the amplified PCR product (ΔRn). (B, C) PCR fragment length scans analyzed with GeneScan software, comparing segmentally involved and clinically unaffected (nonsegmental) skin regions. (B) Marker D3S3634, nonsegmental skin area (top) and segmental skin area (bottom). (C) Marker D3S1302 nonsegmental skin area (top) and segmental skin area (bottom). This haplotype analysis demonstrated loss of the paternal allele for marker D3S1302 (arrow).
LOH in segmental skin areas.
Using DNA extracted from either excised skin samples or cultured keratinocytes derived from segmental or nonsegmental areas of disease involvement, we again amplified the region of the APT2C1 gene harboring exon 22 and its adjacent splice sites. After subjecting these PCR products to CSGE analysis, we observed a decrease in signal intensity of the wild-type homoduplex band in keratinocytes originating from segmentally affected areas, indicative of LOH (Figure 3A, yellow arrows).
Direct sequencing of newly synthesized PCR fragments revealed only the mutant A allele at the first base of the splice donor site of exon 22 in all skin and keratinocyte samples that were obtained from segmentally affected skin regions, likewise indicating LOH (Figure 3B, bottom lane).
Laser-assisted microdissection.
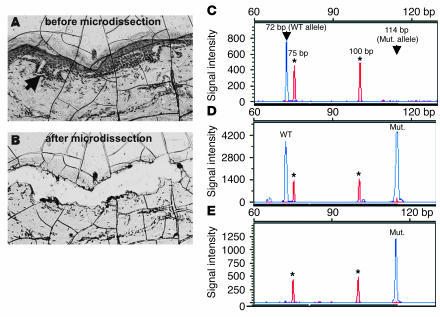
Examination of laser-micro-dissected epidermis from segmental skin areas (Figure 5, A–E) revealed, within epidermal blisters, a population of keratinocytes only harboring the mutant A allele, thus again supporting LOH (Figure 5E).
Figure 5.
Microdissection analysis in a segmental skin area. (A) Liquid coverslip–embedded tissue section displaying intraepidermal blister formation, indicated by an arrow. (B) The same area after laser-assisted microdissection. (C–E) Electropherograms showing restriction fragment length analysis of PCR-amplified DNA encompassing the splice donor site of exon 22 that was cleaved with BsaAI. Asterisks indicate the positions of the 75-bp and 100-bp peaks of the molecular size marker GeneScan-350. y-axis values measured in arbitrary fluorescence intensity units. (C) Control DNA only reveals the 72-bp dye-labeled wild-type fragment. (D) DNA obtained from the patient’s blood displays the 72-bp wild-type fragment and the 114-bp mutant (mut.) fragment. (E) DNA obtained from microdissected tissue only shows the mutant product of 114 bp, which is indicative of LOH.
Haplotype analysis.
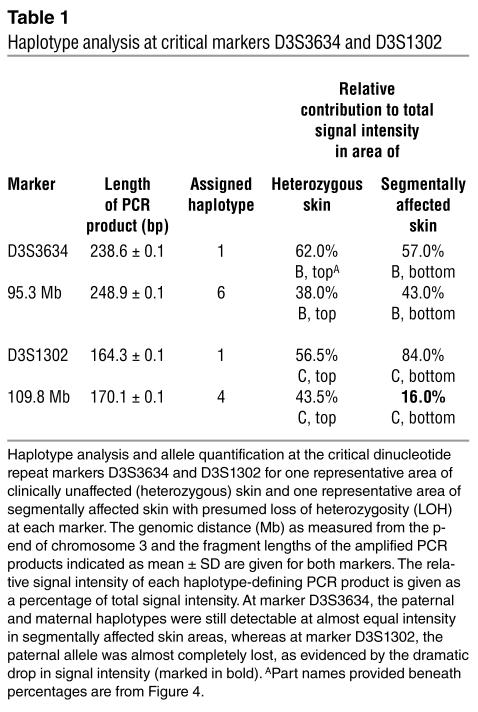
Comparative haplotype analysis in DNA samples obtained from peripheral blood leukocytes, heterozygous keratinocytes, and keratinocytes from segmentally affected skin regions of the index patient revealed consistent loss of one allele, from marker D3S1302 extending distally to marker D3S3568, a chromosomal region also harboring the ATP2C1 gene (Figure 1). Analysis of the maternal and paternal haplotype at these markers indicated that it is the paternal wild-type allele that is consistently lost in more severely affected skin areas.
Furthermore, haplotype analysis revealed that loss of the paternal wild-type allele occurred between markers D3S3634 and D3S1302 when comparing clinically unaffected skin regions with those showing a segmental involvement (Figure 4, B and C).
At marker D3S3634, 2 signals of almost identical intensity were detected in both clinically unaffected (Figure 4B, top panel) and segmentally involved (Figure 4B, bottom panel) skin, whereas at marker D3S1302, there was a marked decrease in signal intensity of the paternal allele in segmentally affected skin (Figure 4C, arrow, and Table 1). In general, a decrease in signal intensity of less than 20% was assumed to reflect loss of the corresponding allele (Table 1, marked in bold).
Table 1.
Haplotype analysis at critical markers D3S3634 and D3S1302
Gene dosage measurements in regions with LOH.
Allele-specific quantification of ATP2C1 gene copies in DNA samples obtained from leukocytes as well as keratinocytes from clinically unaffected and segmentally involved skin areas with presumed LOH revealed an approximately equal number of ATP2C1 and factor VIII (FVIII; used as reference) gene copies (Table 2), indicating that in each sample, 2 copies of the ATP2C1 gene per diploid set of chromosomes were present.
Table 2.
Amount of ATP2C1 gene copies in different tissues
Discussion
It has long been known that autosomal dominant skin disorders, which usually present a diffuse and symmetrical involvement, may also show a segmental arrangement that most likely reflects mosaicism. Evidence supporting this concept was first provided in 1983, when cytogenetic studies revealed a trisomy 18 mosaicism in a case of linear hyperpigmentation (14).
Initially, it was thought that this segmental manifestation always resulted from a somatic mutation (15) and, even today, some authors still adhere to this erroneous concept. Later it was reported that patients with a segmental manifestation of an autosomal dominant skin disease had an increased risk of transmitting the trait in the ordinary, nonsegmental form to their children (16, 17). Such observations implied that the underlying mutation could also affect the germline in the form of gonadal mosaicism. Thus, these mutational events could no longer be attributed to somatic mutations; instead, they seem to result from postzygotic mutations that may or may not affect the germline (18–20).
However, further clinical observations indicated that the origin of segmental genodermatoses was more complex, because the segmental lesions did not always show the same degree of severity as that encountered in the corresponding nonmosaic trait. Sometimes, a rather pronounced involvement was superimposed upon the ordinary, diffuse manifestation of the same disease.
These observations prompted us to postulate a novel genetic concept of dichotomous segmental forms of autosomal dominant skin disorders reflecting different states of zygosity (Figure 1) (4, 5).
Recently, molecular evidence has been provided for several mosaic cutaneous traits representing a type 1 segmental involvement, including epidermolytic hyperkeratosis of Brocq (21), neurofibromatosis type 1 (22), Darier disease (23, 24), and Apert syndrome (25).
By contrast, the concept of type 2 segmental involvement has so far not been investigated at the cellular level, although possible examples of type 2 segmental manifestation now include at least 15 different autosomal dominant skin disorders (26).
Recently, a type 2 segmental manifestation of HHD (6) was reported in a woman of German origin (individual IV-1 in Figure 2A) who had been presented as a child under another diagnosis (7). To analyze the genetic basis of this unusual phenotype, we first amplified all coding exons and the adjacent splice sites of the ATP2C1 gene from genomic DNA. Mutation screening by heteroduplex analysis using CSGE revealed a complex heteroduplex formation in the PCR fragment containing exon 22 of the ATP2C1 gene (Figure 3A). This heteroduplex formation was not observed in 300 chromosomes from unrelated Caucasian individuals studied for control purposes, indicative of a mutation (data not shown). Direct sequencing of newly synthesized PCR fragments containing exon 22 showed a heterozygous G-to-A substitution at the first base of the consensus splice donor site of exon 22, resulting in a splice site mutation designated 2146+1G→A (Figure 3B, middle lane).
The effect of mutation 2146+1G→A was studied on the cDNA level by RT-PCR and by quantitative real-time PCR. Subcloning of the PCR product and subsequent automated sequencing analysis of 20 clones revealed that the splice site mutation resulted in skipping of exon 22 (Figure 3C). Relative quantitation of wild-type and mutant cDNA obtained from a nonsegmental (heterozygous) skin area with quantitative real-time PCR revealed a ratio of wild-type to mutant ATP2C1 cDNA of 3.8 ± 0.2, indicating that the splice site mutation most likely results in nonsense-mediated mRNA decay (Figure 4A) (27).
A relapse of the disease in the index patient in October 2002 (Figure 2B) enabled us to obtain skin fragments from segmental and nonsegmental areas for cellular and molecular studies. Subsequently, DNA and RNA were isolated from cultured keratinocytes. Using keratinocyte DNA from segmental and nonsegmental areas, we again PCR-amplified the region of the APT2C1 gene harboring exon 22 and its adjacent splice sites. After subjecting these PCR products to CSGE analysis, we observed a marked decrease in intensity of the wild-type homoduplex band in those keratinocytes originating from the segmental areas as a first indication of LOH (Figure 3A, yellow arrows). Direct sequencing of newly synthesized PCR fragments revealed either the mutant A allele at the first base of the splice donor site of exon 22 alone, or a very weak and barely visible wild-type G signal along with the much stronger mutant A signal (Figure 3B, bottom lane). This is indicative of an either homozygous or hemizygous state of the mutation and is likewise suggestive of LOH. We further studied laser-microdissected epidermis from segmental skin areas (Figure 5, A–E) and were able to demonstrate, within epidermal blisters, a population of keratinocytes exclusively harboring the mutant A allele, again indicative of LOH (Figure 5E). Hence, all of these 3 lines of evidence, each based on a different experimental approach, suggest that in this case of type 2 segmental HHD, LOH was the underlying mechanism leading to a rather pronounced mosaic arrangement.
This type of mosaicism may originate from one of various mutational events giving rise to either homozygosity or hemizygosity for a mutant allele, including mitotic recombination, localized gene conversion, point mutation, deletion, or nondisjunction with either loss or reduplication of the mutant chromosome (20, 28, 29).
In order to study the genetic mechanism leading to LOH in more detail, we carried out 2 types of experiments. In a first step, haplotype analysis was performed with 23 informative microsatellite markers almost completely spanning chromosome 3, including a subset of markers flanking the ATP2C1 gene in both centromere and telomere directions. For all markers spanning the end of the short arm of chromosome 3 until marker D3S3634, haplotype analysis of skin specimens showed an allele pattern similar to that obtained from leukocyte-derived DNA (Figure 1). Haplotype analysis of the region from marker D3S1302 until D3S3568 near the end of the long arm of chromosome 3, however, revealed consistent loss of one allele in skin specimens and keratinocytes originating from more severely affected segmental skin areas (Figures 1 and 4B; Table 1). Moreover, analysis of markers D3S1589, D3S3568, and others showed that the allelic loss exclusively concerned the paternal allele, whereas the maternal one was maintained (Figure 1 and Table 1). This pattern was consistently observed in different skin and keratinocyte samples derived from various segmental regions, thus indicative of a single postzygotic mutational event that must have occurred at an early developmental stage. The chromosomal region in which we observed loss of the paternal wild-type allele harbors the ATP2C1 gene, indicating that the rather severe segmental involvement indeed originates from LOH (Figure 1). At the same time, these data largely exclude the possibility of an additional de novo mutational event in trans in these skin regions.
Subsequently, we quantified the number of ATP2C1 gene copies per diploid genome (Table 2). The ratio between the number of ATP2C1 gene copies and that of the X-chromosomal FVIII gene, which was used as a reference gene, was approximately 1 in all samples studied, indicating that each sample contained 2 copies of the ATP2C1 gene per diploid set of chromosomes. Since both automated sequencing and haplotype analysis only revealed the presence of the mutated maternal allele within the chromosomal region harboring the ATP2C1 gene, we conclude that 2 mutant copies of the ATP2C1 gene must be present within the region of LOH.
This largely excludes a deletion or mitotic nondisjunction with loss of the homologous wild-type chromosome as the underlying genetic mechanism in LOH regions, because each of these mutational events would result in hemizygosity with only one mutated gene copy being present per diploid set of chromosomes. Instead, our results indicate a homozygous state of the mutation with 2 mutated ATP2C1 gene copies per diploid set of chromosomes, most likely due either to an early event of postzygotic mitotic recombination by crossing-over or to gene conversion.
Interestingly, we did not observe reversion to heterozygosity for all informative markers haplotyped toward the telomere end of chromosome 3. This leads us to the assumption that, in the case presented herein, the genetic mechanism underlying the type 2 segmental manifestation is mitotic recombination rather than gene conversion.
The data presented herein provide the first molecular evidence to our knowledge in support of the recently postulated concept (4, 5) of dichotomous segmental forms of autosomal dominant skin disorders. Further studies, which are currently underway in our laboratory, may show whether similar results can be obtained in other autosomal dominant cutaneous traits.
Methods
Family and control individuals.
We studied 7 individuals from a 4-generation family of German origin (Figure 2A) and 150 unrelated, unaffected control individuals. In 4 family members, a diagnosis of HHD was established on the basis of characteristic clinical and histopathological features. All individuals provided informed consent for inclusion in the study, in accordance with guidelines set forth by the local institutional review board of the Medical Faculty of the RWTH Aachen. Clinical investigation was conducted according to Declaration of Helsinki principles.
Sample collection, primary keratinocyte cell culture, and DNA/RNA extraction.
EDTA anticoagulated blood samples were drawn from family members and controls, in accordance with guidelines set forth by the local institutional review board. Skin samples from the index patient (individual IV-1 in Figure 2A) were obtained after therapeutic surgical excision. Some samples were examined histopathologically, whereas most of the samples from both segmental and nonsegmental areas were subjected to cellular and molecular genetic analysis after epidermis and dermis had been separated as previously described in detail (8).
In brief, skin specimens were washed 3 times in phosphate-buffered saline (PBS) containing penicillin, streptomycin, gentamycin, and amphotericin B (Roche Diagnostics). After having been rinsed with 70% ethanol, they were cut into pieces and incubated overnight at 4°C in 2 U/ml dispase solution (GIBCO BRL; Invitrogen Corp.). In a second step, they were incubated at 37°C for 2 additional hours. The epidermis was then removed from the dermis and incubated for 30 minutes at 37°C in 5 ml trypsin/EDTA (PAA Laboratories GmbH). After stopping the enzymatic digestion by addition of 5 ml trypsin-neutralizing solution (TNS; PAA Laboratories GmbH), the cell suspension was centrifuged for 10 minutes at 250 g, and the cell pellet was resuspended in supplemented keratinocyte growth medium (KGM; Clonetics).
Isolation of genomic DNA from blood specimens was performed according to standard techniques (9). For extraction of DNA and RNA from freshly obtained skin specimens (individual IV-1 in Figure 1A), we used the PUREGENE DNA Purification Kit and PURESCRIPT Total RNA Purification Kit (Gentra Systems) following the manufacturer’s instructions. DNA and RNA from primary keratinocyte cell cultures were isolated with the High Pure PCR Template Preparation Kit (Roche Diagnostics) and the High Pure RNA Isolation Kit (Roche Diagnostics) according to the manufacturer’s instructions.
Laser-assisted microdissection.
Formalin-fixed tissue sections of 10 μm were placed on glass slides, deparaffinized, and stained with H&E. Slides were covered with a 10% bovine albumin and 1% amylopectin solution, which served as a liquid coverslip. Defined areas of affected epidermis were excised by laser beam and subsequently laser-catapulted into Eppendorf caps using the PALM Robot MicroBeam system (PALM GmbH). Eppendorf caps contained 20 μl of ATL tissue lysis buffer (QIAamp DNA Micro Kit; QIAGEN Inc.) with 2.5 mg/ml proteinase K. Tissue digestion was carried out overnight at 37°C in 0.2-ml Eppendorf tubes. Carrier RNA was added, and DNA extraction from this solution was performed with the QIAamp DNA Micro Kit according to the manufacturer’s instructions. DNA was eluted from spin columns in 30 μl of H2O. We used 6 μl of this solution for each PCR.
In order to semiquantitatively analyze the mutant–to–wild type ratio of DNA obtained from microdissected tissue, we carried out PCR amplification and restriction fragment analysis. A PCR fragment of 114 bps in size containing the 3′ end of exon 22 and the adjacent intron encompassing the splice donor site mutation was amplified with the primers 5′-6-FAM-CAGGTCTGCAATCGAAGAGG-3′ (forward) and 5′-TTATCCAGAAGACCCATGGTG-3′ (reverse). PCR was performed in a total volume of 25 μl with 0.5 U Taq polymerase AmpliTaq Gold (Roche Molecular Diagnostics) according to the manufacturer’s recommendations. Amplification was started after Taq-polymerase activation at 95°C for 8 minutes. The following thermal cycle program was used: 45 cycles of 20 seconds at 56°C, 20 seconds at 72°C, 20 seconds at 94°C, and a final elongation step of 5 minutes at 72°C. We digested 10 μl of PCR product for 16 hours with 2.5 U of the restriction endonuclease BsaAI (New England Biolabs). Wild-type DNA was cut into 2 fragments, a 72-bp dye-labelled fragment and an unlabelled 42-bp fragment. The splice site mutation 2146+1G→A abolishes this BsaAI restriction site. Fragment length analysis and quantification of DNA fragments were performed on an ABI PRISM 310 Genetic Analyzer with GeneScan software and GeneScan-350 ROX was used as a molecular size marker (Applied Biosystems Inc.).
Mutation detection.
PCR was performed using DNA extracted from EDTA blood, skin specimens, primary keratinocyte cultures, or keratinocytes excised by laser-assisted microdissection. We amplified all coding exons and the adjacent splice sites of the ATP2C1 gene with primer pairs located in flanking intron regions (primer sequences and annealing conditions available from the authors upon request).
PCR was carried out according to the following program: initial denaturation at 95°C for 5 minutes; followed by 35 cycles of denaturation at 95°C for 45 seconds, annealing at primer-specific temperatures for 1 minute, and extension at 72°C for 1 minute and 15 seconds; followed by a final extension at 72°C for 10 minutes in a Biometra TGradient thermal cycler (Whatman Biometra GmbH). Each amplification reaction contained approximately 100 ng of genomic DNA, 50 ng/μl of each forward and reverse primer, and 32 μl of Platinum Taq PCR Super Mix (Invitrogen Corp.), in a total volume of 35 μl.
For mutation detection, PCR products were analyzed by CSGE, as described by Ganguly et al. (10). PCR products displaying a heteroduplex on CSGE analysis were initially purified with the High Pure PCR Product Purification Kit (Roche Diagnostics). In a second step, PCR fragments were purified on Edge Centriflex columns (Edge BioSystems) and sequenced directly with POP-6 polymer in an ABI Prism 310 Genetic Analyzer (Applied Biosystems Inc.).
To verify the splice site mutation in exon 22 identified in this study, we used a combination of heteroduplex analysis and automated sequencing. We studied 300 chromosomes for the presence of this mutation, none of which showed the nucleotide deviation (data not shown), and thereby excluded it as a common polymorphism.
Subcloning of RT-PCR fragments and quantitative RT-PCR.
To study the functional consequences of the splice site mutation in exon 22, we extracted total RNA from clinically unaffected skin (heterozygous for the mutation) and reverse transcribed it into cDNA using the GeneAmp Reverse Transcription Kit (Applied Biosystems Inc.) according to the manufacturer’s instructions. Amplification of a cDNA fragment containing exons 20–24 of the ATP2C1 gene was performed following the aforementioned PCR protocol with 4 μl of cDNA and primers RT-HH20-24F (5′-CGTCTGGGATTGTATTCCAAAAC-3′) and RT-HH20-24R (5′-CATGTGAAGGTCATTGTTGTGTCTC-3′) in a total volume of 100 μl. RT-PCR products were subcloned into the pCR2.1TOPO vector provided with the TOPO TA Cloning Kit (Invitrogen Corp.) according to the manufacturer’s instructions. Automated sequencing of the cloned wild-type and mutant cDNA fragments was performed with primer RT-HH20-24F.
Haplotype analysis.
To study LOH in keratinocytes originating from segmentally involved skin regions of the index patient, we performed haplotype analysis using microsatellite markers. Fluorescence-labeled forward primers were used for PCR amplification according to conditions outlined elsewhere (http://www.ncbi.nlm.nih.gov:80/entrez/query.fcgi?db=unists). Fragment length analysis was performed on an ABI PRISM 310 Genetic Analyzer with GeneScan software (Applied Biosystems Inc.). At each marker typed, the signal intensity of the respective haplotype-defining PCR product was determined, and the signal of all other PCR products (i.e., stutter bands) was excluded from further analysis. Haplotype-defining PCR products were analyzed for their length (bp) and relative signal intensity compared to the total intensity of haplotype-defining PCR products (excluding stutter bands), as determined by the GeneScan software, thereby normalizing for different total fluorescence of individual PCR reactions. The smallest PCR product detected was designated as haplotype 1, and all other haplotypes were designated according to the number of additional repeats.
Defining bands for each marker were determined and all other bands (i.e., stutter bands) were excluded from further analysis. For microsatellite bands, we documented the length of each band and the ratio of the area under the curve of a detected peak for a specific band to the sum of the area under the curve for all defining bands (excluding stutter bands), as determined by the GeneScan software, thereby normalizing for different total fluorescence of each PCR reaction.
Two subsets of informative microsatellite markers were used (Figure 1). One set flanked the ATP2C1 gene on chromosome 3q21-q24 from centromere to telomere in the following order: cen-D3S1589 (127.2)—D3S3606 (128.5)—D3S3607 (128.6)—ATP2C1 (132.0)—D3S1587 (132.1)—D3S3541 (133.0)—D3S1290 (134.3)-tel (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi). The other set of markers was widely distributed over chromosome 3 and consisted of p-D3S1304 (6.9)—D3S1597 (9.4)—D3S1619 (33.9)—D3S1612 (34.4)—D3S3678 (43.2)—D3S3532 (57.3)—D3S1300 (60.3)—D3S3697 (66.2)—D3S3633 (81.8)—D3S3634 (95.3)—D3S1302 (109.8)—D3S1310 (116.1)—ATP2C1 (132.0)—D3S3684 (135.2)—D3S3586 (140.3)—D3S3694 (143.5)—D3S3575 (160.6)—D3S3568 (165.8)-q, with chromosomal locations shown in parentheses by distance from the p end of chromosome 3 in Mb (http://www.ncbi.nlm.nih.gov/mapview/map_search.cgi).
Gene dosage measurements.
To assess the gene dosage (the number of ATP2C1 genes per diploid set of chromosomes), we determined the number of ATP2C1 gene copies in relation to a reference gene (FVIII) according to methods established and reported previously (12, 13). Briefly, a real-time PCR reaction using the conditions mentioned above was performed with a primer set and a probe detecting both the wild-type and mutant alleles of the ATP2C1 gene (forward primer: 5′-GATCTTGGCTCACTGCAACCT-3′; reverse primer: 5′-GGCTGAGGCAGGAGAATCG-3′; probe: 5′-FAM-CCTCCCGGGTTTAA-MGB-3′).
A similar assay was used for detection of FVIII located on the X chromosome (forward primer: 5′-CTACCATCCAGGCTGAGGTTTATG-3′; reverse primer: 5′-CACCAACAGCATGAAGACTGACA-3′; probe: 5′-VIC-ACAGTGGTCATTACACTTAAGAACATGGCTTCCC-TAMRA-3′).
Both measurements were performed with an external standard consisting of a dilution series of genomic DNA from an affected nonmosaic female family member (individual III-4 in Figure 2A). Subsequently, the ratio of ATP2C1 to FVIII within the different samples was calculated. Taking into consideration the X-chromosomal location of the FVIII gene, this ratio allows for estimation of the number of ATP2C1 gene copies per diploid genome in each analyzed sample. Thus, a ratio of 1 reflects the presence of 2 copies of the ATP2C1 gene per diploid set of chromosomes, and the gene dosage would therefore be 2.
Acknowledgments
We are grateful to the patient and her family for their interest and cooperation in this study. This work was supported in part by a rotation position from the Medical Faculty of the RWTH Aachen (to P. Poblete-Gutiérrez) and START grant 691011 from the Medical Faculty of the RWTH Aachen (to J. Frank). A. König, R. Happle, and J. Frank are members of the Network for Ichthyoses and Related Keratinization Disorders (NIRK), which is supported by the German Federal Ministry of Education and Research (BMBF).
Pamela Poblete-Gutiérrez and Tonio Wiederholt contributed equally to this work.
Jorge Frank’s present address is: Department of Dermatology, University Hospital Maastricht, Maastricht, Germany.
Footnotes
See the related Commentary beginning on page 1407.
Nonstandard abbreviations used: CSGE, conformation-sensitive gel electrophoresis; FVIII, factor VIII; HHD, Hailey-Hailey disease; LOH, loss of heterozygosity; mutation 2146+1G→A, guanine-to-adenine substitution at the first base of the consensus splice donor site of exon 22 of the ATP2C1 gene.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Burge SM. Hailey-Hailey disease: the clinical features, response to treatment and prognosis. Br. J. Dermatol. 1992;126:275–282. doi: 10.1111/j.1365-2133.1992.tb00658.x. [DOI] [PubMed] [Google Scholar]
- 2.Hu Z, et al. Mutations in ATP2C1, encoding a calcium pump, cause Hailey-Hailey disease. Nat. Genet. 2000;24:61–65. doi: 10.1038/71701. [DOI] [PubMed] [Google Scholar]
- 3.Sudbrak R, et al. Hailey-Hailey disease is caused by mutations in ATP2C1 encoding a novel Ca(2+) pump. Hum. Mol. Genet. 2000;9:1131–1140. doi: 10.1093/hmg/9.7.1131. [DOI] [PubMed] [Google Scholar]
- 4.Happle R. Segmental forms of autosomal dominant skin disorders: different types of severity reflect different states of zygosity. Am. J. Med. Genet. 1996;66:241–242. doi: 10.1002/(SICI)1096-8628(19961211)66:2<241::AID-AJMG24>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 5.Happle R. A rule concerning the segmental manifestation of autosomal dominant skin disorders: review of clinical examples providing evidence for dichotomous types of severity. Arch. Dermatol. 1997;133:1505–1509. [PubMed] [Google Scholar]
- 6.Vakilzadeh F, Kolde G. Relapsing linear acantholytic dermatosis. Br. J. Dermatol. 1985;112:349–355. doi: 10.1111/j.1365-2133.1985.tb04864.x. [DOI] [PubMed] [Google Scholar]
- 7.König A, Hörster S, Vakilzadeh F, Happle R. Type 2 segmental manifestation of Hailey-Hailey disease: poor therapeutic response to dermabrasion is due to severe involvement of adnexal structures. Eur. J. Dermatol. 2000;10:265–268. [PubMed] [Google Scholar]
- 8.Hoeller D, et al. An improved and rapid method to construct skin equivalents from human hair follicles and fibroblasts. Exp. Dermatol. 2001;10:264–271. doi: 10.1034/j.1600-0625.2001.100406.x. [DOI] [PubMed] [Google Scholar]
- 9.Sambrook, J., Fritsch, E.F., and Maniatis, T. 1989. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA.
- 10.Ganguly A, Rock MJ, Prockop DJ. Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10325–10329. doi: 10.1073/pnas.90.21.10325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Anhuf D, Eggermann T, Rudnik-Schoneborn S, Zerres K. Determination of SMN1 and SMN2 copy number using TaqMan technology. Hum. Mutat. 2003;22:74–78. doi: 10.1002/humu.10221. [DOI] [PubMed] [Google Scholar]
- 13.Wilke K, Duman B, Horst J. Diagnosis of haploidy and triploidy based on measurement of gene copy number by real-time PCR. Hum. Mutat. 2000;16:431–436. doi: 10.1002/1098-1004(200011)16:5<431::AID-HUMU8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 14.Chemke J, Rappaport S, Etrog R. Aberrant melanoblast migration associated with trisomy 18 mosaicism. J. Med. Genet. 1983;20:135–137. doi: 10.1136/jmg.20.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung EG. Erbprognose in der Dermatologie. Z. Hautkr. 1977;52:443–449. [PubMed] [Google Scholar]
- 16.Boltshauser E, Stocker H, Machler M. Neurofibromatosis type 1 in a child of a parent with segmental neurofibromatosis (NF-5) Neurofibromatosis. 1989;2:244–245. [PubMed] [Google Scholar]
- 17.Nazzaro V, Ermacora E, Santucci B, Caputo R. Epidermolytic hyperkeratosis: generalized form in children from parents with systematized linear form. Br. J. Dermatol. 1990;122:417–422. doi: 10.1111/j.1365-2133.1990.tb08292.x. [DOI] [PubMed] [Google Scholar]
- 18.Bernards A, Gusella JF. The importance of genetic mosaicism in human disease. N. Engl. J. Med. 1994;331:1447–1449. doi: 10.1056/NEJM199411243312110. [DOI] [PubMed] [Google Scholar]
- 19.Happle R. Mosaicism in human skin. Understanding the patterns and mechanisms. Arch. Dermatol. 1993;129:1460–1470. [PubMed] [Google Scholar]
- 20.Hall JG. Review and hypotheses: somatic mosaicism: observations related to clinical genetics. Am. J. Hum. Genet. 1988;43:355–363. [PMC free article] [PubMed] [Google Scholar]
- 21.Paller AS, et al. Genetic and clinical mosaicism in a type of epidermal nevus. N. Engl. J. Med. 1994;331:1408–1415. doi: 10.1056/NEJM199411243312103. [DOI] [PubMed] [Google Scholar]
- 22.Tinschert S, et al. Segmental neurofibromatosis is caused by somatic mutation of the neurofibromatosis type 1 (NF1) gene. Eur. J. Hum. Genet. 2000;8:455–459. doi: 10.1038/sj.ejhg.5200493. [DOI] [PubMed] [Google Scholar]
- 23.Sakuntabhai A, Dhitavat J, Burge S, Hovnanian A. Mosaicism for ATP2A2 mutations causes segmental Darier’s disease. J. Invest. Dermatol. 2000;115:1144–1147. doi: 10.1046/j.1523-1747.2000.00182.x. [DOI] [PubMed] [Google Scholar]
- 24.Wada T, et al. A Japanese case of segmental Darier’s disease caused by mosaicism for the ATP2A2 mutation. Br. J. Dermatol. 2003;149:185–188. doi: 10.1046/j.1365-2133.2003.05412.x. [DOI] [PubMed] [Google Scholar]
- 25.Munro CS, Wilkie AO. Epidermal mosaicism producing localised acne: somatic mutation in FGFR2. Lancet. 1998;352:704–705. doi: 10.1016/S0140-6736(05)60820-3. [DOI] [PubMed] [Google Scholar]
- 26.Happle R. Dohi Memorial Lecture. New aspects of cutaneous mosaicism. J. Dermatol. 2002;29:681–692. doi: 10.1111/j.1346-8138.2002.tb00204.x. [DOI] [PubMed] [Google Scholar]
- 27.Hentze MW, Kulozik AE. A perfect message: RNA surveillance and nonsense-mediated decay. Cell. 1999;96:307–310. doi: 10.1016/s0092-8674(00)80542-5. [DOI] [PubMed] [Google Scholar]
- 28.German J. Genetic disorders associated with chromosomal instability and cancer. J. Invest. Dermatol. 1973;60:427–434. [Google Scholar]
- 29.Happle R. Loss of heterozygosity in human skin. J. Am. Acad. Dermatol. 1999;41:143–164. doi: 10.1016/s0190-9622(99)70042-3. [DOI] [PubMed] [Google Scholar]