Abstract
Genetic and environmental factors contribute to age-dependent susceptibility to type 2 diabetes. Recent studies have reported reduced expression of PPARγ coactivator 1α (PGC-1α) and PGC-1β genes in skeletal muscle from type 2 diabetic patients, but it is not known whether this is an inherited or acquired defect. To address this question we studied expression of these genes in muscle biopsies obtained from young and elderly dizygotic and monozygotic twins without known diabetes before and after insulin stimulation and related the expression to a Gly482Ser variant in the PGC-1α gene. Insulin increased and aging reduced skeletal muscle PGC-1α and PGC-1β mRNA levels. This age-dependent decrease in muscle gene expression was partially heritable and influenced by the PGC-1α Gly482Ser polymorphism. In addition, sex, birth weight, and aerobic capacity influenced expression of PGC-1α in a complex fashion. Whereas expression of PGC-1α in muscle was positively related to insulin-stimulated glucose uptake and oxidation, PGC-1β expression was positively related to fat oxidation and nonoxidative glucose metabolism. We conclude that skeletal muscle PGC-1α and PGC-1β expression are stimulated by insulin and reduced by aging. The data also suggest different regulatory functions for PGC-1α and PGC-1β on glucose and fat oxidation in muscle cells. The finding that the age-dependent decrease in the expression of these key genes regulating oxidative phosphorylation is under genetic control could provide an explanation by which an environmental trigger (age) modifies genetic susceptibility to type 2 diabetes.
Introduction
Skeletal muscle is a tissue of high energy demands and important for insulin-stimulated glucose disposal, and thus it is a major site of insulin resistance. A reduction in oxidative type 1 muscle myofibers and an increase in glycolytic type 2 muscle myofibers have been associated with type 2 diabetes (1). PPARγ coactivator 1α (PGC-1α) was recently found to drive the formation of oxidative type 1 myofibers and activate genes of mitochondrial oxidative metabolism when expressed in muscle of transgenic mice (2). In addition, the expression of PGC-1α and a set of genes involved in oxidative phosphorylation were found to be decreased not only in muscle from patients with type 2 diabetes but also in muscle from individuals with impaired glucose tolerance (IGT) or normal glucose tolerance but with a family history of type 2 diabetes, suggesting that this could represent an early inherited trait (3, 4). PGC-1α, which is a master regulator of a number of metabolic pathways in skeletal muscle, adipose tissue, liver, and pancreas, belongs to a small family of transcriptional coactivators also including PGC-1β and the PGC-1–related coactivator (5–9). The biological and molecular functions of PGC-1β and PGC-1–related coactivator are less well characterized compared with PGC-1α. Nevertheless, PGC-1β gene expression was recently found to be decreased in muscle of patients with type 2 diabetes and in nondiabetic subjects with a family history of type 2 diabetes (4). PGC-1β, like PGC-1α, induces expression of genes involved in oxidative phosphorylation in both muscle and liver cells (7, 10). Mice overexpressing PGC-1β had an increased energy expenditure, and PGC-1β was suggested to antagonize obesity by increasing fat oxidation in the transgenic animals (11).
Both genetic and environmental factors contribute to increased susceptibility to type 2 diabetes (12). Among environmental factors, obesity, reduced physical activity, age, and intrauterine environment as reflected by birth weight are generally accepted risk factors for type 2 diabetes. Common variants in the PPARγ, calpain 10, and PGC-1α genes have been associated with increased risk of type 2 diabetes (13–15). Particularly, a Gly482Ser polymorphism in the PGC-1α gene has been associated with increased risk of type 2 diabetes in Danish (15) and Japanese populations (16) and with abdominal obesity and reduced lipid oxidation in Pima Indians (17).
To examine the influence of genetic and environmental factors on the expression of PGC-1α and PGC-1β in human skeletal muscle, we studied mRNA expression of these two transcriptional coactivators in muscle biopsies from young (n = 86, age 28 ± 0.2 years) and elderly (n = 68, age 62.4 ± 0.2 years) monozygotic (MZ) and dizygotic (DZ) twins before and after a hyperinsulinemic euglycemic clamp. We used a generalized estimating equation (GEE) model to test the influence of PGC-1α and PGC-1β gene expression on insulin-stimulated glucose disposal rate (Rd), glucose oxidation, fat oxidation, and nonoxidative glucose metabolism (NOGM) in skeletal muscle. Using a classical twin approach, we could study heritability of skeletal muscle PGC-1α and PGC-1β expression. In addition, we also examined whether variation in the PGC-1α gene influenced expression of PGC-1α and PGC-1β in muscle and whether this was modified by age. Finally, we analyzed if muscle mRNA levels of PGC-1α and PGC-1β correlate with the expression of a target gene, glucose transporter 4 (GLUT4).
Results
Clinical characteristics.
The younger twins had significantly lower BMI, waist-to-hip ratio (WHR), fat body mass, percentage of body fat, and fat oxidation during clamp, as well as higher lean body mass (LBM), total body aerobic capacity (VO2max), insulin-stimulated Rd, glucose oxidation, and NOGM than the elderly twins (Table 1). The young and elderly twins had similar rates of basal fat oxidation and birth weight.
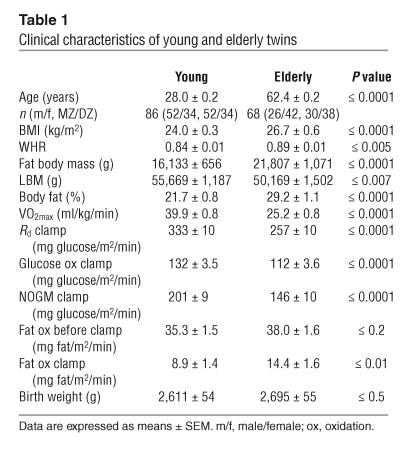
Table 1.
Clinical characteristics of young and elderly twins
The influence of insulin and age on skeletal muscle PGC-1α and PGC-1β mRNA levels.
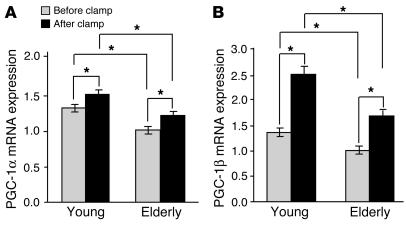
To study the influence of insulin and age on PGC-1α and PGC-1β expression in human skeletal muscle, we measured the mRNA levels of these two transcriptional coactivators in muscle biopsies taken from young and elderly twins before and after a hyperinsulinemic euglycemic clamp (see Figure 1, A and B; values normalized to the mRNA level of cyclophilin A). In muscle from young twins, insulin increased the PGC-1α mRNA expression ratio 15% from 1.29 ± 0.046 to 1.48 ± 0.057 (P < 0.001) and PGC-1β mRNA 84% from 1.36 ± 0.086 to 2.50 ± 0.16 (P < 0.0001). In the elderly twins, insulin increased muscle PGC-1α mRNA expression 20% from 0.99 ± 0.049 to 1.19 ± 0.060 (P < 0.001) and muscle PGC-1β mRNA 66% from 1.01 ± 0.076 to 1.68 ± 0.11 (P < 0.0001). The level of PGC-1α and PGC-1β mRNA was reduced about 20–30% in muscle specimens from elderly compared with young twins, both in the basal (P < 0.0001 and P < 0.001 for PGC-1α and PGC-1β, respectively) and insulin-stimulated steady state (P < 0.001 and P < 0.0001 for PGC-1α and PGC-1β, respectively) (Figure 1, A and B). In young twins, there was a significant correlation between basal PGC-1α and PGC-1β mRNA levels (r = 0.25, P < 0.05). Among the elderly twins, both basal (r = 0.33, P < 0.005) and insulin-stimulated PGC-1α and PGC-1β levels (r = 0.27, P < 0.05) correlated significantly.
Figure 1.
Effects of insulin and age on human skeletal muscle PGC-1α and PGC-1β mRNA levels. Skeletal muscle specimens were taken from young (n = 86) and elderly (n = 68) twins before and after a hyperinsulinemic clamp. RNA was prepared and analyzed for PGC-1α (A) and PGC-1β (B) mRNA expression, together with the internal standard cyclophilin A using TaqMan Real-Time PCR with an ABI 7900 system. The PGC-1α/cyclophilin A ratio and the PGC-1β/cyclophilin A ratio were calculated for each sample, and the ratios are presented in the figure. Results are expressed as the mean ± SEM. *P ≤ 0.05.
Correlation between skeletal muscle PGC-1α mRNA and protein level.
To evaluate whether PGC-1α mRNA correlates with its protein level we analyzed PGC-1α protein in muscle biopsies from eight young and eight elderly twins before and after insulin stimulation. There was a significant correlation between the level of PGC-1α mRNA and protein (r = 0.42, P < 0.05). In these muscle specimens, the level of PGC-1α protein was significantly reduced in the basal state in elderly (0.25 ± 0.03) compared with young twins (0.42 ± 0.07, P < 0.05). While in the insulin-stimulated state the age-related reduction in PGC-1α protein level was not significant (elderly 0.28 ± 0.03 versus young 0.39 ± 0.05, P = 0.1).
Association between PGC-1α Gly482Ser polymorphism and skeletal muscle PGC-1α and PGC-1β mRNA expression, VO2max, percentage of body fat, and birth weight.
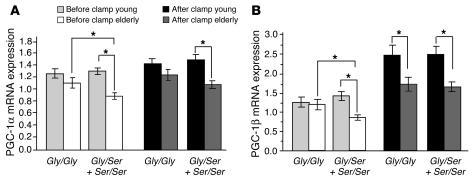
The PGC-1α Gly482Ser polymorphism had a significant effect on basal skeletal muscle PGC-1α mRNA expression in the elderly twins in whom carriers of more Ser alleles (Gly/Ser + Ser/Ser) had lower PGC-1α mRNA levels compared with carriers of the Gly/Gly genotype (0.90 ± 0.06 versus 1.12 ± 0.08, P < 0.05) (Figure 2A). The age-related decrease in muscle PGC-1α expression was only significant in carriers of the PGC-1α 482Ser allele (Figure 2A). Elderly twins carrying more Ser alleles had significantly lower basal (elderly 0.90 ± 0.06 versus young 1.32 ± 0.06, P < 0.0001) and insulin-stimulated (elderly 1.09 ± 0.06 versus young 1.50 ± 0.08, P < 0.0001) muscle PGC-1α mRNA levels compared with young twins. In contrast, there was no significant age-related reduction in muscle PGC-1α expression in carriers of the Gly/Gly genotype, neither in basal state (young 1.27 ± 0.07 versus elderly 1.12 ± 0.09; NS) nor after the clamp (young 1.44 ± 0.08 versus elderly 1.25 ± 0.09; NS).
Figure 2.
The effect of age on the association between skeletal muscle PGC-1α and PGC-1β mRNA expression and the PGC-1α Gly482Ser polymorphism. (A) PGC-1α and (B) PGC-1β mRNA expression, as measured by TaqMan Real-Time PCR (ABI 7900), in skeletal muscles of young and elderly twins before and after a hyperinsulinemic clamp, representing Gly/Gly (n = 38 and n = 29 for young and elderly, respectively) and Gly/Ser + Ser/Ser (n = 48 and n = 39 for young and elderly, respectively) PGC-1α-482 genotypes. The level of PGC-1α and PGC-1β transcripts are normalized to the mRNA level of endogenous cyclophilin A. The PGC-1α/cyclophilin A ratio and the PGC-1β/cyclophilin A ratio were calculated for each sample, and the ratios are presented in the figure. Results are expressed as the mean ± SEM. *P ≤ 0.05.
The PGC-1α Gly482Ser polymorphism also affected the level of skeletal muscle PGC-1β mRNA expression (Figure 2B). Elderly but not young carriers of more PGC-1α 482Ser alleles had reduced basal skeletal muscle PGC-1β mRNA expression compared with carriers of the Gly/Gly genotype (0.86 ± 0.07 versus 1.20 ± 0.1, P < 0.05) (Figure 2B). There was no age-related decrease in basal muscle PGC-1β expression in carriers homozygous for the Gly allele (young 1.25 ± 0.13 versus elderly 1.20 ± 0.14; NS). In contrast, carriers of PGC-1α 482Ser allele showed an age-related decrease in PGC-1β expression both during the basal (young 1.43 ± 0.11 versus elderly 0.86 ± 0.068, P < 0.0001) and insulin-stimulated states (young 2.49 ± 0.20 versus elderly 1.65 ± 0.13, P < 0.005) (Figure 2B).
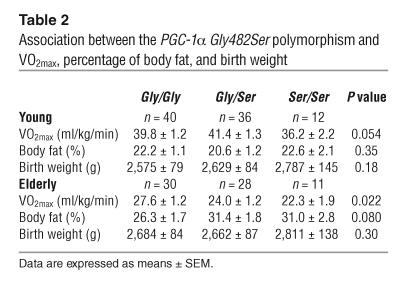
Elderly twins with the Ser/Ser genotype had reduced VO2max compared with carriers of the Gly/Gly genotype (22.3 ± 1.9 versus 27.6 ± 1.2, P = 0.022) (Table 2), while there was only a borderline effect of this PGC-1α polymorphism on VO2max in young twins (Ser/Ser 36.2 ± 2.2 versus Gly/Gly 39.8 ± 1.2, P = 0.054). The PGC-1α Gly482Ser polymorphism did not affect the percentage of body fat or birth weight (Table 2).
Table 2.
Association between the PGC-1α Gly482Ser polymorphism and VO2max, percentage of body fat, and birth weight
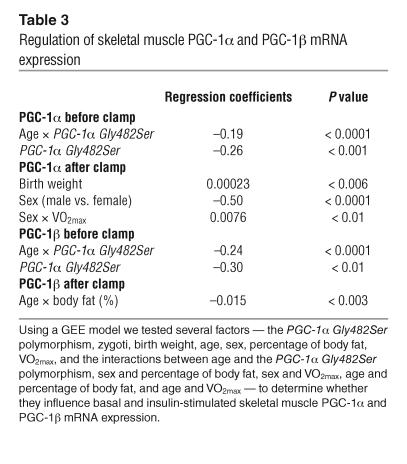
Factors influencing expression of PGC-1α and PGC-1β in skeletal muscle.
A GEE model was used to test whether any of the following parameters influence the basal and insulin-stimulated PGC-1α and PGC-1β mRNA levels in skeletal muscle: the PGC-1α Gly482Ser polymorphism, zygosity, birth weight, age, sex, percentage of body fat, and VO2max, as well as the interactions between sex and percentage of body fat, sex and VO2max, age and PGC-1α Gly482Ser polymorphism, age and percentage of body fat, and age and VO2max (Table 3). The final models were reached using backward selection regression. Both basal PGC-1α and PGC-1β mRNA levels were inversely related to the interaction between age and PGC-1α Gly482Ser polymorphism (P < 0.0001) and to the PGC-1α Gly482Ser polymorphism (P < 0.001 and P < 0.01, respectively). Insulin-stimulated PGC-1α expression was positively related to birth weight (P < 0.006) and an interaction between sex and VO2max (P < 0.01), and inversely related to sex (reduced expression in females, P < 0.0001), while insulin-stimulated PGC-1β expression was inversely related to an interaction between age and percentage of body fat (P < 0.003).
Table 3.
Regulation of skeletal muscle PGC-1α and PGC-1β mRNA expression
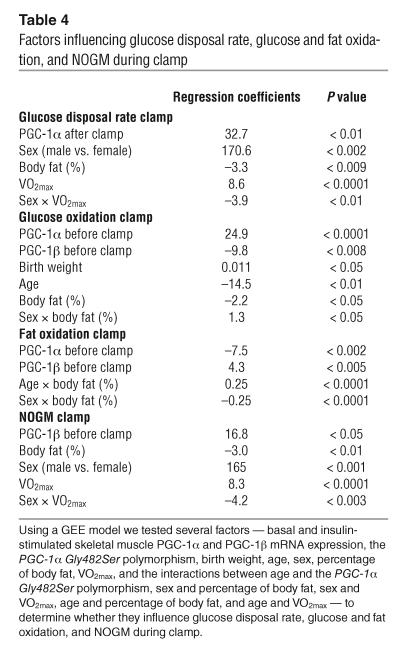
Impact of muscle PGC-1α and PGC-1β expression on glucose and lipid metabolism.
Again, the GEE model was used to identify factors, including PGC-1α and PGC-1β expression, influencing insulin-stimulated glucose disposal, glucose oxidation, fat oxidation, and NOGM in skeletal muscle. Independent variables included in the models were as follows: basal and insulin-stimulated PGC-1α and PGC-1β mRNA levels in muscle, PGC-1α Gly482Ser polymorphism, zygosity, birth weight, age, sex, percentage of body fat, and VO2max. Also included were the interactions between age and PGC-1α Gly482Ser polymorphism, sex and percentage of body fat, sex and VO2max, age and percentage of body fat, and age and VO2max (Table 4). Insulin-stimulated glucose disposal was positively related to insulin-stimulated PGC-1α expression (P < 0.01), sex (increased in females, P < 0.002), and VO2max (P < 0.0001), and inversely related to percentage of body fat (P < 0.009) and the interaction between sex and VO2max (P < 0.01). Insulin-stimulated glucose oxidation was positively related to basal PGC-1α expression (P < 0.0001), birth weight (P < 0.05), and the interaction between sex and percentage of body fat (P < 0.05). In contrast, glucose oxidation was inversely related to basal PGC-1β expression (P < 0.008), age (P < 0.01), and percentage of body fat (P < 0.05). Consequently, fat oxidation during the clamp was positively related to basal PGC-1β expression (P < 0.005) and the interaction between age and percentage of body fat (P < 0.0001), while it was inversely related to basal PGC-1α expression (P < 0.002) and to the interaction between sex and percentage of body fat (P < 0.0001). NOGM was positively related to basal PGC-1β expression (P < 0.05), sex (increased in females, P < 0.001), and VO2max (P < 0.0001), and inversely related to percentage of body fat (P < 0.01) and the interaction between sex and VO2max (P < 0.003).
Table 4.
Factors influencing glucose disposal rate, glucose and fat oxidation, and NOGM during clamp
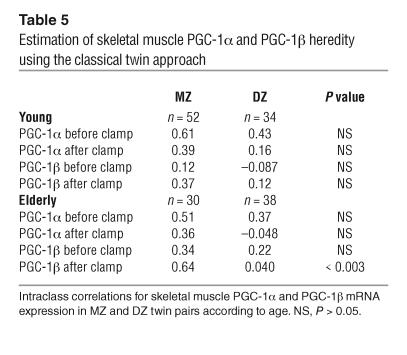
Heritability of skeletal muscle PGC-1α and PGC-1β expression.
Intraclass correlations for muscle PGC-1α and PGC-1β mRNA expression were calculated in young and elderly MZ and DZ twins in order to estimate heredity of the expression in skeletal muscle (Table 5). No statistical significant heritability was observed for skeletal muscle PGC-1α expression in either young or elderly twins. Likewise, there were no significant differences in intraclass correlations for basal and insulin-stimulated PGC-1β levels in MZ and DZ young twins. In the elderly twins, however, the intraclass correlation for insulin-stimulated muscle PGC-1β expression was significantly different between the MZ and DZ twins (P < 0.003), demonstrating a genetic component (Table 5).
Table 5.
Estimation of skeletal muscle PGC-1α and PGC-1β heredity using the classical twin approach
The impact of birth weight on skeletal muscle PGC-1α and PGC-1β expression.
MZ twins are genetically identical and significant intrapair correlations between differences in two phenotypic variables, including birth weight, are therefore determined solely by environmental factors. In contrast, significant intrapair correlations between differences in two phenotypic variables in DZ twins may be determined either by environmental or by genetic factors. Importantly, the intrapair analysis allows for control of common environmental and maternal factors with a putative influence on the outcome among both MZ and DZ twins. In young twins, there were no significant phenotypic correlations between muscle PGC-1α expression and birth weight. In elderly twins, however, there was a significant positive correlation between insulin-stimulated PGC-1α expression and birth weight among DZ twins (r = 0.34, P < 0.05), but not among MZ twins. While no significant intrapair correlations were demonstrated among the MZ twins, there was a positive intrapair correlation between birth weight and insulin-stimulated PGC-1α (r = 0.40, P < 0.05) among the younger DZ twins. Furthermore, there were positive intrapair correlations between birth weight and the insulin-stimulated increase in PGC-1α expression both among the young (r = 0.47, P < 0.01) and elderly (r = 0.50, P < 0.01) DZ twins. No significant absolute or intrapair correlations were seen between birth weight and PGC-1β expression in the basal or clamp state.
Skeletal muscle GLUT4 mRNA expression.
GLUT4 mRNA expression was analyzed in biopsies from young and elderly twins in order to evaluate whether PGC-1α and PGC-1β expression is related to GLUT4 expression in human muscle in vivo. In young twins, basal GLUT4 mRNA expression correlated positively to both basal and insulin-stimulated PGC-1α (r = 0.40, P < 0.001 and r = 0.32, P < 0.005, respectively) and PGC-1β expression (r = 0.33, P < 0.005 and r = 0.22, P < 0.05, respectively), while insulin-stimulated GLUT4 correlated significantly only to insulin-stimulated PGC-1α (r = 0.36, P < 0.001). Among the elderly twins, basal and insulin-stimulated PGC-1α levels, but not PGC-1β, correlated to basal GLUT4 expression (r = 0.46, P < 0.001, and r = 0.34, P < 0.005), while insulin-stimulated GLUT4 expression correlated to both basal and insulin-stimulated PGC-1α (r = 0.37, P < 0.005 and r = 0.52, P < 0.001, respectively) and PGC-1β expression (r = 0.42, P < 0.001 and r = 0.41, P < 0.001, respectively). The level of GLUT4 mRNA expression was reduced in muscle biopsies obtained from elderly compared with young twins, both in the basal (elderly, 1.61 ± 0.12, versus young, 1.90 ± 0.060, P < 0.001) and insulin-stimulated state (elderly, 1.71 ± 0.10, versus young, 2.05 ± 0.074, P < 0.001). The GEE model was used to test if any of the following parameters influence insulin-stimulated glucose disposal in skeletal muscle: basal and insulin-stimulated PGC-1α, PGC-1β, and GLUT4 mRNA levels in muscle, PGC-1α Gly482Ser polymorphism, zygosity, birth weight, age, sex, percentage of body fat, VO2max, as well as the interactions between age and PGC-1α Gly482Ser polymorphism, sex and percentage of body fat, sex and VO2max, age and percentage of body fat, and age and VO2max. When using this model, insulin-stimulated glucose disposal was positively related to insulin-stimulated PGC-1α expression (P < 0.02), basal GLUT4 expression (P < 0.002), VO2max (P < 0.05), and to the interaction between sex and percentage of body fat (P < 0.001), and was negatively related to percentage of body fat (P < 0.0001).
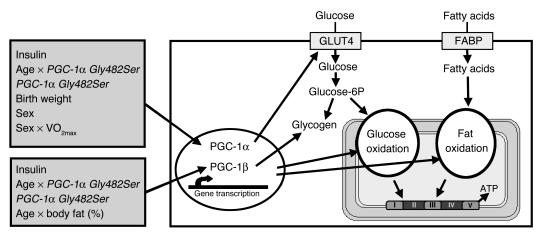
Schematic model.
The findings of the present study have been summarized in a model (Figure 3).
Figure 3.
Schematic model demonstrating factors influencing the mRNA expression of PGC-1α and PGC-1β in skeletal muscle, as well as the role of these two transcriptional coactivators on glucose and lipid metabolism. Glucose 6P, glucose-6-phosphate.
Discussion
The transcriptional coactivator PGC-1α and, possibly, PGC-1β are key regulators of genes involved in oxidative phosphorylation in skeletal muscle, and their expression is reduced in patients with type 2 diabetes (2–4, 10, 18). The question thus rises whether this is an inherited or acquired defect. The finding of decreased expression of these genes in skeletal muscle from individuals with IGT and in nondiabetic individuals with a family history of type 2 diabetes (4) would support the view that the defect is inherited. These individuals also show severe insulin resistance, however, and it can therefore not be deduced whether downregulation of gene expression precedes or follows the impaired glucose metabolism. To circumvent these problems, we adopted the classical twin approach, e.g., we studied expression of PGC-1α and PGC-1β genes in muscle biopsies from MZ and DZ twins. Importantly, the detailed metabolic characterization of this study design enabled us to determine whether this gene expression is influenced by insulin and rate of glucose metabolism as well as by age and known variants in the PGC-1α gene.
Insulin enhanced the expression of PGC-1α and PGC-1β in muscle of all subjects, but the level of expression decreased significantly with age. Interestingly, the age-related decline in gene expression was clearly influenced by a common Gly482Ser polymorphism in the PGC-1α gene. Expression of PGC-1α also correlated with VO2max, suggesting that the observed findings have implications on mitochondrial functions. Our data suggest, however, that these two coactivators may have different effects on genes involved in oxidation of glucose and fat.
Insulin resistance in skeletal muscle increases with age, and since skeletal muscle is a major site of insulin-stimulated glucose disposal, aging is a key risk factor for type 2 diabetes (19). Aging is also associated with a reduction in VO2max, and a low VO2max predicts subsequently diabetes. Factors that would influence the age-related decline in VO2max could thus be of importance in the pathogenesis of type 2 diabetes. This study provides evidence that expression of the transcriptional coactivators PGC-1α and PGC-1β in skeletal muscle could represent such factors. We found decreased levels of muscle PGC-1α and PGC-1β in elderly twins, and this phenotype was partially under genetic control. In contrast to our results, Short et al. did not observe an effect of age on the abundance of PGC-1α mRNA in skeletal muscle (20). They analyzed muscle PGC-1α expression in males and females aged 21–87 years using regression analysis. Of note, the age-related decrease in PGC-1α mRNA expression found in our study was only in the magnitude of 20–30%, which is in the same range as reported for the coordinated decrease in expression of genes involved in oxidative phosphorylation in type 2 diabetes (3). Our study design, including a total of 154 individual twins, was optimized to increase the sensitivity of detecting subtle changes in gene expression, and we compared the expression in muscle biopsies from a group of young twins (28 ± 0.2 years) with a group of elderly twins (62.4 ± 0.2 years). In the GEE model, an interaction between age and the PGC-1α Gly482Ser polymorphism was inversely related to both basal PGC-1α and PGC-1β expression, while the interaction between age and percentage of body fat influenced insulin-stimulated PGC-1β levels. Also in support of our findings of an age-related decline in gene expression, Patti et al. found reduced PGC-1α and PGC-1β mRNA levels in skeletal muscle from elderly individuals with insulin resistance and type 2 diabetes compared with younger controls. It was therefore not possible to exclude that the age difference contributed to the observed difference in gene expression (4). We also found, using the GEE model, that birth weight, sex, and the interaction between sex and VO2max influenced insulin-stimulated PGC-1α, but not PGC-1β, expression in muscle. Our results demonstrating that VO2max is associated with muscle PGC-1α expression is in agreement with other studies showing that exercise induces expression of PGC-1α, but not PGC-1β, in muscle (21–25). Birth weight and sex have not been shown previously to influence PGC-1α expression in human skeletal muscle. Uteroplacental insufficiency and low birth weight increase the risk of insulin resistance and type 2 diabetes (26–30). Insulin is an important growth factor in utero. Low birth weight is to some extent genetically determined, and it cannot be excluded that the association between low birth weight and risk of type 2 diabetes may be explained partly by genetic variant(s) influencing birth weight through reduced insulin secretion and/or insulin action. In this study birth weight was positively related to muscle PGC-1α expression and our findings of significant intrapair correlations between birth weight and PGC-1α expression in DZ but not in MZ twins indicate that the relation between birth weight and PGC-1α expression may be influenced by genetic factors.
Both PGC-1α and PGC-1β regulate mitochondrial metabolism and oxidative phosphorylation in muscle cells, and these two transcriptional coactivators stimulate expression of many mitochondrial genes (2, 3, 10, 11, 18). PGC-1α has been shown to activate a large number of nuclear receptors and transcription factors in muscle cells, however, suggesting that PGC-1α can function as a wide-range nuclear-receptor coactivator, while PGC-1β was shown to more specifically coactivate estrogen receptor–related receptors (ERRs) (11). Furthermore, PGC-1β transgenic mice exhibited increased muscle expression of medium-chain acyl CoA dehydrogenase, a known ERR target and a pivotal enzyme of mitochondrial β oxidation in muscle (11), while PGC-1α was found to regulate GLUT4 expression and muscle fiber type determination (2, 31, 32). These studies show that PGC-1α and PGC-1β profoundly alter mitochondrial metabolism, but also suggest distinct regulatory functions for these two coactivators in muscle cells. Indeed, when we used the GEE model to study the influence of a number of factors on glucose and lipid metabolism in muscle, our results indicated that PGC-1α and PGC-1β expression could be involved in the differential regulation of these metabolic pathways. Interestingly, PGC-1α expression was positively related to glucose uptake and oxidation and negatively related to lipid oxidation. In contrast, PGC-1β expression was positively associated with lipid oxidation and NOGM and negatively related to glucose oxidation. The stimulatory effect on NOGM could possibly be explained by feedback inhibition of 6-phosphofructo-2-kinase by end-products of β oxidation, e.g., acetyl-CoA, which would further reduce flux through the glycolytic pathway and divert glucose-6-phosphate to glycogen (33). In contrast to our data, there are cellular and animal studies showing that PGC-1α induces expression of genes involved in lipid oxidation. An explanation for this could be differences between species. Another reason could be that in a complex human in vivo situation PGC-1α and PGC-1β may have different roles in glucose and lipid metabolism mediated by activation of different transcription factors, whereas when PGC-1α is overexpressed in vitro, it might activate a large number of transcription factors and by redundancy activate pathways normally induced by PGC-1β in vivo. It should also be kept in mind that lipid oxidation measured by indirect calorimetry is a relatively crude measure of fat oxidation, including both oxidation of plasma-derived FFA as well as of intracellular FFA (34).
In the present study, both PGC-1α and PGC-1β expression correlated positively to the mRNA level of GLUT4, suggesting that both these transcriptional coactivators regulate the expression of GLUT4 in muscle. To our knowledge, previous studies have only suggested PGC-1α as a regulator of GLUT4 expression. Furthermore, our results suggest PGC-1α to have an impact on insulin-stimulated glucose disposal that seems to go beyond the impact of GLUT4 mRNA levels.
The induction of PGC-1β transcripts by insulin was greater than the induction of PGC-1α. It is possible that this difference could have consequences in insulin-resistant subjects where skeletal muscle PGC-1α and PGC-1β expression is reduced and that, for example, PGC-1β expression could be more affected by insulin resistance than PGC-1α. The study by Patti et al. (4), however, found reduced levels of both PGC-1α and PGC-1β in muscle biopsies from insulin-resistant subjects taken before the clamp.
Twin studies are useful when analyzing the genetic and environmental determinants of a particular trait. If the concordance rate is higher in MZ than in DZ twins, genetic factors play a more important role than the environment in causing the trait. By means of the classical twin approach, we investigated whether skeletal muscle PGC-1α and PGC-1β expression are inherited traits and found only heritability of insulin-stimulated PGC-1β expression in elderly twins. Common variants including noncoding variants in genes often have a modest effect on gene expression in target tissues (35). We thus hypothesized that the common PGC-1α Gly482Ser polymorphism could influence expression of PGC-1α and possibly the PGC-1β gene in skeletal muscle. It was also the case that carriers of more Ser alleles showed a greater age-related decline in gene expression than carriers of the Gly/Gly genotype. These results demonstrate that the age-dependent decrease in gene expression is under genetic control, and they also present a mechanism whereby a gene-environment interaction could increase susceptibility to type 2 diabetes. Heritability is influenced by the sum of genetic and environmental factors, and the latter seemed to have a clear effect on expression of these coactivators in muscle. A genetic variant in the PGC-1α gene modified the environmental effects, however. The complexity of the interaction between age and the PGC-1α Gly482Ser polymorphism may explain why the impact of heritability was only statistically significant for PGC-1β and not PGC-1α expression using the classical twin approach.
Skeletal muscle PGC-1α has previously been demonstrated to regulate its own promoter through interactions with components of the calcium-signaling pathway, resulting in an autoregulatory loop (36). It is therefore possible that the PGC-1α Gly482Ser polymorphism, which results in changed PGC-1α amino acid sequence, could affect the efficiency of PGC-1α as a transcriptional coactivator and by way of the autoregulatory loop cause altered muscle PGC-1α expression. Furthermore, we found significant correlations between skeletal muscle PGC-1α and PGC-1β expression, suggesting a potential transcriptional regulation of PGC-1β by PGC-1α and vice versa. Because the PGC-1α Gly482Ser polymorphism affects muscle PGC-1α expression in elderly subjects, we investigated if this genotype also influences the level of PGC-1β expression. Indeed, elderly but not young carriers of more PGC-1α 482Ser alleles had reduced basal levels of PGC-1β in muscle. Our results in combination with the study by Handschin et al. support a model whereby PGC-1α is involved in regulating both its own as well as PGC-1β expression in muscle by an autofeedback loop, and the PGC-1α Gly482Ser polymorphism seems to affect the efficiency of this regulatory loop.
While the aim of this study was primarily to investigate the impact of genetic versus nongenetic factors on PGC-1α and PGC-1β expression on the transcriptional level, we also measured muscle PGC-1α protein levels in a subset of young and elderly twins to determine the extent to which differences in muscle PGC-1α mRNA levels reflected differences in PGC-1α protein levels. Indeed, we found a statistically significant correlation between PGC-1α mRNA and protein expression. In addition, PGC-1α protein level decreased with age. Thus, our data certainly do indicate that the demonstrated regulatory mechanisms and interactions operating at the transcriptional mRNA level also — within a certain timeframe — relate to the translational PGC-1α protein level. The fact that insulin increased PGC-1α mRNA but not protein level is likely to be explained by the short (2 hours) insulin infusion not being a sufficient time period for both mRNA and subsequently protein synthesis to occur.
In summary, insulin increased and aging reduced skeletal muscle PGC-1α and PGC-1β levels in vivo as determined during the euglycemic clamp in young and elderly MZ and DZ twins. A common variant, Ser482Gly, in the PGC-1α gene influenced the age-related decline in gene expression. Our data also suggest that PGC-1α and PGC-1β may have different effects on glucose and lipid metabolism. Taken together the data provide a scenario by which genetic and environmental factors such as aging could interact to influence genes regulating oxidative phosphorylation and thereby increase susceptibility to type 2 diabetes.
Methods
Subjects.
Subjects were identified through The Danish Twin Register (37). A random sample of young and elderly same-sex MZ and DZ twin pairs born in Funen County during 1966–1975 (22–31 years) and 1931–1940 (57–66 years) with available original midwife records, including birth weight, were initially included in the study. All potential subjects identified according to these inclusion criteria were contacted and interviewed in order to exclude subjects fulfilling the exclusion criteria. Exclusion criteria were as follows: either twin from a pair not willing to participate, information of pre- or postmaturity (birth more than or less than 3 weeks from expected time point), known diabetes, serious heart, liver or kidney disease, use of medication known to influence glucose or lipid metabolism, including oral anticonception, that could not be withdrawn, and pregnancy/lactation.
A total of 98 twin pairs (33 young MZ; 22 young DZ; 21 elderly MZ; 22 elderly DZ) were enrolled in the clinical examination, including an oral glucose tolerance test (OGTT), and gene expression was analyzed in skeletal muscle biopsies from 77 twin pairs (26 young MZ; 17 young DZ; 15 elderly MZ; 19 elderly DZ). The clinical characteristics of these subjects are described in Table 1. Zygosity was determined by serological testing, the most valid method, with a success rate exceeding 99%. Among elderly twins 74.5% had normal glucose tolerance (NGT), 22.0% had IGT, and 3.5% had previously unknown type 2 diabetes. Of the young twins, 98.5% had NGT and 1.5% had IGT. The present study was approved by the Scientific Ethical Committees for the counties of Fyn and Vejle located at Odense University Hospital according to the Helsinki Declaration.
Clinical examination.
Subjects underwent a 2-day clinical examination separated by 1–2 weeks. Each twin pair was investigated simultaneously. The subjects were instructed to abstain from strenuous physical activity for 24 hours and to fast overnight (10–12 hours) before both examination days.
Day 1 included a standard 75-g OGTT. Peripheral venous blood was drawn before oral glucose ingestion and thereafter at 30, 60, and 120 minutes. Weight and height were measured with the subjects in lightweight clothes without shoes, and BMI was calculated using the following formula: weight (kg)/height2 (m2). Waist circumference was measured using a soft tape on standing subjects midway between the lowest rib and the iliac crest. Hip circumference was measured over the widest part of the gluteal region, and the WHR was calculated accordingly. Body composition, i.e., LBM and fat mass, was determined by dual-energy x-ray absorptiometry (DEXA) scanning.
On day 2 subjects underwent a 2-hour hyperinsulinemic euglycemic clamp as previously described (37). Polyethylene catheters were placed in the antecubital vein for infusion and in the contralateral dorsal hand or antecubital vein for blood sampling. This “sampling” hand was placed in a heated Plexiglas box to ensure arterialization of the venous blood sample. A primed continuous infusion of constant [3-3H]-tritiated glucose (bolus 22 μCi, 0.22 μCi/min) was initiated at 0 minutes and continued throughout the basal period (120 minutes) and during the clamp period (120 minutes). Basal steady state was defined as the last 30 minutes of the 120-minute basal period. Subsequently, a primed continuous insulin infusion (40 mU/m2/min) was initiated and continued for 120 minutes. Insulin-stimulated steady state was defined as the last 30 minutes of the 2-hour clamp period when tracer equilibrium, i.e., constant specific activity, was anticipated. A variable infusion of glucose (180 g/l) enriched with tritiated glucose (100 μCi/500 ml) maintained euglycemia during insulin infusion. Plasma glucose concentration was monitored every 5–10 minutes during the basal and clamp periods using an automated glucose oxidation method (Glucose Analyzer 2; Beckman Instruments Inc.). Blood samples were drawn for measurements of glucose and insulin every 10–30 minutes during the basal and clamp steady-state periods. Indirect calorimetry was performed during both steady state periods using a computerized flow-through canopy gas analyzer system (Deltatrac; Datex Inc.). After an equilibrium period of 10 minutes, the average gas exchange rates recorded over the steady-state periods were used to calculate rates of glucose and lipid oxidation.
Muscle biopsies.
Muscle biopsy tissue was obtained from the vastus lateralis muscle under local anesthesia (lidocaine) using a Bergstöm needle with suction applied. The biopsy specimens were quickly blotted on filter paper and frozen in liquid nitrogen. The tissues were stored at –80°C until further processed.
Analytical methods.
Plasma glucose concentrations during the OGTT and the hyperinsulinemic euglycemic clamp were analyzed by the glucose dehydrogenase oxidation method. Plasma insulin concentrations were measured using a two-site, two-step, time-resolved, immunofluoremetric assay (DELFIA) as previously described (37, 38). Cross-reactivities with proinsulin, C-peptide, and Des(31, 32) split product in the insulin assay were all less than 0.4%. Intra-assay coefficients of variation in the physiological ranges were 3.6–4.3% for plasma insulin. Inter-assay coefficients of variation were 1.7–3.4% for plasma insulin. Tritiated water was measured as described by Hother-Nielsen and Beck-Nielsen (39).
Calculations of basal and insulin-stimulated Rd.
Rd was calculated at 10-minute intervals during the steady-state periods using Steele’s non–steady-state equations (40). In the calculations, distribution volume of glucose was assumed to be 200 ml per kilogram of body weight and the pool fraction to be 0.65. NOGM was calculated as Rd minus glucose oxidation as determined by indirect calorimetry. Rd, glucose oxidation, and NOGM are expressed as milligrams glucose per square meter per minute and are presented as mean values of the 30-minute steady-state periods.
Analysis of PGC-1α, PGC-1β, and GLUT4 mRNA levels in skeletal muscle.
Total RNA was extracted from frozen skeletal muscle specimens using the Tri Reagent kit according to the manufacturer’s instructions (Sigma-Aldrich). The cDNA was synthesized using Superscript II RNase H– Reverse Transcriptase and random hexamer primers (Invitrogen Corp.). PGC-1α, PGC-1β, and GLUT4 mRNA levels were quantified using TaqMan Real-Time PCR with an ABI 7900 system (Applied Biosystems). Gene-specific probes and primer pairs for PGC-1α (Assays-on-Demand, Hs00173304_m1; Applied Biosystems), PGC-1β (Assays-on-Demand, Hs00370186_m1; Applied Biosystems), and GLUT4 (Assays-on-Demand, Hs00168966_m1; Applied Biosystems) were used. For each probe/primer set, a standard curve was generated, which was confirmed to increase linearly with increasing amounts of cDNA. Each sample was run in duplicate, the transcript quantity was calculated according to the standard curve method based on a 2-step serial dilution on each plate with RNA content ranging from 100 to 1.5625 ng, and the transcript quantity was normalized to the mRNA level of cyclophilin A (4326316E; Applied Biosystems).
Analysis of PGC-1α protein levels in skeletal muscle using the Western blot technique.
Protein lysate was prepared from muscle specimens as previously described (41), and the protein content was measured using the bicinchoninic acid method (Pierce Chemical Co.). Twenty-five micrograms of lysate proteins were separated using Criterion 4–15% Tris-HCl linear gradient gels (Bio-Rad Laboratories), and then transferred to PVDF membranes (Bio-Rad Laboratories). After blocking (TBS, 0.1% Tween-20, and 0.5% gelatin, G-9391; Sigma-Aldrich) at 4°C overnight, the membranes were incubated with primary Ab’s for PGC-1α (dilution 1:200, sc-5816; Santa Cruz Biotechnology Inc.) and the endogenous control, cyclophilin B (dilution 1:50,000, PA1-027; Affinity BioReagents Inc.), for 2 hours, followed by incubation with secondary Ab’s linked to HRP, anti-goat (dilution 1:50,000, sc-2020; Santa Cruz Biotechnology Inc.) and anti-rabbit (dilution 1:100,000, I1904-41P; USBiological Inc.), detecting the primary Ab’s for PGC-1α and cyclophilin B, respectively. Immunoreactive proteins were visualized by chemiluminescence using SuperSignal West Dura Extended Duration Substrate (Pierce Chemical Co.) and the Fujifilm Luminescent Image Analyzer LAS-3000 with a charged-coupled device camera (Science Imaging Scandinavia AB). The protein levels of PGC-1α and cyclophilin B were then quantified using Multi Gauge software (version 2.2; Fujifilm), and in each sample the level of PGC-1α was normalized to the level of cyclophilin B. A blocking peptide (sc-5816-P; Santa Cruz Biotechnology Inc.) was used in a control analysis to verify the approximately 91-kDa PGC-1α protein.
Genotyping.
Genomic DNA was extracted from blood using conventional methods (42). The Gly482Ser (GGTØAGT) polymorphism of PGC-1α was genotyped using an allelic discrimination assay performed with an ABI 7900 system (Applied Biosystems Inc.) using the PCR primers, 5′-CACTTCGGTCATCCCAGTCAA-3′ (forward) and 5′-TTATCACTTTCATCTTCGCTGTCATC-3′ (reverse) and TaqMan MGB probes, Fam-5′-AGACAAGACCGGTGAA-3′ and Vic-5′-CAGACAAGACCAGTGAA-3′ (17).
Statistical methods.
Differences in PGC-1α, PGC-1β, and GLUT4 mRNA and PGC-1α protein levels between the different groups studied were analyzed using the Student’s t test or nonparametric Mann-Whitney test where appropriate. Correlations were calculated using Pearson correlation coefficients for normally distributed values and Spearman correlation coefficients when normality was rejected. All P values were 2-tailed, and P values less than 0.05 were considered significant. Statistical calculations were performed by NCSS software (NCSS Statistical Software).
GEEs were used to fit general linear models for the twin data using the approach of Zeger and Liang (43). We modeled the within twin pair association as a correlation, where we allowed the correlation to be different for the MZ and DZ twins. We reached the final models using backward selection regression. The significance level for variable elimination was set to 0.05.
MZ twins have identical genotypes, and any differences are theoretically due to environmental factors, whereas DZ twins, on average, share 50% of their genes. The extent to which MZ twins are more alike than DZ twins is therefore presumed to reflect a genetic influence on the phenotype in question. Heritability (expressed as h2) gives the proportion of the total variation of a trait attributable to genetic variation and can be estimated by comparing the similarity of a given phenotype within MZ twin pairs with the similarity within DZ twin pairs. Intraclass correlation is a method to measure resemblance within twin pairs: r = cov (twin 1, twin 2)/– var (twin 1) × var (twin 2) (44). Statistical comparisons of intraclass correlations were made after transformation using the Fisher z transformation. The heritability is expressed as twice the difference of the intraclass correlation of MZ and DZ twins [h2 = 2(rMZ – rDZ)] (44).
Acknowledgments
This investigation was funded by the following foundations: Crafoord, Malmö University Hospital, Swegene, the Diabetes Programme at Lund University, Påhlsson, Magnus Bergvall, Thuring, and Borgström; as well as by grants from Region Skåne, Juvenile Diabetes Research Foundation — Wallenberg Center of Excellence, the Swedish Research Council, Novo Nordisk, the Danish Medical Research Council, and the Danish Diabetes Association. We thank Margareta Svensson and Marianne Modest for excellent technical assistance.
Footnotes
See the related Commentary beginning on page 1414.
Charlotte Ling and Pernille Poulsen contributed equally to this work.
Nonstandard abbreviations used: DZ, dizygotic; ERR, estrogen receptor–related receptor; GEE, generalized estimating equation; GLUT4, glucose transporter 4; IGT, impaired glucose tolerance; LBM, lean body mass; MZ, monozygotic; NGT, normal glucose tolerance; NOGM, nonoxidative glucose metabolism; OGTT, oral glucose tolerance test; PGC-1, PPARγ coactivator 1; Rd, glucose disposal rate; VO2max, total body aerobic capacity; WHR, waist-to-hip ratio.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Nyholm B, et al. Evidence of an increased number of type IIb muscle fibers in insulin-resistant first-degree relatives of patients with NIDDM. Diabetes. 1997;46:1822–1828. doi: 10.2337/diab.46.11.1822. [DOI] [PubMed] [Google Scholar]
- 2.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 3.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 4.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 6.Yoon JC, et al. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Dev. Cell. 2003;5:73–83. doi: 10.1016/s1534-5807(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 7.Lin J, et al. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J. Biol. Chem. 2003;278:30843–30848. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 8.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 9.Andersson U, Scarpulla RC. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell. Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.St-Pierre J, et al. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J. Biol. Chem. 2003;278:26597–26603. doi: 10.1074/jbc.M301850200. [DOI] [PubMed] [Google Scholar]
- 11.Kamei Y, et al. PPAR(gamma) coactivator 1(beta)/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koistinen HA, Zierath JR. Regulation of glucose transport in human skeletal muscle. Ann. Med. 2002;34:410–418. doi: 10.1080/078538902321012351. [DOI] [PubMed] [Google Scholar]
- 13.Altshuler D, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 14.Horikawa Y, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 15.Ek J, et al. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia. 2001;44:2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 16.Hara K, et al. A genetic variation in the PGC-1 gene could confer insulin resistance and susceptibility to type II diabetes. Diabetologia. 2002;45:740–743. doi: 10.1007/s00125-002-0803-z. [DOI] [PubMed] [Google Scholar]
- 17.Muller YL, Bogardus C, Pedersen O, Baier L. A Gly482Ser missense mutation in the peroxisome proliferator-activated receptor gamma coactivator-1 is associated with altered lipid oxidation and early insulin secretion in Pima Indians. Diabetes. 2003;52:895–898. doi: 10.2337/diabetes.52.3.895. [DOI] [PubMed] [Google Scholar]
- 18.Wu Z, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 19.Petersen KF, et al. Mitochondrial dysfunction in the elderly: possible role in insulin resistance. Science. 2003;300:1140–1142. doi: 10.1126/science.1082889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Short KR, et al. Impact of aerobic exercise training on age-related changes in insulin sensitivity and muscle oxidative capacity. Diabetes. 2003;52:1888–1896. doi: 10.2337/diabetes.52.8.1888. [DOI] [PubMed] [Google Scholar]
- 21.Pilegaard H, Saltin B, Neufer PD. Exercise induces transient transcriptional activation of the PGC-1alpha gene in human skeletal muscle. J. Physiol. 2003;546:851–858. doi: 10.1113/jphysiol.2002.034850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goto M, et al. cDNA cloning and mRNA analysis of PGC-1 in epitrochlearis muscle in swimming-exercised rats. Biochem. Biophys. Res. Commun. 2000;274:350–354. doi: 10.1006/bbrc.2000.3134. [DOI] [PubMed] [Google Scholar]
- 23.Terada S, et al. Effects of low-intensity prolonged exercise on PGC-1 mRNA expression in rat epitrochlearis muscle. Biochem. Biophys. Res. Commun. 2002;296:350–354. doi: 10.1016/s0006-291x(02)00881-1. [DOI] [PubMed] [Google Scholar]
- 24.Norrbom J, et al. PGC-1(alpha) mRNA expression is influenced by metabolic perturbation in exercising human skeletal muscle. J. Appl. Physiol. 2003;96:189–194. doi: 10.1152/japplphysiol.00765.2003. [DOI] [PubMed] [Google Scholar]
- 25.Meirhaeghe A, et al. Characterization of the human, mouse and rat PGC1 beta (peroxisome-proliferator-activated receptor-gamma co-activator 1 beta) gene in vitro and in vivo. Biochem. J. 2003;373:155–165. doi: 10.1042/BJ20030200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hales CN, et al. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 1991;303:1019–1022. doi: 10.1136/bmj.303.6809.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phipps K, et al. Fetal growth and impaired glucose tolerance in men and women. Diabetologia. 1993;36:225–228. doi: 10.1007/BF00399954. [DOI] [PubMed] [Google Scholar]
- 28.Lithell HO, et al. Relation of size at birth to non-insulin dependent diabetes and insulin concentrations in men aged 50–60 years. BMJ. 1996;312:406–410. doi: 10.1136/bmj.312.7028.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poulsen P, Vaag AA, Kyvik KO, Moller Jensen D, Beck-Nielsen H. Low birth weight is associated with NIDDM in discordant monozygotic and dizygotic twin pairs. Diabetologia. 1997;40:439–446. doi: 10.1007/s001250050698. [DOI] [PubMed] [Google Scholar]
- 30.Ravelli AC, et al. Glucose tolerance in adults after prenatal exposure to famine. Lancet. 1998;351:173–177. doi: 10.1016/s0140-6736(97)07244-9. [DOI] [PubMed] [Google Scholar]
- 31.Michael LF, et al. Restoration of insulin-sensitive glucose transporter (GLUT4) gene expression in muscle cells by the transcriptional coactivator PGC-1. Proc. Natl. Acad. Sci. U. S. A. 2001;98:3820–3825. doi: 10.1073/pnas.061035098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miura S, Kai Y, Ono M, Ezaki O. Overexpression of peroxisome proliferator-activated receptor gamma coactivator-1alpha down-regulates GLUT4 mRNA in skeletal muscles. J. Biol. Chem. 2003;278:31385–31390. doi: 10.1074/jbc.M304312200. [DOI] [PubMed] [Google Scholar]
- 33.Groop LC, Ferrannini E. Insulin action and substrate competition. Baillieres Clin. Endocrinol. Metab. 1993;7:1007–1032. doi: 10.1016/s0950-351x(05)80243-5. [DOI] [PubMed] [Google Scholar]
- 34.Groop LC, Bonadonna RC, Shank M, Petrides AS, DeFronzo RA. Role of free fatty acids and insulin in determining free fatty acid and lipid oxidation in man. J. Clin. Invest. 1991;87:83–89. doi: 10.1172/JCI115005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science. 2002;297:1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 36.Handschin C, Rhee J, Lin J, Tarr PT, Spiegelman BM. An autoregulatory loop controls peroxisome proliferator-activated receptor gamma coactivator 1alpha expression in muscle. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7111–7116. doi: 10.1073/pnas.1232352100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poulsen P, Levin K, Beck-Nielsen H, Vaag A. Age-dependent impact of zygosity and birth weight on insulin secretion and insulin action in twins. Diabetologia. 2002;45:1649–1657. doi: 10.1007/s00125-002-0983-6. [DOI] [PubMed] [Google Scholar]
- 38.Hemmila I, Dakubu S, Mukkala VM, Siitari H, Lovgren T. Europium as a label in time-resolved immunofluorometric assays. Anal. Biochem. 1984;137:335–343. doi: 10.1016/0003-2697(84)90095-2. [DOI] [PubMed] [Google Scholar]
- 39.Hother-Nielsen O, Beck-Nielsen H. On the determination of basal glucose production rate in patients with type 2 (non-insulin-dependent) diabetes mellitus using primed-continuous 3-3H-glucose infusion. Diabetologia. 1990;33:603–610. doi: 10.1007/BF00400204. [DOI] [PubMed] [Google Scholar]
- 40.Steele R. Influences of glucose loading and of injected insulin on hepatic glucose output. Ann. N. Y. Acad. Sci. 1959;82:420–430. doi: 10.1111/j.1749-6632.1959.tb44923.x. [DOI] [PubMed] [Google Scholar]
- 41.Markuns JF, Wojtaszewski JF, Goodyear LJ. Insulin and exercise decrease glycogen synthase kinase-3 activity by different mechanisms in rat skeletal muscle. J. Biol. Chem. 1999;274:24896–24900. doi: 10.1074/jbc.274.35.24896. [DOI] [PubMed] [Google Scholar]
- 42.Vandenplas S, et al. Blot hybridisation analysis of genomic DNA. J. Med. Genet. 1984;21:164–172. doi: 10.1136/jmg.21.3.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 44.Neale M.C, and Cardon, L.R. 1992. Methodology for genetic studies of twins and families. Kluwer Academic Publishers. Dordrecht, The Netherlands. 35–53.