SUMMARY
Amino acid taste is expected to be a universal property among animals. Although sweet, bitter, salt, and water tastes have been well characterized in insects, the mechanisms underlying amino acid taste remain elusive. From a Drosophila RNAi screen we identify an ionotropic receptor, Ir76b, as necessary for yeast preference. Using calcium imaging, we identify Ir76b+ amino acid taste neurons in legs, overlapping partially with sweet neurons but not those that sense other tastants. Ir76b mutants have reduced responses to amino acids, which are rescued by transgenic expression of Ir76b, and a mosquito ortholog AgIr76b. Co-expression of Ir20a with Ir76b is sufficient for conferring amino acid responses in sweet taste neurons. Notably, Ir20a also serves to block salt response of Ir76b. Our study establishes the role of a highly conserved receptor in amino acid taste, and suggests a mechanism for mutually exclusive roles of Ir76b in salt and amino acid-sensing neurons.
ETOC BLURB
Ganguly et al demonstrate that Ir76b mediates cellular and behavioral responses to amino acids which underlie post-mating yeast and amino acid feeding preferences of Drosophila females. Ir20a, possibly one among many factors, plays a role in changing Ir76b activity from an ungated salt receptor to an amino acid-gated receptor.
INTRODUCTION
The importance of dietary protein and amino acids has been investigated for several insects including Drosophila, and reveals that, like mammals, insects must acquire some essential amino acids via foods (Golberg and De Meillon, 1948; Hinton et al., 1951; House, 1962; Singh and Brown, 1957). Females, in particular, require large supplies of amino acids for synthesizing egg yolk (Dimond et al., 1956). Restriction of amino acids thus has a direct impact on female fecundity (Chang, 2004; Dimond et al., 1956; Fink et al., 2011). Amino acid deprivation also significantly affects larval growth and development, as well as adult life span (Baltzer et al., 2009; Britton and Edgar, 1998; Chang, 2004; Grandison et al., 2009; Vrzal et al., 2010).
Given the importance of amino acids in food sources, it is perhaps not surprising that insects demonstrate taste sensitivity to amino acids. Behavioral analyses in various insects, including honeybees, ants, and the dengue fever vector, Aedes aegypti, show that mixtures of some amino acids and sugar are preferred over sugar alone (Alm et al., 1990; Ignell et al., 2010; Wada et al., 2001). Moreover, electrophysiological recordings show that selected amino acids evoke action potentials in taste hairs of some insects. For instance, in blowflies and fleshflies some individual amino acids were found to activate either sweet- or salt-sensing neurons; others were found to have inhibitory effects on these taste neurons (Shiraishi and Kuwabara, 1970). Studies in blood-feeding tsetse flies identified neurons in tarsal taste hairs that are exquisitely sensitive to several individual amino acids, as well as to a mixture of amino acids that are found in human sweat (van der Goes van Naters and den Otter, 1998). Amino acid-sensing neurons have also been described in cabbage butterflies (Van Loon and Van Eeuwijk, 1989) and Helicoverpa moths (Zhang et al., 2011; Zhang et al., 2010).
Drosophila exhibit strong feeding preference for yeasts and yeast extract, which serve as a major source of protein (Tatum, 1939). Mated females, as well as adult flies fed on a protein deficient diet, can identify and select yeast over sucrose in binary choice assays (Ribeiro and Dickson, 2010; Vargas et al., 2010). A recent study reports behavioral taste sensitivity to free amino acids, albeit only in flies raised on a diet lacking in protein (Toshima and Tanimura, 2012). In these experiments, flies extended their proboscis upon stimulation of taste hairs with amino acid solutions, indicating a role for taste hairs as amino acid sensors. However, little is known about the molecular and cellular basis of amino acid taste.
Many amino acids taste savory or sweet to humans. Mammals detect amino acids using a heteromeric receptor comprised of two subunits, T1R1 and T1R3, expressed in fungiform taste buds (Nelson et al., 2002). The T1R1/T1R3 receptor has broad specificity for L-amino acids and does not respond to the D isomers. T1Rs, which are G protein-coupled receptors related to metabotropic glutamate receptors, have no counterparts in insect genomes.
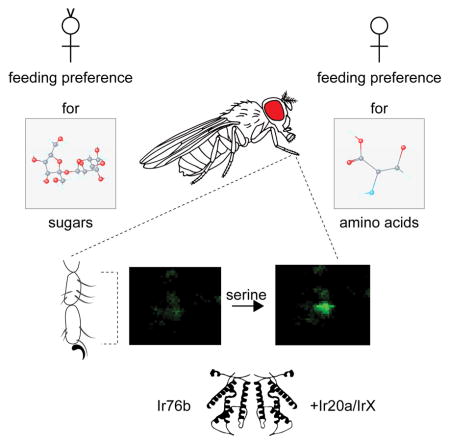
Here, we investigated behavioral and cellular responses in the fly to amino acids, identifying them as critical cues for feeding preference to yeast extract. We find that mated females exhibit feeding preference for individual amino acids, which are preferred to different extents in binary choice experiments with sucrose. From an RNAi screen we identify a requirement for a highly conserved chemosensory ionotropic receptor, Ir76b, in mediating feeding preference for yeast extract. Using genetic silencing and calcium imaging experiments, we characterize the role of Ir76b+ neurons in behavioral and cellular responses to amino acids in mated females. We find that responses to all tested amino acids are abolished in Ir76b mutants, and rescued by transgenic expression of Ir76b. Moreover, Ir76b function is conserved across millions of years of evolution – expression of the Ir76b ortholog from Anopheles gambiae also rescues the behavioral deficits in Ir76b mutant flies. Ir76b has been recently described as a salt taste receptor (Zhang et al., 2013), however we find that amino acid-sensing neurons do not respond to salt. Analysis of additional candidates from our initial RNAi screen reveal additional Irs involved in amino acid taste. Co-expression of one of these, Ir20a, with Ir76b, is sufficient to confer amino acid sensitivity to sweet taste neurons. Moreover, the presence of Ir20a blocks Ir76b-mediated salt response as measured in cellular and behavioral assays. Taken together, our results demonstrate a highly conserved gustatory role for Ir76b in detection of amino acids, in addition to its function as a salt taste receptor. Our studies also identify a potential role for Ir20a in facilitating mutually exclusive functions of Ir76b in salt and amino acid taste neurons.
RESULTS
Amino acids mediate sexually dimorphic feeding preference for yeast extract
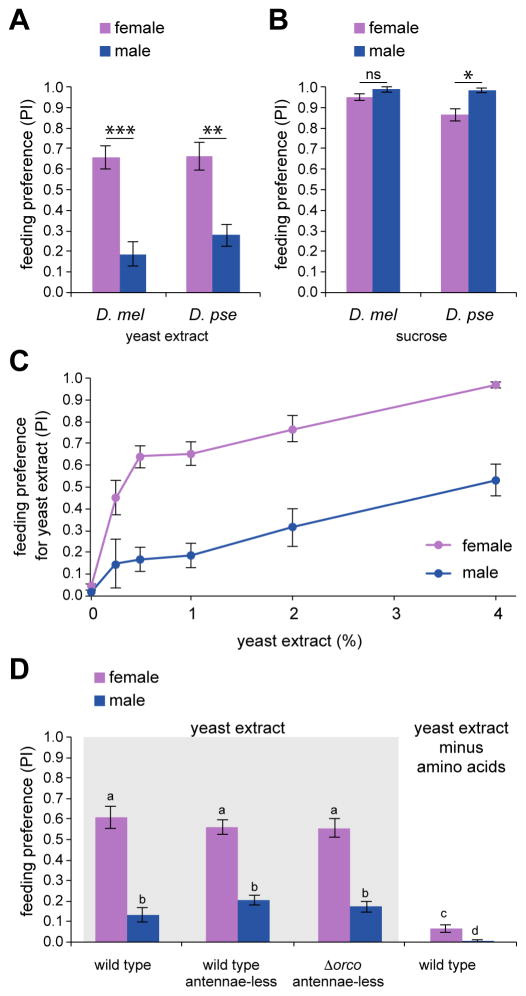
To explore mechanisms underlying yeast taste detection, we first characterized feeding responses to yeast extract, which contains free amino acids, peptides, sugars, and salts as well as various B vitamins. We used binary choice feeding tests in which batches of cohabiting male and female flies were offered a choice between 5 mM sucrose and 1% yeast extract, after which their yeast preference indices (PI) were computed separately. By testing D. melanogaster and a distantly related species, D. pseudoobscura, we found that female flies of both species preferred yeast extract to a greater extent than male flies (Figure 1A). Control experiments with sucrose alone revealed little if any sex-specific variation in sugar feeding (Figure 1B). Relative preference for yeast extract was concentration-dependent in both sexes, but nevertheless lower in males than observed in females (Figure 1C). In addition, flies without antennae (wild type, antennae-less) or without any functional olfactory neurons (Benton et al., 2009; Larsson et al., 2004) (Δorco, antennae-less) showed the same preference for yeast extract as intact wild type flies (Figure 1D), suggesting the capability to evaluate it as a food source even in the absence of olfactory input. Surprisingly, flies failed to display the same behavioral preference for yeast extract depleted of amino acids (Figure 1D). Together, the results suggest that amino acids mediate behavioral responses to yeast extract.
Figure 1. Yeast preference of Drosophila females is driven by amino acids.
(A and B) Mean preference indices (PI) of Drosophila melanogaster (D. mel) and Drosophila pseudoobscura (D. pse) to 1% yeast extract (pink dye; tested against 5 mM sucrose, blue) and 5 mM sucrose (blue dye; tested against water, pink) obtained from binary feeding assays. n=6–7 (D. mel), n=6–12 (D. pse). *P<0.05, **P<0.01, ***P<0.001, Mann-Whitney U test.
(C) Behavioral responses to indicated concentrations of yeast extract (pink dye) tested against 5 mM sucrose (blue dye) in binary choice feeding tests. Genotype was w1118. n=8–13.
(D) Mean PI of intact w1118 (wild type), and w1118 (wild type, antennae-less) or ΔOr83b2 mutants (Δorco, antennae-less) with surgically ablated antennae, obtained from binary feeding tests with choices between 1% yeast extract with or without amino acids (pink dye) as indicated and 5 mM sucrose (blue dye). n=10–18. Different letters indicate significantly different groups, P<0.05, two-way ANOVA with pairwise comparison.
Individual amino acids are preferred to different extents
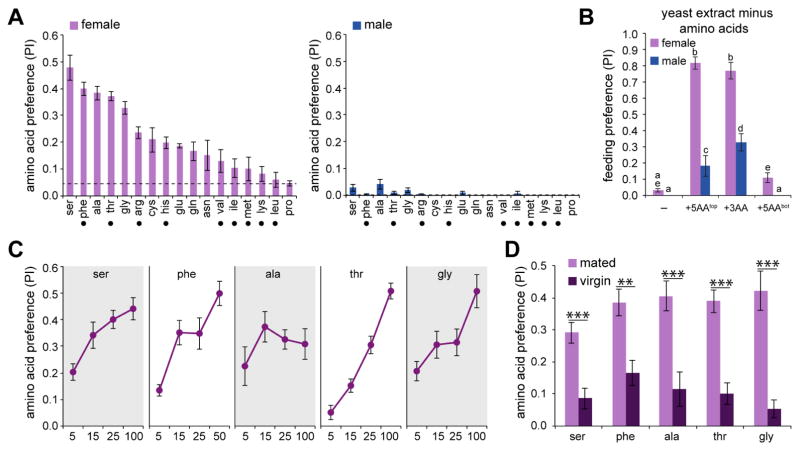
We next characterized behavioral responses to individual amino acids. We elected to test individual L-amino acids in binary choice tests with 5 mM sucrose, which evokes robust feeding responses by itself. Amino acids were tested at 25 mM, which is within the range reported in a number of commercially available yeast extracts, and feeding choice was monitored along with overall feeding participation of both males and females. The test conditions were so chosen to reveal variations in preferences for individual amino acids, which could be more easily observed relative to sucrose. We found that different amino acids were preferred to greatly different extents relative to sucrose (Figure 2A), as compared to the results of a previous study in which mean preferences of individual amino acids were found to be more similar to each other (Toshima and Tanimura, 2012). As observed for yeast extract, male flies tested in parallel showed little or no preference for any of the amino acids when sucrose was offered as an alternative (Figure 2A).
Figure 2. Mated females show increased preference for some amino acids.
(A) Mean PI for 25 mM of indicated amino acid (pink dye; tested against 5 mM sucrose, blue dye). Dashed lines indicate preference for water solvent control (pink dye) tested against 5 mM sucrose (blue dye). Black dots indicate the essential amino acids. n=6–24.
(B) Mean PI for 1% amino acid-deprived yeast extract alone (–) or supplemented with 25 mM of each of the top five amino acids (+5AAtop) from (A), or 25 mM each of serine, threonine, and phenylalanine (+3AA), or 25 mM of each of the bottom five amino acids (+5AAbot) from (A). Each of these combinations (pink dye) were tested against 5 mM sucrose (blue dye). n=7–12. For each stimulus, different letters indicate significantly different groups, P<0.05, two-way ANOVA with pairwise comparison.
(C) Mean PI of mated females for indicated concentrations of each of the top five amino acids (pink dye) tested against 5 mM sucrose (blue dye). For each concentration, n=5–11.
(D) Mean PI of mated or virgin females for 25 mM of named amino acid (pink dye) tested against 5 mM sucrose (blue dye). n=6. **P<0.01, ***P<0.001, two-way ANOVA with pairwise comparison. For all experiments, genotype was w1118. See also Figures S1 and S2.
The strongest behavioral responses were elicited by serine, phenylalanine, alanine, threonine, and glycine. We performed additional control experiments to examine the validity of these responses. First, we tested L- and D-phenylalanine in parallel and found that, by contrast to L-phenylalanine, flies exhibit no preference for the D-isomer over sucrose (Figure S1). We then tested the L and D isomers of phenylalanine against each other, and as predicted, found that flies preferred L-phenylalanine over D-phenylalanine (Figure S1). Simultaneous tests in which L-phenylalanine was added to both dyes yielded an iso-preference index (PI = 0.4563±0.0353, mean±s.e.m., n=6). We then carried out a series of experiments in which we tested serine against one of three other amino acids. The results from these experiments were consistent with the preferences derived from binary choice assays with sucrose, and showed that serine was preferred to the same extent as phenylalanine, slightly preferred over glycine, and strongly preferred over proline (Figure S2). Flies showed equal preference (PI = 0.5083±0.0083, mean±s.e.m., n=6) in control experiments in which serine was added to both dyes (Figure S2).
The five amino acids that elicited the strongest preference were together sufficient to restore behavioral activity to amino acid-deprived yeast extract (Figure 2B). Among these, phenylalanine and threonine are essential dietary amino acids (Sang and King, 1961). Glycine, although not essential, is required for normal growth and development (Sang and King, 1961). The activity of the five amino acids was mimicked by a subset of three, which included two essential amino acids, phenylalanine and threonine, along with serine (Figure 2B). In fact, males exhibited a higher preference for this mixture than to yeast extract alone, possibly due to differences in relative amounts of these and other amino acids in yeast extract. Using the same paradigm, we tested a mixture of the five amino acids that elicited the weakest preference and found that this mixture did not confer any change in palatability (Figure 2B).
Mean feeding preferences elicited by each of the top five amino acids typically increased in a concentration dependent manner, across a range up to 100 mM (50 mM for phenylalanine, due to limitations of solubility) (Figure 2C). Moreover, experiments to compare behavioral preferences of mated and virgin females revealed that the choice to feed on amino acids was significantly elevated upon mating (Figure 2D), consistent with previous studies that showed increased yeast preference in mated females (Ribeiro and Dickson, 2010; Vargas et al., 2010).
An RNAi screen identifies a requirement for Ir76b in mediating feeding preference for yeast extract
To identify receptors involved in mediating feeding preference for yeast extract, we used RNAi to knock down expression of candidate genes using the pan-neuronal elav-GAL4 driver and tested adult flies in binary choice assays with sucrose and yeast extract. We focused on Ionotropic receptor (Ir) genes (Benton et al., 2009), which have been associated with amine and acid sensing in the fly olfactory system (Ai et al., 2010; Min et al., 2013; Silbering et al., 2011), and more recently have been found to be expressed in taste neurons as well (Koh et al., 2014). We found that preference for yeast extract was weakest in Ir76b-RNAi females (Figure S3A). Notably Ir76b, whose expression was previously reported in both olfactory and gustatory tissues (Benton et al., 2009), was among chemoreceptor genes expressed at high levels in taste tissue transcriptomes (rpkm=16.49 in female proboscis, rpkm=5.92 in female legs, n=2). Mean preference for yeast extract was reduced for a number of other Ir-RNAi lines, including for some reported to be expressed in taste neurons, such as Ir20a, Ir47a, Ir52a, Ir52d, Ir56a, and Ir56d (Koh et al., 2014). Although a significant reduction was observed only for Ir20a-RNAi, a few other candidates yielded PI values for most independent trials that were lower than the value of mean PI–standard deviation of the GAL4 control (Figure S3B).
We confirmed the absence of a role for Ir8a and Ir25a, the other broadly expressed receptors, by testing available null mutants, neither of which showed significant reduction in preference for yeast extract (Figure S4). Interestingly, in a few instances Ir-RNAi lines caused significant increases in preference for yeast extract as compared to elav-GAL4 controls. These results raise the possibility that some Irs may be involved in detecting components of yeast extract, either volatile or non-volatile, that are repulsive to some degree. Alternatively, since flies were always tested in the context of a choice between sucrose and yeast extract, Ir genes associated with this phenotype may be involved in sucrose response.
Tarsal Ir76b+ neurons respond to amino acids
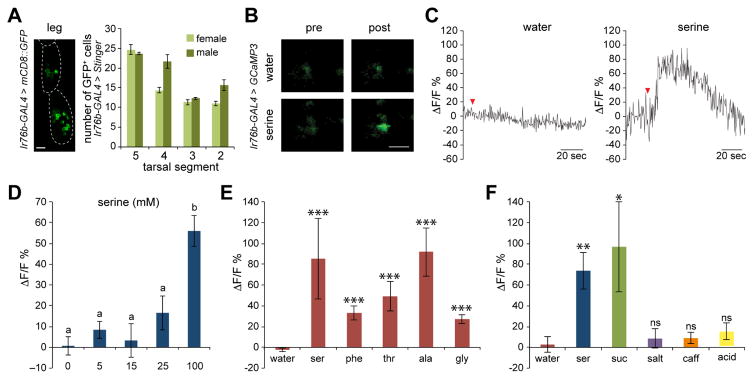
We found no evidence for amino acid sensitivity in a previous electrophysiological analysis of L-type taste hairs (Dahanukar et al., 2007), which are among the extensively characterized taste sensilla of the labellum. We therefore focused on characterizing the role of tarsal Ir76b+ neurons in detection of amino acids. Ir76b-GAL4 is broadly expressed in the tarsi, and reporter expression could be visualized in ~10–25 neurons in each of the four distal tarsal segments in both sexes (Figure 3A), suggesting that Ir76b may label multiple cells in each sensillum. We expressed GCaMP3 under the control of Ir76b-GAL4 and measured tastant-evoked changes in calcium activity in neurons of the fourth and fifth tarsal segments. We first measured responses to serine, the amino acid that evoked the strongest behavioral response. Tarsi of mated female flies were stimulated with 100 mM serine and changes in GCaMP3 fluorescence were monitored in Ir76b-GAL4 cells in the fourth and fifth segments of the tarsi. Application of 100 mM serine resulted in a significant increase in fluorescence, which was not observed with water alone (Figures 3B and 3C, Movies S1 and S2), and mean change in fluorescence increased with serine concentrations between 15 and 100 mM (Figure 3D). The threshold concentration of serine for visualizing a single cell response was higher than that observed for a behavioral response, as has been previously reported for sugars (Dahanukar et al., 2007). The number of serine-activated neurons ranged from 1–9 in the different samples; differences in the number and intensity of labeled cells, along with differences in alignment of individual preparations, likely contribute to some of the observed variability. A larger fraction of serine-responsive cells were obtained from the 5th tarsal segment (18 of 21) as compared to the 4th segment (3 of 21), but we could not map the identity of hairs innervated by the activated cells.
Figure 3. Ir76b-GAL4 neurons in female tarsi are activated by amino acids.
(A) Representative image of GFP+ cells in distal segments of female tarsi (left) and mean numbers of GFP+ cells in indicated tarsal segments in female and male flies (right). n=3; error bars indicate s.e.m. Genotypes were Ir76b-GAL4RB; UAS-mCD8::GFP (left) and Ir76b-GAL4RB; UAS-Stinger (right). Scale bar=20 μM.
(B) Images of GCaMP3 fluorescence in a representative cell before (pre) and after (post) application of water control or 100 mM serine. Genotype was Ir76b-GAL4RB; UAS-GCaMP3. Scale bar=10 μM.
(C) Mean change in fluorescence (ΔF/F %) in representative Ir76b-GAL4RB; UAS-GCaMP3 cells in forelegs of female flies. Red arrowheads denote application of water control or 100 mM serine as indicated.
(D) Mean percent changes in GCaMP3 fluorescence after application of indicated concentration of serine. n=10–21 cells from 2–5 flies tested per concentration. Measurements were taken from cells that responded to 100 mM serine. For each stimulus, different letters indicate significantly different groups, P<0.05, one-way ANOVA with Tukey’s post hoc test.
(E) Mean percent changes in GCaMP3 fluorescence after application of indicated stimuli. Amino acids were tested at 100 mM, except phenylalanine, which was tested at 50 mM. ***P<0.001, Mann-Whitney U tests versus water. n=8–41 cells from 3–10 flies per stimulus.
(F) Mean percent changes in GCaMP3 fluorescence after application of indicated stimuli. Measurements were taken from 100 mM serine-responsive cells. *P<0.05, **P<0.01, Mann-Whitney U tests versus water. n=10–13 cells from 4–5 flies.
See also Figure S5.
We next measured responses to each of the top five amino acids that evoked strong behavioral responses, and observed stimulus-evoked increases in fluorescence in each case (Figure 3E). Each amino acid was tested at a concentration of 100 mM (except phenylalanine at 50 mM). Interestingly, Ir76b-GAL4 neurons in male tarsi did not show strong responses to the five amino acids (Figure S5), suggesting that sex-specific differences in peripheral sensitivities may account for differences in taste preference, at least in part.
Taste neurons are typically divided into sub-populations that are selective for a single taste category (Freeman and Dahanukar, 2015). To determine the specificity of Ir76b-GAL4 cells we measured activity of serine-activated cells to other categories of tastants. First, we identified neurons activated by 100 mM serine. For these experiments, the focal plane was selected to visualize such cells that could be easily identified by their position and relative arrangement among labeled cells for sequential application and imaging using other stimuli. We then applied other tastants and measured calcium activity in these identified cells. We found that serine-responsive cells were not activated by 50 mM NaCl, 10 mM caffeine, or pH 2 HCl (Figure 3F). However, 10 of 13 such neurons were activated by sucrose (Figure 3F), revealing overlap between serine- and sucrose-sensing neurons in the tarsi. It is possible that this may have some functional significance, as interactions between sweet and amino acid taste have been reported previously (Alm et al., 1990; Wada et al., 2001). By contrast, the detection of other categories of tastants appears to occur independently.
Ir76b is necessary for cellular responses to amino acids
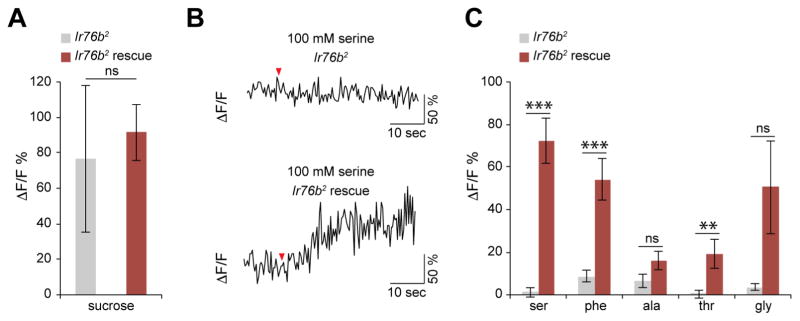
To investigate the role of Ir76b in cellular responses to amino acids, we measured responses in Ir76b mutant flies. We obtained a null allele, Ir76b2, which was generated by imprecise excision of a P-element inserted in the third intron of Ir76b (Zhang et al., 2013). We compared responses of Ir76b2 mutants with those of flies in which Ir76b expression was rescued using the Ir76b-GAL4 driver. Imaging analysis revealed that response to 100 mM sucrose was not significantly different between the mutant and rescue flies (Figure 4A). However, expression of UAS-Ir76b resulted in significantly increased responses to serine, phenylalanine and threonine (Figures 4B and 4C). Although not significant, mean fluorescence changes in response to alanine and glycine were also higher in the rescue flies than those observed in the mutant (P=0.0881, n=11–16 and P=0.0554, n=25–30 respectively, Mann-Whitney U tests versus Ir76b2 mutant). These results demonstrate that Ir76b is necessary for taste neuron responses to amino acids.
Figure 4. Ir76b is necessary for cellular responses to amino acids.
(A) Mean percent changes in GCaMP3 fluorescence in Ir76b2 (Ir76b-GAL4RB; Ir76b2, UAS-GCaMP3) and Ir76b2 rescue (Ir76b-GAL4RB; Ir76b2, UAS-Ir76b/Ir76b2, UAS-GCaMP3) flies upon application of 100 mM sucrose. n=18–32 cells from 3–5 flies, Mann-Whitney U tests.
(B) Representative ΔF/F traces showing changes in GCaMP3 fluorescence in Ir76b2 (Ir76b-GAL4RB; Ir76b2, UAS-GCaMP3) and Ir76b2 rescue (Ir76b-GAL4RB; Ir76b2, UAS-Ir76b/Ir76b2, UAS-GCaMP3) flies upon application of 100 mM serine.
(C) Mean percent changes in GCaMP3 fluorescence in Ir76b2 (Ir76b-GAL4RB; Ir76b2, UAS-GCaMP3) and Ir76b2 rescue (Ir76b-GAL4RB; Ir76b2, UAS-Ir76b/Ir76b2, UAS-UGCaMP3) flies upon application of indicated stimuli. Amino acids were tested at 100 mM, except phenylalanine at 50 mM. n=11–32 cells from 2–7 flies (amino acids). **P<0.01, ***P<0.001, Mann-Whitney U tests.
Ir76b is necessary for behavioral responses to amino acids
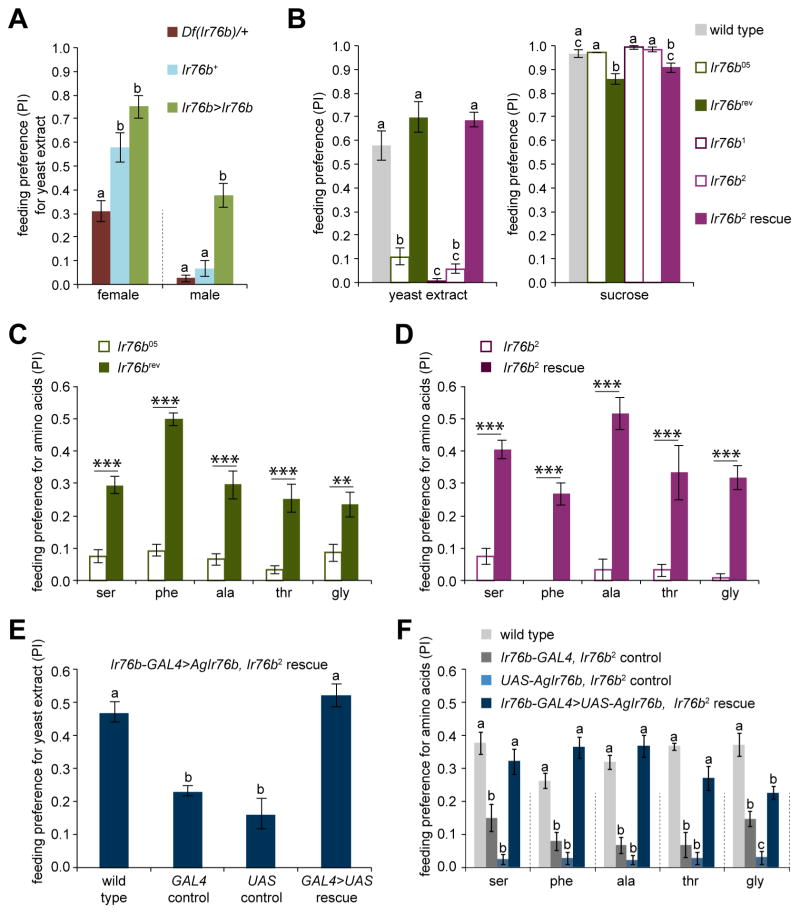
To begin to examine the contribution of Ir76b in driving behavioral responses to yeast extract and amino acids we investigated the effect of varying the gene dosage of Ir76b. Females with one copy of Ir76b (Df(Ir76b)/+) showed a reduced mean preference for yeast extract as compared to control flies bearing two copies of the gene (Ir76b+) (Figure 5A). Similarly, overproduction of Ir76b via the GAL4/UAS system (Ir76b>Ir76b) caused an increase in mean response to yeast extract in both sexes, and significantly so in male flies (Figure 5A). Together, these results support a role for Ir76b in the behavioral switch from sucrose to yeast preference.
Figure 5. Ir76b is necessary for behavioral responses to amino acids.
(A) Mean PI for 1% yeast extract (pink) tested against 5 mM sucrose (blue) of Df(3L)XS533/+ (Df(Ir76b)/+), w1118 (Ir76b+) and Ir76b-GAL4RB; UAS-Ir76b (Ir76b>Ir76b) flies. n=5–9. For each sex, different letters indicate significantly different groups two-way ANOVA with pairwise comparison.
(B) Mean PI of mated female flies for 1% yeast extract (pink; tested against 5 mM sucrose, blue; left) and 5 mM sucrose (pink; tested against water, blue; right) obtained from binary feeding tests. Genotypes were as follows: w1118 (wild type), Ir76b05/Ir76b05 (Ir76b05), Ir76b05 precise excision (Ir76brev), Ir76b1/Ir76b1 (Ir76b1), Ir76b2/Ir76b2 (Ir76b2), Ir76b-GAL4RB/Ir76b-GAL4RB; Ir76b2, UAS-Ir76b/Ir76b2, UAS-Ir76b (Ir76b2 rescue). n=6–10 (yeast extract) and n=8–12 (sucrose). For each stimulus, different letters indicate significantly different groups, P<0.05, one-way ANOVA with Tukey’s post hoc test.
(C) Behavioral responses of mated females of Ir76b05 mutant and precise excision revertant (Ir76brev) genotypes to indicated amino acids (25 mM, pink) tested in binary choice tests with 5 mM sucrose (blue). n=5–12. **P<0.01, ***P<0.001, two-way ANOVA with pairwise comparison.
(D) Behavioral responses of mated females of Ir76b2 mutants and transgenic rescue flies (Ir76b-GAL4RB; Ir76b2, UAS-Ir76b) to indicated amino acids (25 mM, pink) tested in binary choice tests with 5 mM sucrose (blue). n=5–9. ***P<0.001, two-way ANOVA with pairwise comparison.
(E) Mean PI of mated females for 1% yeast extract (pink; tested against 5mM sucrose, blue) obtained from binary feeding tests. Genotypes were as follows: w1118 (wild type); Sp/CyO; Ir76b1, Ir76b-GAL4 (GAL4 control); UAS-AgIr76b; Ir76b1 (UAS control); UAS-AgIr76b; Ir76b1, Ir76b-GAL4 (GAL4>UAS rescue). n=6–18. The different letters indicate significantly different groups, P<0.05, one-way ANOVA with Tukey’s post hoc test.
(F) Behavioral responses of mated females to indicated amino acids (25 mM, pink) in binary choice tests with 5 mM sucrose (blue). Genotypes were as in (E). n=6–8. The different letters indicate significantly different groups, P<0.05, one-way ANOVA with Tukey’s post hoc test. Dotted lines delineate groups for ANOVA. See also Figure S6.
We next tested Ir76b mutant females and found that they showed a drastic reduction in mean preference for yeast extract as compared to Ir76b+ control flies (Figure 5B). We confirmed that the deficit in yeast extract preference was due to the loss of Ir76b by two independent rescue experiments. First, precise excision of the Ir76b05 P element showed that preference for yeast extract was restored in the revertant (Ir76brev) flies (Figure 5B). Second, expression of UAS-Ir76b using Ir76b-GAL4 restored behavioral preference for yeast extract in Ir76b2 mutants (Figure 5B). There were only minor differences in sucrose feeding behavior across all the genotypes tested (Figure 5B).
Given that the selection of yeast extract in feeding choice experiments is dependent on the presence of amino acids, we tested the role of Ir76b in behavioral responses to amino acids. In a series of feeding choice experiments with each of the top five amino acids we found that behavioral responses in Ir76b05 and Ir76b2 mutants were greatly reduced (Figures 5C and 5D). Moreover, those of Ir76brev flies were significantly higher than those in Ir76b05 flies (Figure 5C). Similarly, transgenic rescue in Ir76b2 flies by expressing UAS-Ir76b via Ir76b-GAL4 rescued behavioral responses to each of the five amino acids (Figure 5D). Based on these results, the simplest interpretation is that Ir76b is necessary for taste acceptance of amino acids.
Ir76b function is evolutionarily conserved
All insect genomes sequenced to date reveal one or more orthologs of Ir76b, which belongs to a group designated as “antennal Irs” whose expression in the antenna is potentially conserved in all insects (Croset et al., 2010). Although gustatory expression of Ir76b has not been investigated in other insects, we were curious whether a distantly related Ir76b ortholog could substitute for fly Ir76b function. We elected to test Ir76b from the malaria vector Anopheles gambiae, which is separated from D. melanogaster by ~260 million years of evolution (Moreno et al., 2010). We constructed UAS-AgamIr76b and tested behavioral responses to yeast extract in animals in which the UAS-AgamIr76b transgene was the only source of Ir76b. Results of feeding experiments showed that AgamIr76b restored preference for yeast extract in Ir76b2 mutants (Figure 5E). Additional experiments showed that behavioral responses to each of the top five amino acids were also rescued by AgamIr76b (Figure 5F). Thus, Ir76b function in mediating taste responses to amino acids appears to be evolutionarily conserved in flies and mosquitoes.
Ir76b marks functionally distinct subsets of taste neurons
Two lines of evidence suggest that Ir76b is associated with multiple functional categories of taste neurons. First, we found that Ir76b-GAL4 labeled multiple neurons per sensillum in tarsi, and these are known to be functionally distinct (Ling et al., 2014). Second, Ir76b is involved in taste responses to salt (Zhang et al., 2013), polyamines (Hussain et al., 2016), as well as to amino acids (Figures 4 and 5). We were therefore prompted to further characterize expression of Ir76b-GAL4. In addition to the Ir76b-GAL4 driver used above (Silbering et al., 2011), hereafter referred to as Ir76b-GAL4RB, we obtained a second Ir76b-GAL4 line (Zhang et al., 2013), named Ir76b-GAL4CM for comparison. We observed that both drivers were broadly expressed in external and internal taste organs, and there was little difference in expression in tarsi (61±0.7 and 62±1.73 cells respectively, mean±s.e.m., n=3) and pharyngeal taste organs (not shown) between them. Both drivers also labeled taste pegs that line the oral surface of the labial palps (Figure 6A). However, Ir76b-GAL4RB appeared to be excluded from labellar taste hairs that house salt-sensing neurons, which are labeled by Ir76b-GAL4CM. We confirmed this difference by creating an Ir76b-LexARB transgene and performing double labeling experiments with Ir76b-GAL4CM, which revealed the presence of numerous cells in the labellum, and axonal projections in the subesophageal zone (SEZ) labeled exclusively by Ir76b-GAL4CM (Figure 6A and 6B).
Figure 6. Different subsets of Ir76b-GAL4 neurons mediate behavioral responses to amino acids and salt.
(A) Confocal images (left) and schematic (right) of Ir76b-LexARB (red) and Ir76b-GAL4CM (green) in the labellum. Neurons labeled exclusively in green innervate taste hairs. Genotype was Ir76b-LexARB/UAS-mCD8::GFP; Ir76b-GAL4CM/lexAop-mCherry::HA. Scale bar=20 μM.
(B) Confocal images (left) and schematic (right) of axonal projections of Ir76b-LexARB (red) and Ir76b-GAL4CM (green) neurons in the subesophageal zone (SEZ) visualized using anti-HA (red) and anti-GFP (green). Genotype as in (A). Scale bar=20 μM.
(C) Confocal images of Ir76b-LexARB (red) and Gr64f-GAL4 or Gr89a-GAL4 (green) neurons in the tarsi. Genotypes were Ir76b-LexARB/Gr64f-GAL4; UAS-mCD8::GFP/lexAop-mCherry::HA and Ir76b-LexARB/Gr89a-GAL4; UAS-mCD8::GFP/lexAop-mCherry::HA. Arrowheads mark Ir76b+ cells that are also positive for Gr64f or Gr89a. Asterisks mark Ir76b+ that do not overlap with the indicated markers. Scale bar=10 μM.
(D) Confocal images of axonal projections of Ir76b-LexARB (red) and Gr64f-GAL4 or Gr89a-GAL4 (green) neurons in the SEZ visualized using anti-HA (red), anti-GFP (green) and anti-nc82 (blue). Genotypes as in (C). Scale bar=20 μM.
(E) Mean PI of mated females of indicated genotypes from binary feeding tests. Stimuli presented in each set of binary choice trials are listed above (pink) and below (blue) the graphs. n=6–11. For each stimulus, different letters indicate significantly different groups, P<0.05, one-way ANOVA with Tukey’s post hoc test. Dotted lines delineate separation of groups for ANOVA. Genotypes were: UAS-Kir2.1/UAS-Kir2.1 (UAS-Kir); Ir76b-GAL4RB/CyO; TM2/TM6b (Ir76b-GAL4RB); Ir76b-GAL4RB/+; UAS-Kir2.1/TM2 (Ir76b-GAL4RB > Kir); Ir76b-GAL4CM/TM6b (Ir76b-GAL4CM); Ir76b-GAL4CM/UAS-Kir2.1 (Ir76b-GAL4CM > Kir).
We next determined the overlap of Ir76b-LexARB with markers for sweet (Gr64f-GAL4) and bitter (Gr89a-GAL4) taste neurons. Consistent with the results of our calcium imaging experiments (Figure 3F), we found overlap between Gr64f-GAL4 and Ir76b-LexA (Figure 6C). The Ir76b+/Gr64f+ cells are likely those activated by both serine and sucrose. Interestingly, Ir76b-LexA expression also partially overlapped with that of Gr89a-GAL4. Although serine-activated neurons did not respond to a bitter tastant (Figure 3F), the expression analysis predicts that one or more bitter compounds would activate a distinct sub-population of Ir76b+ cells. Visualization of axonal projection patterns in the subesophageal zone revealed patterns of overlap between Ir76b+ termini and those of sweet and bitter neurons (Figure 6D), as observed in the periphery.
Given our observation that serine-responsive Ir76b+ tarsal neurons were not activated by salt (Figure 3F), we wanted to explore the idea that different subsets of Ir76b+ neurons may be involved in appetitive responses to salt and amino acids. While Ir76b-GAL4CM and Ir76b-GAL4RB were co-expressed in tarsal neurons, the main difference between them appears to be that Ir76b-GAL4CM labeled salt-sensing neurons in the labellum, whereas Ir76b-GAL4RB did so weakly, if at all. We therefore took advantage of the two drivers to perform two sets of experiments. First, we rescued Ir76b function in an Ir76b2 mutant background using either of the two drivers, which showed that both are sufficient to drive expression in a pattern that rescues salt and amino acid responses (Figure S6). Second, we silenced the two populations of Ir76b+ neurons using the inwardly rectifying potassium channel Kir2.1 and compared consequences on behavioral responses to various tastants. As expected, Kir2.1 expression under the control of either driver resulted in a significant loss of preference for yeast extract as compared to the level observed in GAL4 or UAS control flies (Figure 6E). By contrast, only silencing of Ir76b-GAL4CM neurons caused significant defects in behavioral preference for salt (Figure 6E). In both cases, minor but significant reductions were observed for behavioral responses to sucrose as compared to control flies (Figure 6E), likely stemming from sucrose sensitivity of tarsal Ir76b+ neurons (Figure 3F). There was no effect on rejection of caffeine (Figure 6E), suggesting that Ir76b may not be associated with caffeine-sensing class of bitter taste neurons. Alternatively, changes in caffeine-sensitivity may need to be evaluated across a range of concentrations. Overall, these results support the idea that Ir76b is expressed in multiple functional categories of taste neurons, and largely distinct sets of Ir76b+ taste neurons mediate appetitive responses to low salt and amino acids.
Ir20a alters Ir76b response from salt to amino acids
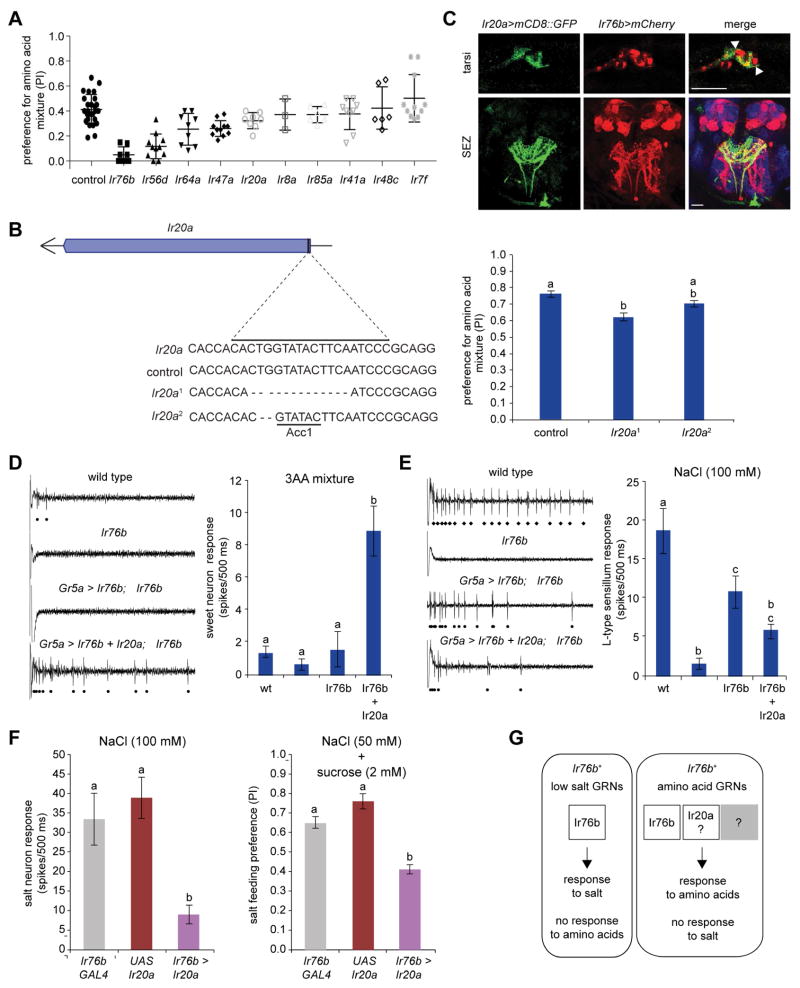
We next wished to investigate molecular mechanisms that underlie the difference in Ir76b function in salt and amino acid neurons. Based on previous findings that implicate Ir76b as an independently functioning Na+ channel in salt taste neurons (Zhang et al., 2013), and as a co-receptor in olfactory neurons (Abuin et al., 2011), we hypothesized that other members of the Ir family expressed in amino acid sensing neurons may serve to gate the conductance of Ir76b. To identify candidate co-receptors, we returned to the results of our initial RNAi screen and re-tested several candidate Ir-RNAi lines (Figure S3B), including Ir76b-RNAi as a positive control. Binary feeding choice experiments were performed using sucrose and amino acid-depleted yeast extract with a mixture of serine, threonine and phenylalanine. As expected, Ir76b-RNAi yielded a dramatic loss of preference for the amino acid mixture (Figure 7A). Four other RNAi lines, for Ir20a, Ir47a, Ir56d, and Ir64a respectively, showed significant reductions in amino acid preference as compared to wild type flies (Figure 7A), suggesting that one or more of these receptors may function with Ir76b to mediate amino acid taste. As would be expected, the expression of at least three of these candidates (Ir20a, Ir47a, Ir56d) has been reported in tarsal taste neurons (Koh et al., 2014). RNAi knock-down of Ir7f, Ir8a, Ir41a, Ir48c, and Ir85a had no effect in this assay, suggesting that these receptors may be involved in sensing other amino acids or other cues in yeast extract.
Figure 7. Ir76b is not sufficient for conferring amino acid response.
(A) Scatter plots showing PIs of mated female flies for 1% yeast extract–amino acids supplemented with 25 mM each of serine, threonine and phenylalanine. Genotypes were elav-GAL4/UAS-Ir-RNAi; UAS-Dcr2 or elav-GAL4; UAS-Ir-RNAi/UAS-Dcr2. F1 progeny from elav-GAL4; UAS-Dcr2 crossed to w1118 were used as control. n=6–26 except for Ir8a (n=3). *P<0.05, **P<0.01, ***P<0.001, Mann-Whitney U tests versus the control.
(B) Schematic for generation of Ir20 mutants using CRISPR/Cas9-mediated gene disruption (left). Mean PI of mated females of the indicated genotypes for 1% yeast extract–amino acids supplemented with 25 mM each of serine, threonine and phenylalanine. n=11–17. Different letters indicate statistically different groups, P<0.05, one-way ANOVA.
(C) Confocal images of Ir76b-LexARB (red) and Ir20a-GAL4 (green) neurons in tarsi (top) and SEZ (bottom). Axonal projections in the SEZ were visualized using anti-HA (red), anti-GFP (green) and anti-nc82 (blue). Genotype was Ir76b-LexARB/Ir20a-GAL4; UAS-mCD8::GFP/lexAop-mCherry::HA. Scale bar=20 μM.
(D) Representative traces (left) and mean responses (right) obtained from L-type labellar sensilla of the first 500 ms upon stimulation with a mixture of 100 mM serine, 50 mM phenylalanine and 100 mM threonine (3AA mixture). Black dots indicate action potentials assigned to the sweet taste neuron and counted. Genotypes: w1118 (wild type); Sp/CyO; Ir76b2 (ΔIr76b); Gr5a-GAL4; Ir76b2, UAS-Ir76b (Gr5a > Ir76b; ΔIr76b); Gr5a-GAL4/UAS-Ir20a; Ir76b2, UAS-Ir76b/Ir76b2 (Gr5a > Ir76b + Ir20a; ΔIr76b). n=9–18 sensilla from 3–4 flies. For each stimulus, different letters indicate significantly different groups, P<0.05, one-way ANOVA.
(E) Representative traces (left) and mean responses (right) obtained from L-type labellar sensilla of the first 500 ms upon stimulation with 100 mM NaCl. Only spikes of the larger amplitude were counted, representing the salt neuron in wild type (diamonds), and the sweet neuron in the Ir76b1 mutant background (dots). Genotypes as in (C). n=9–18 sensilla from 3–4 flies. For each stimulus, different letters indicate significantly different groups, P<0.05, one-way ANOVA.
(F) Mean responses in the first 500 ms upon stimulation obtained from L-type sensilla with a stimulus of 100 mM NaCl (left). n=10–11 sensilla from 3 flies. Mean PI of mated females (7–10 day old) for 50 mM NaCl mixed with 2 mM sucrose (tested against 2 mM sucrose) obtained from binary choice tests (right). Results are pooled from pink/blue dye swap experiments. n= 12–14. Genotypes were: Ir76b-GAL4CM/TM6b (Ir76b-GAL4); UAS-Ir20a/CyO; Dr/TM3 (UAS-Ir20a); and UAS-Ir20a/+; Ir76b-GAL4CM/TM3 (Ir76b > Ir20a). For each stimulus, different letters indicate significantly different groups, P<0.05, one-way ANOVA.
(G) Cartoon illustrating possible mechanisms by which distinct subsets of Ir76b+ taste neurons mediate salt and amino acid taste.
We next wished to test the possibility that Ir76b functions along with one of the other Irs to mediate amino acid response by expression in sweet taste neurons. We selected Ir20a for this analysis, because Ir47a and Ir56d are likely to already be present in Gr5a+ sweet neurons based on reporter analysis (Koh et al., 2014). We first confirmed the role of Ir20a in detecting amino acids by generating CRISPR/Cas9-mediated mutants (Figure 7B). Of the alleles recovered, we selected two for further analysis: Ir20a1 had a 1-nt deletion, which predicted a truncated protein product of 53 amino acids; Ir20a2 had a 2-nt deletion, which predicted a protein product of 74 amino acids. We also tested a control in which the Ir20a sequence had not been altered. Ir20a1 mutants showed a significant reduction in feeding preference for the amino acid mixture as compared to control flies (P<0.001 versus control, n=11). Although not statistically significant, mean feeding preference was also reduced in Ir20a2 mutants (P=0.136 versus control, n=13). Consistent with the Ir20a-RNAi phenotype, the magnitude of the defect is small, which may account for the discrepancy between Ir20a1 and Ir20a2 mutants. As expected, Ir20a-GAL4 expression overlaps with Ir76b-LexA in tarsi and axonal projections from tarsal and pharyngeal neurons (Figure 7C). Together, these results are consistent with the model that amino acid detection is at least partially dependent on Ir20a. The observation that Ir20a is expressed only in a small subset of Ir76b neurons, and that loss of Ir20a has a weaker consequence than that of Ir76b, also suggests that Ir20a may have some functional redundancies with other Irs.
We used Gr5a-GAL4 to ectopically express Ir76b, either by itself or in combination with Ir20a, in sweet taste neurons. Ectopic experiments were conducted in an Ir76b2 mutant background to eliminate the activity of the salt taste neuron (Zhang et al., 2013), which is housed along with the Gr5a+ sweet taste neuron in L-type sensilla. We used single sensillum recordings to measure responses of sweet taste neurons in L-type sensilla to a mixture of serine, threonine and phenylalanine. While Ir76b alone did not confer sensitivity to the amino acid mixture, we found that co-expression of Ir20a and Ir76b together was sufficient to do so in the milieu of the sweet taste neuron (Figure 7D).
Given our observations that responses to salt and amino acids appear to be mutually exclusive, we next tested whether co-expression of Ir20a affected the Ir76b-mediated response to salt. As reported before (Zhang et al., 2013), we found that expression of Ir76b in Gr5a+ neurons was sufficient to confer a salt response, although the strength of the response was somewhat lower than that observed in the endogenous context (Figure 7E). However, sweet taste neurons in which both Ir20a and Ir76b were expressed showed a reduced mean response to NaCl (Figure 7E). To further test the idea that the presence of Ir20a can block salt response of Ir76b, we expressed Ir20a using Ir76b-GAL4 and measured responses to salt. As predicted, expression of Ir20a caused a significant reduction in cellular and behavioral responses to salt as compared to that observed in Ir76b-GAL4 or UAS-Ir20a control flies (Figure 7F). Although we cannot rule out the possibility that Ir20a interferes with Ir76b in a non-physiological manner, the simplest interpretation of our results is that Ir76b activity is gated by Ir20a to mediate amino acid taste.
DISCUSSION
Here we report the identification of the cellular and molecular basis of amino acid detection in Drosophila taste neurons. Genetic analyses, combined with behavior assays and calcium imaging studies reveal that Ir76b, an ionotropic receptor previously found to mediate salt taste (Zhang et al., 2013), is necessary for amino acid detection by tarsal taste neurons. Analysis of Ir76b expression and function is consistent with a model (Figure 7G) in which this receptor marks two functionally exclusive populations of cells, one that responds to salt and another that responds to amino acids. In the latter, Ir76b combines with Ir20a, and possibly other Irs, which gate its activity to amino acid ligands.
Ir genes encode proteins related to ionotropic glutamate receptors and represent an ancient family of chemoreceptors, based on their occurrence in genomes of all protostomes (Croset et al., 2010). Their expression and function has been extensively characterized in the fly olfactory system, in which they are expressed in combinations of up to four receptors in olfactory receptors neurons (Abuin et al., 2011). In keeping with their ancient origin, Irs have been associated with detection of broadly appealing or noxious stimuli, including acids, amines, and ammonia (Abuin et al., 2011; Ai et al., 2010; Grosjean et al., 2011; Min et al., 2013). More recently, Ir gene expression has been analyzed in gustatory neurons of both adult and larval stages, and accords possible roles in taste recognition to several members of the family (Koh et al., 2014; Stewart et al., 2015). However, with the exception of Ir76b, taste functions of Ir proteins remain to be characterized. Given that many Ir genes are co-expressed with either Gr5a or Gr66a in sweet or bitter taste neurons (Koh et al., 2014), another open question is whether, and if so how, Ir proteins coordinate with other classes of receptors.
Ir76b has been proposed to function as a Na+ leak channel that is fixed in a permeable state (Zhang et al., 2013). In this model, Ir76b-mediated sodium conductance remains low until contact with salt-laced foods, because the sensillar lymph is rich in potassium but contains low sodium. Ectopic expression of Ir76b yields the predicted outcome – sensitivity to sodium chloride in a concentration dependent manner (Zhang et al., 2013). This was surprising because Ir76b is expressed in a variety of neurons that do not respond to salt, including amino acid-sensing neurons in tarsi. The identification of Ir20a as one co-receptor that promotes amino acid response and blocks salt response is consistent with the idea that Ir76b conductance is regulated differently in salt and amino acid taste neurons by other members of the Ir family (Figure 7F). Notably, although expression of Ir20a blocked salt response of Ir76b+ neurons in L-type sensilla, it was not sufficient to confer sensitivity to amino acids (not shown). Moreover, Ir candidates may have been missed within the limited scope of the initial RNAi screen using yeast extract, which could have several redundant attractive cues. Thus, in all likelihood additional Irs operate in combination with Ir76b and Ir20a to form amino acid receptors. The presence of Ir47a and Ir56d in tarsal neurons as well as labellar sweet taste neurons makes them appealing candidates for such roles. It is also possible that different Irs fulfill the role of Ir20a in other amino acid-sensing neurons. A few observations support this idea. First, Ir20a mutants do not phenocopy Ir76b mutants (Figure 7B). Second, Ir20a displays a restricted pattern of expression in 2–3 neurons in the fifth segment (Figure 7C), representing only a small fraction of Ir76b+ neurons. Third, there appears to be some diversity in amino acid responses across taste neurons, invoking differences in receptor repertoires. Notably, there is precedent for participation of Ir76b in functional heteromeric receptors with two other Irs in olfactory neurons (Abuin et al., 2011; Benton et al., 2009; Silbering et al., 2011). An appealing hypothesis is that Ir76b might operate likewise in taste neurons, in complexes with combinations of Irs that may have distinct amino acid recognition properties. The occurrence of receptor combinations may also explain why different amino acids evoke responses of different strengths.
Sex-dependent variations in food choice have been described previously (Ribeiro and Dickson, 2010), but the extent to which they depend on variation in sensitivity of taste neurons remains to be examined. The results of our calcium imaging experiments suggest that differences in tarsal sensitivity to amino acids may underlie sexual dimorphism in yeast and amino acid preference. Moreover, the observation that overexpression of Ir76b caused an increase in the preference for yeast extract implies that levels of Ir76b are limiting, particularly in male flies. We therefore expected sexual dimorphism in expression levels of Ir76b. However, transcriptome analysis revealed otherwise (not shown). Moreover, neither Ir76b-GAL4 nor Ir20a-GAL4 showed any sexual dimorphism in expression in tarsal or pharyngeal neurons, where both are expressed (not shown). Thus, the mechanisms by which amino acid taste and yeast preference are enhanced in females as compared to males are likely to be dependent on as yet unknown sex-specific factors in Ir76b+ neurons. Interestingly, Ir76b-GAL4 is not expressed in fru+ neurons (not shown), suggesting that fru circuitry may not underlie the sex-specificity of peripheral amino acid responses.
Amino acid and yeast preferences are also upregulated in females upon mating. We and others have found that virgin females behave much like males in binary choice assays. Interestingly, preliminary explorations with calcium imaging showed that amino acid responses are present in tarsi of virgin females (not shown), indicating that the low preference for yeast in virgins does not arise from an inability to sense amino acids. Previous studies have shown that the post-mating shift in food preference depends on sex peptide (Ribeiro and Dickson, 2010), which is synthesized by male accessory glands and transferred to the female reproductive tract during copulation, although the manner in which sex peptide receptor (SPR) circuitry impinges on taste circuitry is not known. A recent study found that SPR function in Ir76b+ neurons plays a role in sexually dimorphic responses to polyamines (Hussain et al., 2016). However, we found that RNAi-mediated knockdown of SPR in Gr5a+ or Ir76b+ neurons did not affect the behavioral shift to yeast extract in mated females (not shown). Thus the functional overlap between SPR+ and amino acid-sensing circuitry is likely to occur downstream of the sensory neuron. Consistent with this model, a role has been identified for fru+/dsx+/ppk+/SPR+ neurons in the reproductive tract that convey information either directly or indirectly to the subesophageal zone (Rezaval et al., 2012).
In mammals, amino acids are detected by a dedicated population of taste receptor cells (Nelson et al., 2002). By contrast, we found that amino acid-sensing neurons overlap with sucrose-sensing neurons in fly tarsi. However, behavioral experiments show that the fly can differentiate between sucrose and amino acids, supporting the idea that the two have distinct percepts in the brain. The lack of amino acid sensitivity in labellar sweet taste neurons might provide one avenue with which to distinguish the two categories of tastants. Furthermore, previous studies in other insects suggest possible synergistic interactions between sugars and amino acids when presented in mixtures (Alm et al., 1990; Wada et al., 2001). Such interactions may be achieved, at least in part, via the co-expression of amino acid and sweet taste receptors in a subset of neurons. Indeed, this appears to be the case in fleshflies and blowflies that detect some amino acids via sweet-sensing neurons (Shiraishi and Kuwabara, 1970).
Ir76b is highly conserved in insect genomes (Croset et al., 2010), and the functional substitution of DmIr76b with AgIr76b suggests that its role in taste detection is conserved as well. Although our study highlights the importance of Ir76b and amino acid detection for selection of proteinaceous food sources by phytophagous insects like Drosophila, free amino acids are also found in human sweat (Hier et al., 1946) and may serve as critical cues for blood-feeding disease vectors such as mosquitoes and tsetse flies. The identification of Ir76b as a receptor for amino acid taste invites further exploration of molecular mechanisms of amino acid taste in human disease vectors and may lead to targets for control of insect feeding behaviors.
EXPERIMENTAL PROCEDURES
Fly stocks and constructs
Flies were raised on standard cornmeal-dextrose media at 22–25°C. Unless otherwise indicated, wild type flies were w1118 (BL 5905). Ir76b-LexA was created using a 916bp promoter fragment. Full-length coding sequences for AgIr76b and Ir20a were either synthesized or amplified with PCR and used to generate UAS-Ir constructs. Ir20a mutants were generated using CRISPR/Cas9. A complete list of fly stocks is given in supplemental methods.
Behavior assays
Binary choice feeding assays were performed as previously described (Wisotsky et al., 2011). Briefly, 5–7 day old flies were wet-starved for 24–26 hours and tested in tight-fit Petri dish arenas containing agarose droplets with tastant-dye mixtures. Flies were scored for abdomen color within 24 hours, and preference index for each trial was calculated as: [Npink + 0.5Npurple] / [Npink + Nblue + Npurple]. Trials in which less than 50% of the flies participated were discarded. Catalog numbers for all tastants are provided in supplemental methods.
Immunohistochemistry
Fly brains were dissected and fixed in paraformaldehyde and blocked using normal goat serum. Primary antibodies were mouse α-nc82, rat α-CD8a, rabbit αHA and chick α-GFP; secondary antibodies were Alexa-488 α-rat, Alexa-568 α-mouse, Alexa-488 α-chick, Alexa-568 α-rabbit, and Alexa-647 α-mouse. Catalog numbers for all antibodies are provided in supplemental methods. Confocal z-stack images were acquired using a Leica SP5 confocal microscope and analyzed using Image J.
Calcium imaging
GCaMP3 fluorescence was imaged in distal segments of the tarsi using flies aged ≥ 7days, which were maintained at 29°C for at least 4 days prior to imaging. Decapitated flies with their prothoracic legs extended were immobilized on falcon plates with double-sided sticky tape, and a 100-μl water drop was added on the tarsal segments. 100 μl of tastant solution was added in the water droplet and changes in GCaMP3 fluorescence were monitored using a Leica SP5 confocal microscope. ΔF/F % values were calculated separately for each cell body, and background regions, using the mean intensity value of all frames in the 5-second period prior to addition of the stimulus (Fpre(cell)) and mean intensity value of all frames in the 5-second period around the peak response (Fpost(cell)). ΔF/F % was calculated with the following formula:
Additional details about the fly preparation and data acquisition and analysis are given in supplemental methods.
Electrophysiological recordings
Extracellular tip recordings were obtained from L-type labellar sensilla as described previously (Dahanukar and Benton, 2010). Female flies aged 8–10 days were used for recordings. All chemicals were dissolved in 30 mM tricholine citrate, which served as the electrolyte. Neuronal responses were quantified by counting the number of spikes in the first 500 ms upon contact with the stimulus.
Statistical analyses
Behavioral preference indices were compared using the Mann-Whitney U test or ANOVA with pairwise comparisons using Bonferroni adjustment for multiple comparisons or Tukey’s post hoc analysis. Changes in calcium activity were compared using Mann-Whitney U tests. Sample sizes and statistical tests were chosen on the basis of previously published studies, and are cited in all figure legends. For all column and line graphs, error bars indicate s.e.m.; error bars in scatter plots indicate s.d.
Supplementary Material
HIGHLIGHTS.
Drosophila females display amino acid feeding preference after mating
Taste neurons in female tarsi are activated by amino acids
A highly conserved receptor, Ir76b, is required for amino acid taste
Ir20a mediates amino acid taste and blocks salt taste dependent on Ir76b
Acknowledgments
We thank W. Carabajal, B. Guzman, P. Ehtiyatkar, K. Huynh, S. Liu, M. Luth, I. Naik, Y. Patel, and J. Viduya for participating in the Ir-RNAi screens, B. Jablonska for technical assistance, D. Carter for help with calcium imaging, K. Lung and S. Charlu for initial behavioral investigations, A. Lomeli for transcriptome analysis of taste tissues, G. Tauxe for help with statistical analyses, B. Baker, R. Benton, C. Montell, and K. Scott for sharing flies, and A. Ray and members of the Dahanukar and Ray labs for helpful comments. Stocks obtained from the Bloomington Drosophila Stock Center (NIH P40OD018537) were used in this study. This work was partly funded by an NIH R21 grant (R21DC012408) and an NSF-CAREER award (NSF 1149667) to A.D.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, A.G. and A.D.; Methodology, A.G., L.P. and A.D.; Investigation, A.G., L.P., V.-K.D., A.L., H.S., and E.V.; Writing – Original Draft, A.D., Writing – Reviewing and Editing, A.G. and A.D.; Visualization, A.G., L.P. and A.D.; Supervision, A.G., L.P. and A.D., Funding Acquisition, A.D.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011;69:44–60. doi: 10.1016/j.neuron.2010.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai M, Min S, Grosjean Y, Leblanc C, Bell R, Benton R, Suh GS. Acid sensing by the Drosophila olfactory system. Nature. 2010;468:691–695. doi: 10.1038/nature09537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alm J, Ohnmeiss TE, Lanza J, Vriesenga L. Preference of cabbage white butterflies and honey bees for nectar that contains amino acids. Oecologia. 1990;84:53–57. doi: 10.1007/BF00665594. [DOI] [PubMed] [Google Scholar]
- Baltzer C, Tiefenbock SK, Marti M, Frei C. Nutrition controls mitochondrial biogenesis in the Drosophila adipose tissue through Delg and Cyclin D/Cdk4. PLoS ONE. 2009;4:e6935. doi: 10.1371/journal.pone.0006935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell. 2009;136:149–162. doi: 10.1016/j.cell.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britton JS, Edgar BA. Environmental control of the cell cycle in Drosophila: nutrition activates mitotic and endoreplicative cells by distinct mechanisms. Development. 1998;125:2149–2158. doi: 10.1242/dev.125.11.2149. [DOI] [PubMed] [Google Scholar]
- Chang CL. Effect of amino acids on larvae and adults of Ceratitis capitata (Diptera: Tephritidae) Annals of the Entomological Society of America. 2004;97:529–535. [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Benton R. Chemosensory coding in single sensilla. In: Zhang B, Freeman MR, Waddell S, editors. Drosophila neurobiology: A laboratory manual. CSHL Press; 2010. pp. 247–276. [Google Scholar]
- Dahanukar A, Lei YT, Kwon JY, Carlson JR. Two Gr genes underlie sugar reception in Drosophila. Neuron. 2007;56:503–516. doi: 10.1016/j.neuron.2007.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimond JB, Lea AO, Hahnert WF, Jr, DeLong DM. The amino acids required for egg production in Aedes aegypti. The Canadian Entomologist. 1956;88:57–62. [Google Scholar]
- Fink P, Pflitsch C, Marin K. Dietary essential amino acids affect the reproduction of the keystone herbivore Daphnia pulex. PLoS ONE. 2011;6:e28498. doi: 10.1371/journal.pone.0028498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman EG, Dahanukar A. Molecular neurobiology of Drosophila taste. Curr Opin Neurobiol. 2015;34:140–148. doi: 10.1016/j.conb.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golberg L, De Meillon B. The nutrition of the larva of Aedes aegypti Linnaeus. 4. Protein and amino-acid requirements. Biochemical Journal. 1948;43:379. [PMC free article] [PubMed] [Google Scholar]
- Grandison RC, Piper MD, Partridge L. Amino-acid imbalance explains extension of lifespan by dietary restriction in Drosophila. Nature. 2009;462:1061–1064. doi: 10.1038/nature08619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean Y, Rytz R, Farine JP, Abuin L, Cortot J, Jefferis GS, Benton R. An olfactory receptor for food-derived odours promotes male courtship in Drosophila. Nature. 2011;478:236–240. doi: 10.1038/nature10428. [DOI] [PubMed] [Google Scholar]
- Hier SW, Cornbleet T, Bergeim O. The amino acids of human sweat. J Biol Chem. 1946;166:327–333. [PubMed] [Google Scholar]
- Hinton T, Noyes DT, Ellis J. Amino acids and growth factors in a chemically defined medium for Drosophila. Physiological Zoology. 1951:335–353. [Google Scholar]
- House HL. Insect nutrition. Annu Rev Biochem. 1962;31:653–672. doi: 10.1146/annurev.bi.31.070162.003253. [DOI] [PubMed] [Google Scholar]
- Hussain A, Zhang M, Ucpunar HK, Svensson T, Quillery E, Gompel N, Ignell R, Grunwald Kadow IC. Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 2016;14:e1002454. doi: 10.1371/journal.pbio.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignell R, Okawa S, Englund JE, Hill SR. Assessment of diet choice by the yellow fever mosquito Aedes aegypti. Physiol Entomol. 2010;35:274–286. [Google Scholar]
- Koh TW, He Z, Gorur-Shandilya S, Menuz K, Larter NK, Stewart S, Carlson JR. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron. 2014;83:850–865. doi: 10.1016/j.neuron.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson MC, Domingos AI, Jones WD, Chiappe ME, Amrein H, Vosshall LB. Or83b encodes a broadly expressed odorant receptor essential for Drosophila olfaction. Neuron. 2004;43:703–714. doi: 10.1016/j.neuron.2004.08.019. [DOI] [PubMed] [Google Scholar]
- Ling F, Dahanukar A, Weiss LA, Kwon JY, Carlson JR. The molecular and cellular basis of taste coding in the legs of Drosophila. J Neurosci. 2014;34:7148–7164. doi: 10.1523/JNEUROSCI.0649-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Ai M, Shin SA, Suh GS. Dedicated olfactory neurons mediating attraction behavior to ammonia and amines in Drosophila. Proc Natl Acad Sci U S A. 2013;110:E1321–1329. doi: 10.1073/pnas.1215680110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno M, Marinotti O, Krzywinski J, Tadei WP, James AA, Achee NL, Conn JE. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar J. 2010;9:127. doi: 10.1186/1475-2875-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. An amino-acid taste receptor. Nature. 2002;416:199–202. doi: 10.1038/nature726. [DOI] [PubMed] [Google Scholar]
- Rezaval C, Pavlou HJ, Dornan AJ, Chan YB, Kravitz EA, Goodwin SF. Neural circuitry underlying Drosophila female postmating behavioral responses. Curr Biol. 2012;22:1155–1165. doi: 10.1016/j.cub.2012.04.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro C, Dickson BJ. Sex peptide receptor and neuronal TOR/S6K signaling modulate nutrient balancing in Drosophila. Curr Biol. 2010;20:1000–1005. doi: 10.1016/j.cub.2010.03.061. [DOI] [PubMed] [Google Scholar]
- Sang JH, King RC. Nutritional requirements of axenically cultured Drosophila melanogaster adults. J Exp Biol. 1961;38:793–809. [Google Scholar]
- Shiraishi A, Kuwabara M. The effects of amino acids on the labellar hair chemosensory cells of the fly. J Gen Physiol. 1970;56:768–782. doi: 10.1085/jgp.56.6.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silbering AF, Rytz R, Grosjean Y, Abuin L, Ramdya P, Jefferis GS, Benton R. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J Neurosci. 2011;31:13357–13375. doi: 10.1523/JNEUROSCI.2360-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KRP, Brown AWA. Nutritional requirements of Aedes aegypti L. J Insect Physiol. 1957;1:199–220. [Google Scholar]
- Stewart S, Koh TW, Ghosh AC, Carlson JR. Candidate ionotropic taste receptors in the Drosophila larva. Proc Natl Acad Sci U S A. 2015;112:4195–4201. doi: 10.1073/pnas.1503292112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatum EL. Nutritional requirements of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1939;25:490–497. doi: 10.1073/pnas.25.9.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshima N, Tanimura T. Taste preference for amino acids is dependent on internal nutritional state in Drosophila melanogaster. J Exp Biol. 2012;215:2827–2832. doi: 10.1242/jeb.069146. [DOI] [PubMed] [Google Scholar]
- van der Goes van Naters W, den Otter CJ. Amino acids as taste stimuli for tsetse flies. Physiol Entomol. 1998;23:278–284. [Google Scholar]
- Van Loon JJA, Van Eeuwijk FA. Chemoreception of amino acids in larvae of two species of Pieris. Physiol Entomol. 1989;14:459–469. [Google Scholar]
- Vargas MA, Luo N, Yamaguchi A, Kapahi P. A role for S6 kinase and serotonin in postmating dietary switch and balance of nutrients in D. melanogaster. Curr Biol. 2010;20:1006–1011. doi: 10.1016/j.cub.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrzal EM, Allan SA, Hahn DA. Amino acids in nectar enhance longevity of female Culex quinquefasciatus mosquitoes. J Insect Physiol. 2010;56:1659–1664. doi: 10.1016/j.jinsphys.2010.06.011. [DOI] [PubMed] [Google Scholar]
- Wada A, Isobe Y, Yamaguchi S, Yamaoka R, Ozaki M. Taste-enhancing effects of glycine on the sweetness of glucose: a gustatory aspect of symbiosis between the ant, Camponotus japonicus, and the larvae of the lycaenid butterfly, Niphanda fusca. Chem Senses. 2001;26:983–992. doi: 10.1093/chemse/26.8.983. [DOI] [PubMed] [Google Scholar]
- Wisotsky Z, Medina A, Freeman E, Dahanukar A. Evolutionary differences in food preference rely on Gr64e, a receptor for glycerol. Nat Neurosci. 2011;14:1534–1541. doi: 10.1038/nn.2944. [DOI] [PubMed] [Google Scholar]
- Zhang YF, Huang LQ, Ge F, Wang CZ. Tarsal taste neurons of Helicoverpa assulta (Guenee) respond to sugars and amino acids, suggesting a role in feeding and oviposition. J Insect Physiol. 2011;57:1332–1340. doi: 10.1016/j.jinsphys.2011.06.009. [DOI] [PubMed] [Google Scholar]
- Zhang YF, van Loon JJA, Wang CZ. Tarsal taste neuron activity and proboscis extension reflex in response to sugars and amino acids in Helicoverpa armigera (Hubner) J Exp Biol. 2010;213:2889–2895. doi: 10.1242/jeb.042705. [DOI] [PubMed] [Google Scholar]
- Zhang YV, Ni J, Montell C. The molecular basis for attractive salt-taste coding in Drosophila. Science. 2013;340:1334–1338. doi: 10.1126/science.1234133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.