Abstract
The Randle cycle, which has been invoked to explain the reciprocal relationship between fatty acid oxidation and glucose oxidation, has long been implicated as a potential mechanism for hyperglycemia and type 2 diabetes mellitus (T2DM). Now genetic, functional genomic, and transgenic approaches have identified PPARγ coactivators (PGC-1α and PGC-1β) as key regulators of mitochondrial number and function. They regulate adaptive thermogenesis as well as glucose and fat oxidation in muscle and fat tissue, gluconeogenesis in liver, and even glucose-regulated insulin secretion in β cells. PGC-1α and PGC-1β mRNA levels and the mitochondrial genes they regulate are decreased in muscle of people with prediabetes and T2DM. A new report indicates that PGC-1α and PGC-1β mRNA levels decrease with age in individuals with a genetic variant in PGC-1α, and these decreases correlate with alterations in whole-body glucose and fatty acid oxidation. These findings provide insights into how aging modifies genetic susceptibility to alterations in oxidative phosphorylation and T2DM.
Type 2 diabetes mellitus (T2DM), considered a rare disease no more than 100 years ago, is now an epidemic in the United States and other industrialized countries. Obesity and advancing age are potent risk factors for T2DM, pointing to lifestyle changes of the 20th century that are responsible for the current epidemic. However, despite our diabetogenic environment, some individuals develop diabetes and others do not. Multiple studies provide evidence that genetic factors are important contributors to the large inter-individual variation in diabetes susceptibility (1, 2). Identification of T2DM susceptibility genes has proven challenging, in part due to the heterogeneous and polygenic nature of the condition and due to our limited understanding of its underlying pathophysiology. In the past decade, new and powerful tools for probing the molecular, genetic, and pathophysiological basis of glucose and energy homeostasis have provided key insights into the molecular basis of diabetes. Some of these insights have proven quite surprising based upon the current state of knowledge, while others have been logical extensions of the state of the field.
Genetics of diabetes: what we do know
Simply put, diabetes occurs as a result of an absolute or relative deficiency of insulin. The former occurs in autoimmune forms of diabetes, e.g., type 1 diabetes mellitus, or latent autoimmune diabetes in adults, in which progressive destruction of insulin-secreting β cells leads to an absolute deficiency of insulin. Relative insulin deficiency is far more pervasive and in its most common form, T2DM, is caused by insulin resistance (most often due to obesity) coupled with progressive failure of the β cell to secrete sufficient insulin to compensate for the increased insulin resistance (3). In addition to typical T2DM, we now know of several monogenic diabetes syndromes that lead to a relative deficiency of insulin (reviewed in ref. 2). These include autosomal dominant T2DM (also known as maturity onset diabetes of the young), autosomal recessive syndromes of extreme insulin resistance, maternally inherited diabetes and deafness (MIDD), and partial and complete lipoatrophic diabetes syndromes. Although relatively rare, these syndromes have provided important insights into the molecular and cellular basis of glucose homeostasis. For example, some forms of autosomal dominant T2DM are due to defects in transcription factors necessary for normal β cell growth and differentiation (e.g., hepatocyte nuclear factors [HNFs], β2/neuroD, insulin promoter factor-1 [IPF-1]) (4), while others are due to mutations in molecules involved in glucose-regulated insulin secretion (e.g., glucokinase or the regulatory subunit of the ATP-sensitive potassium channel) (4, 5). MIDD, caused by mutations in mitochondrial DNA, is associated with defective insulin secretion and also some element of insulin resistance (6). Although the specific mechanisms whereby mitochondrial DNA mutations lead to MIDD have not been fully elucidated, this rare syndrome points to mitochondrial function as a key factor in glucose homeostasis that may be relevant to the more common forms of T2DM (also see below).
Other monogenic diabetes syndromes are associated with insulin resistance. For example, autosomal recessive forms of extreme insulin resistance are due to mutations in the insulin receptor gene (7). Genetic syndromes of lipodystrophy are typically associated with insulin resistance and diabetes. Dunnigan-type autosomal dominant familial partial lipoatrophic diabetes is due to mutations in the nuclear envelope protein lamin A/C (encoded by LMNA) (8) and dominant negative mutations in PPARγ (encoded by PPARG) (9). The mechanism whereby mutations in LMNA lead to this syndrome is unknown but may be due to disruption in nuclear function and resulting adipocyte death. PPARγ is well known to play a pivotal role in adipogenesis and insulin signaling, and thus it is logical that functional abnormalities would result in lipodystrophy and diabetes. Complete congenital lipoatrophic diabetes (also known as Berardinelli-Seip syndrome) is caused by mutations in 1-α-acylglycerol-3-phosphate O-acyltransferase 2 (encoded by AGPAT2) (10, 11) or seipin (encoded by BSCL2) (10, 12). Defects in AGPAT2 are likely to affect triacylglycerol synthesis in adipose tissue, resulting in triglyceride-depleted adipocytes and lipodystrophy. Seipin is a protein of unknown function expressed in the brain. Insulin resistance and diabetes are common phenotypes of lipodystrophy syndromes, despite quite disparate genetic etiologies, which highlights the critical role of adipose tissue in glucose homeostasis. Two common mechanisms for insulin resistance in the lipodystrophy syndromes are the deposition of triglycerides in extra-adipose tissues such as muscle and liver and the lack of key adipokines, particularly leptin and adiponectin, that influence glucose and fatty acid homeostasis.
New insights into the molecular basis of typical T2DM
In his 1987 Lilly Lecture, Defronzo elegantly described the triumvirate of β cell, muscle, and liver as a collusion responsible for T2DM (13). The idea that T2DM results from insulin resistance in muscle (causing decreased glucose uptake) and liver (causing increased gluconeogenesis), combined with declining β cell function is now widely accepted. As mentioned above, adipose tissue is now also recognized as an important player in glucose homeostasis. This conceptual framework has shaped how physicians treat patients with diabetes and has guided the identification of drug targets that reduce insulin resistance in muscle and liver and enhance and/or preserve β cell function.
Until recently, the genetic, molecular, and cellular basis for Defronzo’s triumvirate has remained unknown. Indeed, a triumvirate of new and powerful approaches — genetic, functional genomic, and transgenic — has begun to uncover the molecular and cellular basis of insulin resistance, the hallmark of T2DM. Multiple groups have utilized genome-wide and candidate gene–based approaches to begin to identify the genetic underpinnings of T2DM (1, 2, 14). To date, several chromosomal regions, most notably chromosomes 1q21–q24, 2q37, 3q24–q27, 4q32–q33, 11q24, 12q, and 20q have been identified as regions likely to harbor T2DM susceptibility genes. Furthermore, a few common variants in specific genes, each with modest effect, appear to be reproducibly associated with T2DM across several studies. These include calpain 10 (CAPN10) (15), Pro12Ala PPARγ (PPARG) (16), Glu23Lys potassium inward rectifying channel (KCNJ11) (17), and perhaps common variants in the islet-specific promoter of HNF4α (HNF4A) (18, 19) (other less-well-replicated candidate genes are reviewed in ref. 14). Although the biological basis for their associations with T2DM continue to be elucidated, variants in CAPN10 and PPARG appear to influence insulin sensitivity, while variants in KCNJ11 and HNF4A appear to influence β cell function and insulin secretion.
Beyond the Randle cycle
For several decades, it was proposed that the biochemical basis of hyperglycemia in T2DM could be explained by the Randle cycle, by which, simply stated, increased fatty acid oxidation causes a commensurate decrease in glucose oxidation, leading to decreased glucose uptake and hyperglycemia. Recently, functional genomics and transgenic approaches have identified a key common pathway that may play an important role in the pathophysiology of T2DM. Two studies utilized cDNA microarrays to examine differences in gene expression profiles among muscle of humans with T2DM, impaired glucose tolerance (prediabetes), and normal glucose tolerance (20, 21). Both studies found subtle decreases in expression of a subset of genes involved in mitochondrial oxidative phosphorylation (OXPHOS) in T2DM and prediabetic subjects. The subset of downregulated OXPHOS genes is known to be coordinately regulated by the PPARγ coactivators–1α and –1β (PGC-1α and PGC-1β), which were also subtly decreased in muscle tissue from diabetic and prediabetic subjects. Indeed, overexpression of PGC-1α in a mouse skeletal muscle cell line resulted in upregulation of the same set of genes found to be downregulated in human T2DM muscle (20). Transgenic mice overexpressing PGC-1α in muscle had increased formation of mitochondria-rich oxidative type 1 myofibers and reduced formation of glycolytic type 2 myofibers (22). In mice, PGC-1α has been shown to be a major mediator of cold-induced mitochondrial biogenesis and adaptive thermogenesis in brown fat (23, 24), and in liver, PGC-1α is a pivotal regulator of gluconeogenesis (25). Finally, recent evidence suggests that PGC-1α may also influence β cell energy metabolism and insulin release (26). While the role of PGC-1α in energy homeostasis is reasonably well established, the role of PGC-1β is less clear. Transgenic mice overexpressing PGC-1β have increased systemic fat oxidation and are resistant to diet-induced obesity, which suggests coordinated transcriptional effects on mitochondrial genes involved in fat oxidation and, relative to PGC-1α, little effect on mitochondrial genes involved in glucose metabolism (27).
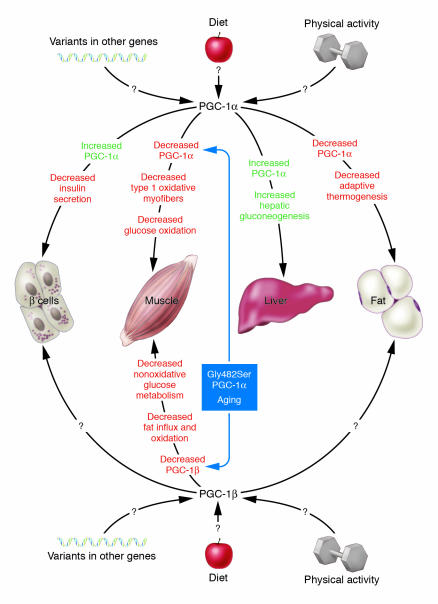
Thus, abnormal expression or function of PGC-1α and PGC-1β can potentially explain the quartet of muscle, liver, fat, and β cell dysfunction in T2DM (Figure 1). The reduction in PGC-1–regulated OXPHOS pathways in the pathogenesis of T2DM is consistent with other observations that implicate this pathway. Abnormalities in mitochondrial structure, number, and oxidative phosphorylation capacity in muscle in T2DM individuals and insulin-resistant offspring of T2DM individuals have been previously described (28, 29). The mechanism whereby exercise has beneficial effects on total body aerobic capacity (VO2max) and prevention and treatment of T2DM may be its well-known ability to increase oxidative phosphorylation capacity in muscle. Finally, as mentioned above, mutations in several genes involved in OXPHOS pathways including mitochondrial DNA (6), HNF4A (4, 18, 19), PPARG (16), and perhaps a common variant in PPARGC1 (the gene for PGC-1α) (ref. 30 and below) have been shown to be associated with T2DM.
Figure 1.
Schematic of the pleiotropic effects of PGC-1α and PGC-1β. Changes in PGC-1α and PGC-1β expression levels in different tissues may explain many of the metabolic abnormalities that accompany T2DM. In muscle: decreased PGC-1α may cause decreased formation of mitochondria-rich oxidative type 1 myofibers and decreased glucose oxidation. Decreased PGC-1β expression may cause decreased fat influx and oxidation and decreased nonoxidative glucose metabolism. Subjects with the Gly482Ser PGC-1α variant appear to be more susceptible to age-related decreases in PGC-1α and PGC-1β. In the β cell: increased PGC-1α expression or function, which is observed in several animal models of diabetes, would be expected to decrease β cell dysfunction and decrease insulin secretion. In liver: increases in PGC-1α expression or function would be expected to cause increased hepatic gluconeogenesis. In fat: decreases in PGC-1α expression or function would be expected to cause decreased mitochondrial biogenesis and defective adaptive thermogenesis, possibly leading to positive energy balance and obesity. Other factors, i.e., physical activity, diet, and other gene variants, may affect PGC-1α and PGC-1β expression or function and thus susceptibility to T2DM.
In this issue of the JCI, Ling and coworkers (31) used an elegant study design comparing young and old monozygotic and dizygotic twins in order to dissect genetic and nongenetic influences on PGC-1α and PGC-1β expression and biology. They found that muscle PGC-1α and PGC-1β mRNA levels decrease with aging, which suggests a potential link between this pathway and age-related decline in glucose homeostasis (Figure 1). Interestingly, only subjects with the 482Ser allele of PPARGC1 were found to have this age-related decrease in PGC-1α and PGC-1β mRNA levels, which may provide a partial explanation for the large inter-individual variation in susceptibility to age-related decline in glucose homeostasis. Decreased PGC-1α and PGC-1β levels in elderly 482Ser PPARGC1 homozygotes were associated with decreased VO2max, which may provide a mechanistic connection between PGC-1α and PGC-1β and mitochondrial number or function. In addition, muscle PGC-1α and PGC-1β mRNA levels were found to be determinants of glucose transporter 4 expression and glucose and fat metabolism during a hyperinsulinemic euglycemic clamp study. PGC-1α predominantly influenced glucose disposal and oxidation, while PGC-1β predominantly influenced fat oxidation and nonoxidative glucose metabolism. Thus, while from a clinical standpoint, we have known that both genetic and environmental influences are important determinants of insulin resistance and T2DM, this study provides mechanistic molecular insights into this complex interaction.
The future: what we don’t know
The results reported by Ling et al. (31) are, for the most part, consistent with and extend the findings of others regarding the importance of PGC-1–regulated mitochondrial pathways in glucose homeostasis and the pathophysiology of T2DM. However, there are several caveats that are worth mention and further consideration. For these physiological studies it was necessary to exclude twins with known diabetes. However, this might have preferentially excluded those individuals with a high genetic burden of T2DM susceptibility genes or individuals exposed to a diabetogenic environment from the older group compared with the younger group, since the older group would have had more time to express the diabetic (exclusionary) phenotype. This potential ascertainment bias might be expected to underestimate any differences in diabetes-related traits between young and old groups. Second, direct comparisons of young and old groups are potentially confounded by a cohort effect. For example, nutritional or other environmental factors that may have differed over the approximately 40-year span between the birth of the young and old cohorts could have important influences on the outcomes measured, particularly by virtue of birth weight and intrauterine effects on adult glucose metabolism (32). Third, although decreased PGC-1α and PGC-1β expression are likely to be causally related to T2DM pathogenesis, the correlative cross-sectional nature of this study does not prove causality. Fourth, the authors propose that association of the Gly482Ser PPARGC1 variant with decreases in both PGC-1α and PGC-1β mRNA levels may be due to decreased function of the Gly482Ser variant upon activation of transcription of its own gene and PGC-1β. It is also possible that the Gly482Ser variant is in linkage disequilibrium with a 5′ or 3′ polymorphism that affects transcription or mRNA stability. Finally, it is argued that monozygotic twins are genetically identical. Although they are clearly more genetically similar to each other than dizygotic twins, it should be kept in mind that even monozygotic twins have potentially important differences in their genetic makeup — most notably for this study, differences in mitochondrial DNA, as well as differences in genomic DNA methylation that may alter expression of individual genes. These variations could affect the expression of PGC-1α and PGC-1β and other OXPHOS genes and pathways, creating genetic differences even in monozygotic twins.
In summary, genetic, transgenic, and functional genomics approaches have provided fruitful insights into the complex molecular pathways that regulate energy homeostasis. These approaches have identified molecules and pathways that could not have been anticipated from classical clinical investigative studies. The challenge for the future is to integrate these distinct molecular pathways in order to reconstruct the phenotype. Understanding how aging, diet, exercise, and other extrinsic factors affect these pathways poses a special challenge. A more complete understanding of the biology will lead to novel preventive and therapeutic modalities. Studies such as that of Ling and coworkers (31) have taken the first steps toward this goal.
Footnotes
See the related article beginning on page 1518.
Nonstandard abbreviations used: HNF, hepatocyte nuclear factor; IPF-1, insulin promoter factor-1; MIDD, maternally inherited diabetes and deafness; OXPHOS, oxidative phosphorylation; PGC, PPARγ coactivator; T2DM, type 2 diabetes mellitus; VO2max, total body aerobic capacity.
Conflict of interest: Alan R. Shuldiner serves as a consultant for Amgen, Pfizer, Inc., and Serono, Inc. He receives research support from Merck & Co., Inc.
References
- 1.Elbein SC. Perspective: the search for genes for type 2 diabetes in the post-genome era. Endocrinology. 2002;143:2012–2018. doi: 10.1210/endo.143.6.8831. [DOI] [PubMed] [Google Scholar]
- 2.Horenstein RB, Shuldiner AR. Genetics of diabetes. Rev. Endocr. Metab. Disord. 2004;5:25–34. doi: 10.1023/B:REMD.0000016122.84105.75. [DOI] [PubMed] [Google Scholar]
- 3.Weyer C, Bogardus C, Mott DM, Pratley RE. The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes mellitus. J. Clin. Invest. 1999;104:787–794. doi: 10.1172/JCI7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fajans SS, Bell GI, Polonsky KS. Molecular mechanisms and clinical pathophysiology of maturity-onset diabetes of the young. N. Engl. J. Med. 2001;345:971–980. doi: 10.1056/NEJMra002168. [DOI] [PubMed] [Google Scholar]
- 5.Huopio H, et al. A new subtype of autosomal dominant diabetes attributable to a mutation in the gene for sulfonylurea receptor 1. Lancet. 2003;361:301–307. doi: 10.1016/S0140-6736(03)12325-2. [DOI] [PubMed] [Google Scholar]
- 6.Kadowaki T, et al. A subtype of diabetes mellitus associated with a mutation of mitochondrial DNA. N. Engl. J. Med. 1994;330:962–968. doi: 10.1056/NEJM199404073301403. [DOI] [PubMed] [Google Scholar]
- 7.Taylor SI, et al. Mutations in the insulin receptor gene. Endocr. Rev. 1992;13:566–595. doi: 10.1210/edrv-13-3-566. [DOI] [PubMed] [Google Scholar]
- 8.Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in Canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 9.Hegele RA, Cao H, Frankowski C, Mathews ST, Leff T. PPARG F388L, a transactivation-deficient mutant, in familial partial lipodystrophy. Diabetes. 2002;51:3586–3590. doi: 10.2337/diabetes.51.12.3586. [DOI] [PubMed] [Google Scholar]
- 10.Garg A. Acquired and inherited lipodystrophies. N. Engl. J. Med. 2004;350:1220–1234. doi: 10.1056/NEJMra025261. [DOI] [PubMed] [Google Scholar]
- 11.Agarwal AK, et al. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 2002;31:21–23. doi: 10.1038/ng880. [DOI] [PubMed] [Google Scholar]
- 12.Magre J, et al. Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat. Genet. 2001;28:365–370. doi: 10.1038/ng585. [DOI] [PubMed] [Google Scholar]
- 13.DeFronzo RA. Lilly lecture 1987. The triumvirate: beta-cell, muscle, liver. A collusion responsible for NIDDM. Diabetes. 1988;37:667–687. doi: 10.2337/diab.37.6.667. [DOI] [PubMed] [Google Scholar]
- 14.Sabra, M., Shuldiner, A.R., and Silver, K. 2004. Candidate genes for type 2 diabetes mellitus. In Diabetes mellitus: a fundamental and clinical text. 3rd edition. D. LeRoith, S.I. Taylor, and J.M. Olefsky, editors. Lippincott Williams & Wilkins. Philadelphia, Pennsylvania, USA. 1003–1012.
- 15.Horikawa Y, et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat. Genet. 2000;26:163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 16.Altshuler D, et al. The common PPAR-gamma pro12ala polymorphism is associated with decreased risk of type 2 diabetes. Nat. Genet. 2000;26:76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 17.Gloyn AL, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003;52:568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- 18.Love-Gregory LD, et al. A common polymorphism in the upstream promoter region of the hepatocyte nuclear factor-4 alpha gene on chromosome 20q is associated with type 2 diabetes and appears to contribute to the evidence for linkage in an Ashkenazi Jewish population. Diabetes. 2004;53:1134–1140. doi: 10.2337/diabetes.53.4.1134. [DOI] [PubMed] [Google Scholar]
- 19.Silander K, et al. Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53:1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 20.Mootha VK, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 21.Patti ME, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. U. S. A. 2003;100:8466–8471. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 23.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 24.Puigserver P, Spiegelman BM. Peroxisome proliferator-activated receptor-gamma coactivator 1 alpha (PGC-1 alpha): transcriptional coactivator and metabolic regulator. Endocr. Rev. 2003;24:78–90. doi: 10.1210/er.2002-0012. [DOI] [PubMed] [Google Scholar]
- 25.Yoon JC, et al. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–138. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 26.Yoon JC, et al. Suppression of beta cell energy metabolism and insulin release by PGC-1alpha. Dev. Cell. 2003;5:73–83. doi: 10.1016/s1534-5807(03)00170-9. [DOI] [PubMed] [Google Scholar]
- 27.Kamei Y, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl. Acad. Sci. U. S. A. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley DE, He J, Menshikova EV, Ritov VB. Dysfunction of mitochondria in human skeletal muscle in type 2 diabetes. Diabetes. 2002;51:2944–2950. doi: 10.2337/diabetes.51.10.2944. [DOI] [PubMed] [Google Scholar]
- 29.Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N. Engl. J. Med. 2004;350:664–671. doi: 10.1056/NEJMoa031314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ek J, et al. Mutation analysis of peroxisome proliferator-activated receptor-gamma coactivator-1 (PGC-1) and relationships of identified amino acid polymorphisms to type II diabetes mellitus. Diabetologia. 2001;44:2220–2226. doi: 10.1007/s001250100032. [DOI] [PubMed] [Google Scholar]
- 31.Ling C, et al. Multiple environmental and genetic factors influence skeletal muscle PGC-1α and PGC-1β gene expression in twins. J. Clin. Invest. 2004;114:1518–1526. doi:10.1172/JCI200421889. doi: 10.1172/JCI21889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hales CN, et al. Fetal and infant growth and impaired glucose tolerance at age 64. BMJ. 1991;303:1019–1022. doi: 10.1136/bmj.303.6809.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]