SUMMARY
Poly(ADP-ribosyl)ation (PARylation) is a post-translational modification of proteins mediated by PARP family members, such as PARP-1. Although PARylation has been studied extensively, few examples of definitive biological roles for site-specific PARylation have been reported. Here we show that C/EBPβ, a key pro-adipogenic transcription factor, is PARylated by PARP-1 on three amino acids in a conserved regulatory domain. PARylation at these sites inhibits C/EBPβ’s DNA binding and transcriptional activities, and attenuates adipogenesis in various genetic and cell-based models. Interestingly, PARP-1 catalytic activity drops precipitously during the first 48 hours of differentiation, corresponding to a release of C/EBPβ from PARylation-mediated inhibition. This promotes the binding of C/EBPβ at enhancers controlling the expression of adipogenic target genes and continued differentiation. Depletion or chemical inhibition of PARP-1, or mutation of the PARylation sites on C/EBPβ, enhances these early adipogenic events. Collectively, our results provide a clear example of how site-specific PARylation drives biological outcomes.
Keywords: Adipogenesis, C/EBPβ, DNA binding, Gene expression, NIH/3T3 cells, PARP inhibitor, PARP-1, PARylation, Poly(ADP-ribose), Transcription, 3T3-L1 cells, Stromal vascular fraction (SVF)
Graphical Abstract
eTOC Blurb
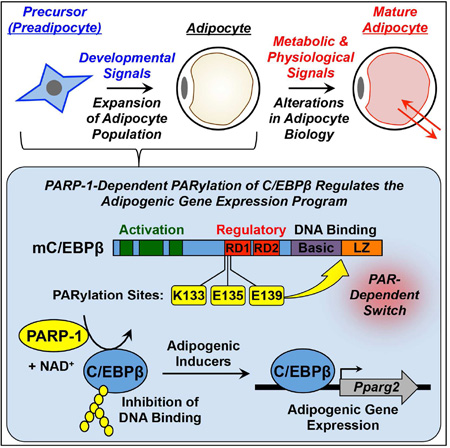
Luo et al. show that PARP-1 attenuates adipogenesis by PARylating C/EBPβ, a proadipogenic transcription factor, on three residues in its regulatory domain. Mutation of these residues produces a PARP-1-resistant superactive transcription factor that promotes adipogenesis in the absence of hormonal inducers.
INTRODUCTION
Poly(ADP-ribose) polymerase-1 (PARP-1) (a.k.a. ARDT1) is an abundant nuclear protein involved in a variety of nuclear processes, including transcription, RNA processing, and DNA repair (Gibson and Kraus, 2012; Ryu et al., 2015). It is the founding member of the PARP family of proteins, which share a conserved “PARP domain” that catalyzes the transfer of ADP-ribose moieties from NAD+ to target proteins, in either poly- or mono(ADP-ribosyl)ation reactions (Gibson and Kraus, 2012). PARP proteins are located in most cellular compartments and play key roles in an array of cellular processes (Gibson and Kraus, 2012; Ryu et al., 2015; Vyas et al., 2013). Among the nuclear PARP proteins, PARP-1 has a well established role in gene regulation, although the mechanisms by which it controls the transcription of target genes remain enigmatic, with both catalytic-dependent and -independent mechanisms described (Kraus and Hottiger, 2013; Krishnakumar and Kraus, 2010a).
Essential steps toward understanding the catalytic-dependent biological roles of PARPs include (1) identifying the target proteins of individual family members, as well as the specific sites at which they are ADP-ribosylated, and (2) determining how ADP-ribosylation of the target proteins affects their biochemical and molecular functions, as well as their biological roles. PARP-1 has been shown to poly(ADP-ribosyl)ate (PARylate) a variety of histone-modifying enzymes and transcription factors to modify their activities (Gao et al., 2009; Gibson et al., 2016; Kanai et al., 2007; Krishnakumar and Kraus, 2010a, b; Olabisi et al., 2008). In most cases, however, detailed analysis of these processes has been hampered by the limited information available on the specific amino acid residues that are PARylated. In this regard, methods for assessing site-specific ADP-ribosylation of proteins have recently been developed (reviewed in (Daniels et al., 2015)) and definitive examples of functional consequences of site-specific ADP-ribosylation are beginning to emerge (Gibson et al., 2016; Kanai et al., 2007; Olabisi et al., 2008).
Previous studies have connected PARP-1 catalytic activity to metabolic outcomes, including adipogenesis, in cell-based model systems, as well as in vivo in mice (Bai et al., 2011; Devalaraja-Narashimha and Padanilam, 2010; Erener et al., 2012a; Erener et al., 2012b; Lehmann et al., 2015; Luo and Kraus, 2011). The site-specific targets of PARP-1 catalytic activity in these processes, however, have not been determined. In the studies described herein, we link PARP-1-mediated site-specific PARylation of C/EBPβ, a key pro-adipogenic transcription factor, to the regulation of adipogenesis. Adipogenesis is controlled by two sequential waves of transcription factor activation (i.e., C/EBPβ and C/EBPδ in the first wave, and C/EBPα and PPARγ2 in the second wave) (Farmer, 2006; Siersbaek et al., 2012). Once expressed and activated, CEBPβ and C/EBPδ drive the expression of the genes encoding the late transcription factors C/EBPα and PPARγ2, which activate downstream adipocyte-specific genes to control terminal differentiation into mature adipocytes (Farmer, 2006; Siersbaek et al., 2012).
Although the transcriptional control of adipogenesis by pro-adipogenic transcription factors is relatively well understood, the molecular mechanisms controlling the sequential waves of transcriptional activity are still poorly understood. Several post-transcriptional modifications of adipogenic transcription factors, such as C/EBPβ, are thought to link hormonal signaling to the transcriptional regulation program (Farmer, 2006; Kowenz-Leutz et al., 1994; Lynch et al., 2011). Here we show that PARylation of mouse C/EBPβ at three amino acids located in a key regulatory domain inhibits C/EBPβ’s DNA binding and transcriptional activities, and attenuates early events in the differentiation of adipocyte precursors. Our results provide a clear example of how PARylation of specific amino acids in a key transcriptional regulatory protein can affect the molecular and biochemical functions of a protein, as well as the biological outcomes that it controls.
RESULTS
Genetic depletion of PARP-1 enhances adipogenesis in primary preadipocytes
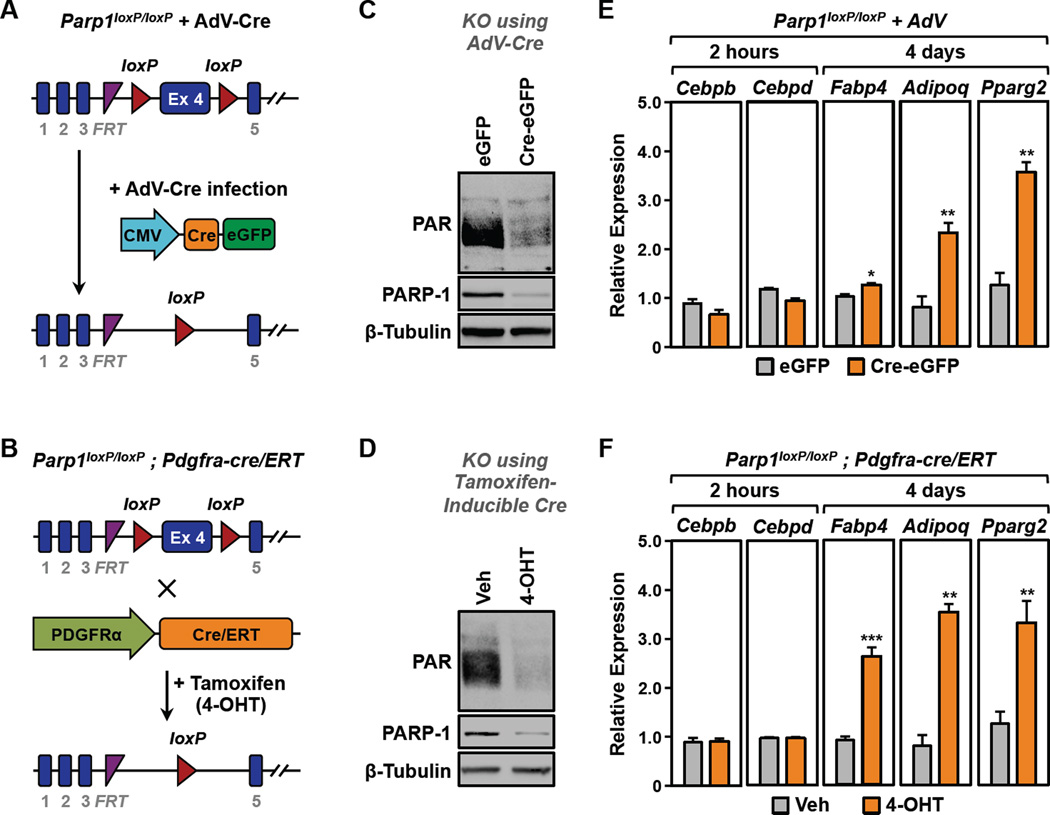
PARP-1 plays important roles in a wide variety of functionally interconnected tissues that control metabolic outcomes and fat metabolism, including liver, muscle, adipose, pancreas, and the central nervous system (Luo and Kraus, 2011, 2012), making it difficult to separate the effects of PARP-1 in one tissue from effects in other tissues. Thus, studies with whole body Parp1 null mice where PARP-1 is depleted throughout development have often yielded conflicting results (Bai et al., 2011; Devalaraja-Narashimha and Padanilam, 2010; Erener et al., 2012b; Lehmann et al., 2015; Luo and Kraus, 2011, 2012). To avoid these complications while examining the specific role of PARP-1 in adipogenesis, we developed a mouse line with a conditional (‘floxed’) allele of Parp1 (Parp1loxP/loxP) (Fig. 1A and S1). We then crossed the Parp1loxP/loxP mice with transgenic mice containing a Pdgfra-cre/ERT cassette to generate a Tamoxifen-inducible conditional allele of Parp1 (Parp1loxP/loxP;Pdgfra-cre/ERT) (Fig. 1B).
Figure 1. Genetic depletion of PARP-1 promotes the differentiation of primary preadipocytes from the stromal-vascular fraction (SVF).
(A and B) Primary SVF preadipocytes isolated from (A) Parp1loxP/loxP mice or (B) Parp1loxP/loxP;Pdgfra-cre/ERT mice were subjected to deletion of Parp1 in culture using adenovirus-Cre (AdV-Cre) or 4-OHT, respectively.
(C and D) Western blots showing the relative levels of PARP-1 and PAR in primary SVF preadipocytes with or without deletion of Parp1. β-tubulin was used as a loading control.
(E and F) SVF preadipocytes were subjected to Parp1 deletion using (E) AdV-Cre or (F) 4-OHT as shown in (A) and (B), respectively, prior to differentiation with MDI. Two hours or 4 days later, the relative expression of various adipocyte marker genes was assayed by RT-qPCR. Each bar represents the mean + SEM for three replicates. Bars marked with asterisks are statistically different from the control (Student’s t-test; *** p-value < 0.001 ** p-value < 0.01 or * p-value < 0.05).
[See also Fig. S1]
To explore the specific role of PARP-1 in adipogenesis, separated from any potential complicating developmental effects of Parp1 deletion, we isolated primary preadipocytes from the stromal vascular fraction (SVF) of adipose tissue (Rodeheffer et al., 2008; Van et al., 1976) collected from Parp1loxP/loxP and Parp1loxP/loxP;Pdgfra-cre/ERT mice. When then treated the SVF cells in culture with adenovirus-Cre (Adv-Cre) (Fig. 1A) or 4-hydroxytamoxifen (4-OHT) (Fig. 1B) to induce the depletion of PARP-1, which resulted in a dramatic reduction in total cellular PAR levels, as determined by Western blotting (Fig. 1, C and D). The cells were induced to differentiate with a cocktail of differentiation agents (MDI), containing IBMX, dexamethasone, and insulin. To assess the effects of PARP-1 depletion on adipogenesis, we monitored the expression of genes associated with adipogenesis by RT-qPCR: (1) ‘early’ genes, Cebpb and Cebpd, and (2) ‘late’ genes Fabp4, Adipoq, and Pparg2, which were assessed 2 hours or 4 days after treatment with MDI, respectively. We observed a significant increase in the expression of Fabp4, Adipoq, and Pparg2, which serve as markers of adipogenesis, upon PARP-1 depletion in both genetic models (Fig. 1, E and F). These results indicate that PARP-1 acts to attenuate adipogenesis, a conclusion that differs from previous results using Parp1 null mice and other cell-based models ((Erener et al., 2012a; Erener et al., 2012b; Lehmann et al., 2015); discussed in more detail below).
PARP-1 catalytic activity attenuates adipogenesis
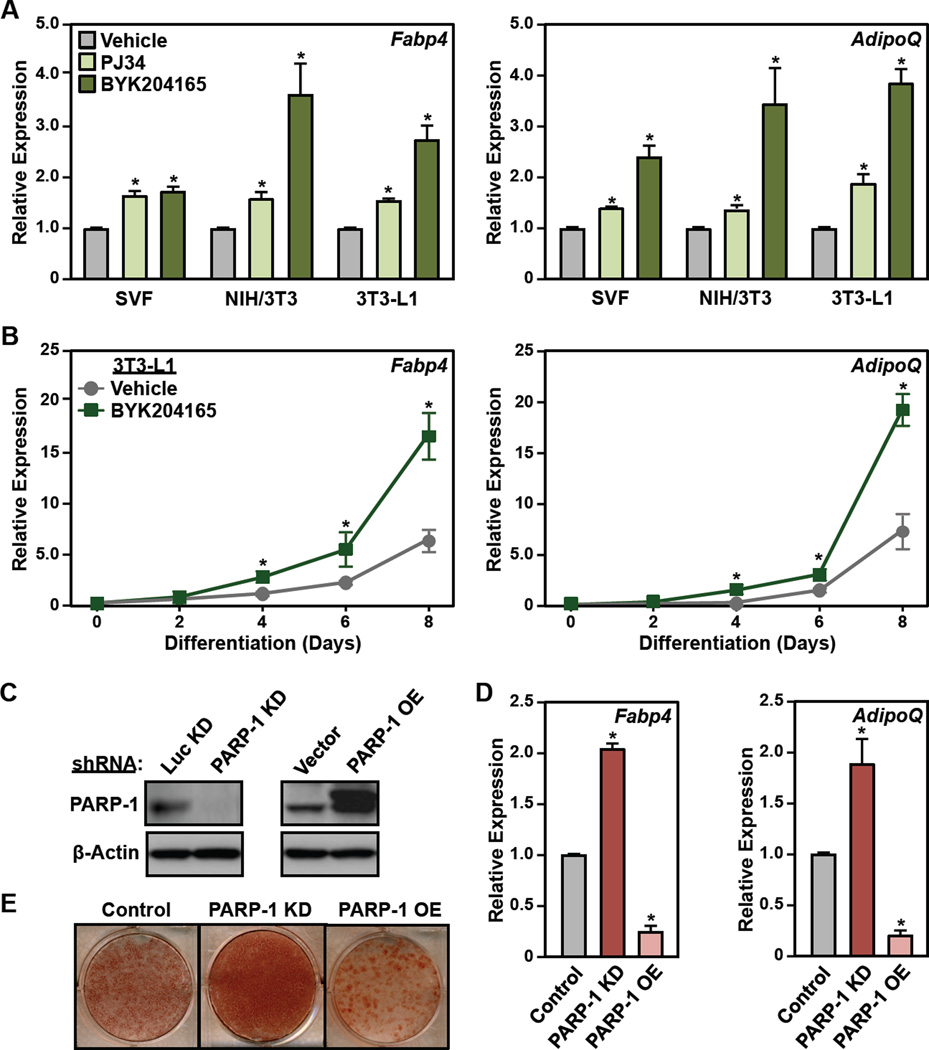
To explore the role of PARP-1 and its catalytic activity in the transcriptional events leading to adipogenesis more broadly, we examined the effect of PARP inhibitors in three mouse cell-based models of adipogenesis: (1) primary SVF preadipocytes (described above), (2) NIH/3T3 fibroblasts (Todaro and Green, 1963), and (3) 3T3-L1 committed preadipocytes (Green and Kehinde, 1975), all of which we differentiated with MDI. Both the PARP-1-selective inhibitor, BYK204165, and a broader spectrum PARP inhibitor, PJ34, promoted adipogenesis in all three cell types, as assessed by increased expression of mRNAs encoding markers of mature adipocytes, Fabp4 and AdipoQ (Figs. 2A, S2A, and S2B). The effect of BYK204165 on Fabp4 and AdipoQ expression was observed by day 4 post-MDI treatment and persisted until day 8 (Fig. 2B), indicating that PARP-1’s effects on early adipogenic events impact the formation of mature adipocytes.
Figure 2. Inhibition or depletion of PARP-1 promotes the differentiation of preadipocytes.
(A) SVF, NIH/3T3, and 3T3-L1 cells were treated with 5 µM PJ34 or 20 µM BYK204165 prior to differentiation with MDI. Four days later, the relative expression of adipocyte marker genes Fabp4 and AdipoQ was assayed by RT-qPCR. Each bar represents the mean + SEM for three replicates. Bars marked with an asterisk are statistically different from the control (Student’s t-test; p-value < 0.05).
(B) Time course of differentiation in 3T3-L1 cells in response to MDI ± 20 µM BYK204165. The relative expression of adipocyte marker genes Fabp4 and AdipoQ was assayed by RT-qPCR every two days. Each point represents the mean ± SEM for three replicates. Points marked with an asterisk are statistically different from the vehicle-treated control (Student’s t-test; p-value < 0.05).
(C) Western blots showing the levels of PARP-1 after knockdown (KD) (left, using shRNAs targeting PARP-1 or luciferase, Luc,) or ectopic expression (right, Flag-tagged PARP-1) in 3T3-L1 cells. β-actin was used as a loading control.
(D and E) 3T3-L1 cells, with or without stable knockdown or ectopic expression of PARP-1, as (C), were differentiated with MDI. (C) Oil Red-O staining at day 8. (D) RT-qPCR for adipocyte marker genes Fabp4 and AdipoQ at day 4. Each bar represents the mean + SEM for three replicates. Bars marked with an asterisk are statistically different from the control (Student’s t-test; p-value < 0.05).
[See also Fig. S2]
In a complementary set of experiments, we observed that shRNA-mediated knockdown (KD) of PARP-1 promotes adipogenesis in 3T3-L1 cells (versus a luciferase shRNA control; Luc) whereas ectopic overexpression (OE) of PARP-1 represses adipogenesis (versus an empty vector control), as assessed by adipogenic marker gene expression (at day 4) and Oil Red O staining (at day 8) (Fig. 1, C – E). As with the PARP inhibitors, the effects of PARP-1 knockdown persisted until the later stages of adipogenesis (Fig. S2C). Re-expression of PARP-1 in cells subjected to shRNA-mediated knockdown of PARP-1 reversed the effects of knockdown (Fig. S2, D and E), indicating that the shRNA was on target. These results indicate that the effects of the PARP inhibitors are mediated by PARP-1. Together, these initial experiments implicate PARP-1 and its catalytic activity in the attenuation of adipogenesis.
Nuclear PAR levels fluctuate during adipogenesis, demarcating the transition between two distinct waves of transcription
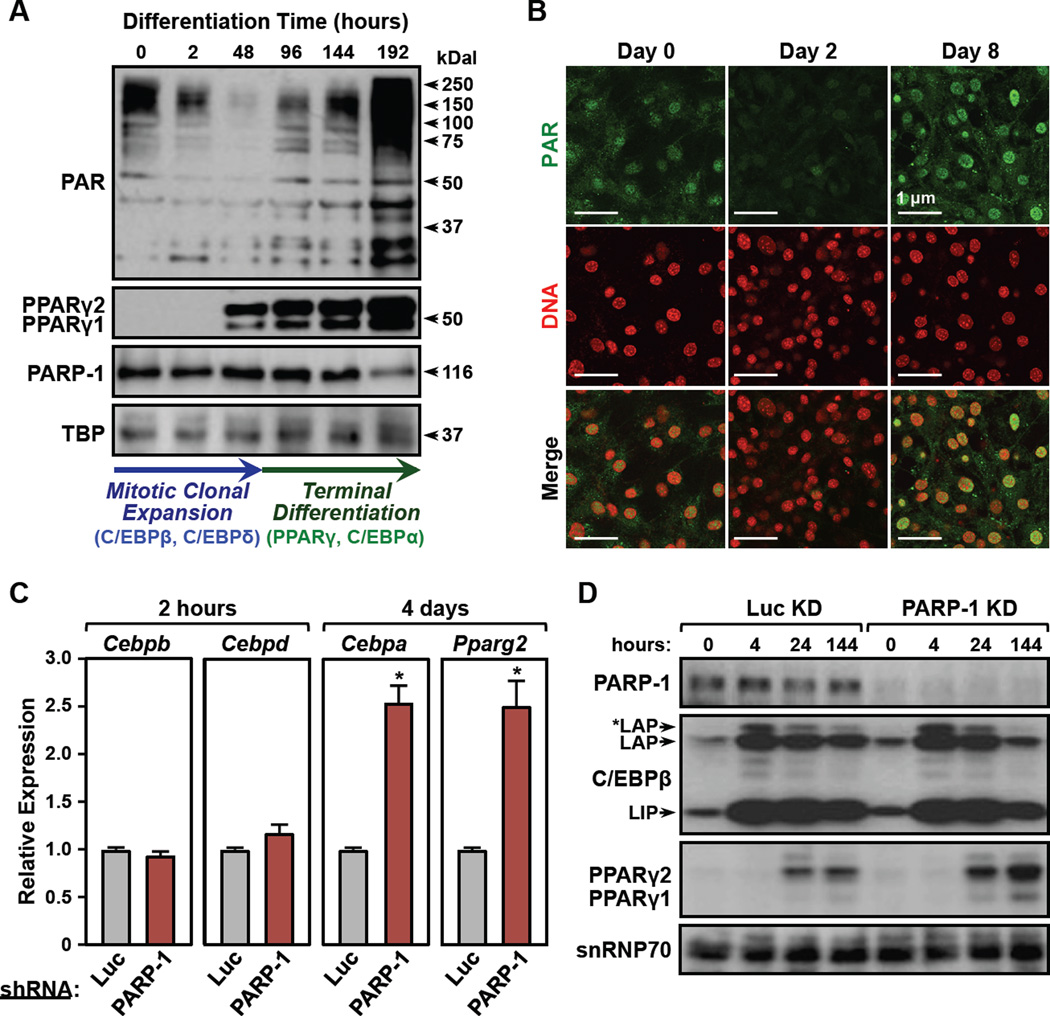
To gain mechanistic insights into the regulation of adipogenesis by PARP-1, we conducted additional experiments using 3T3-L1 cells. Consistent with previous studies (Janssen and Hilz, 1989; Pekala et al., 1981), we observed a reduction in nuclear PAR levels (as detected by Western blotting for PAR) during the first two days of adipogenesis (i.e., post-differentiation with MDI), followed by an increase in PAR levels when the cells undergo terminal differentiation (Fig. 3, A and B). These changes in nuclear PAR levels, however, are not due to alterations in the levels of PARP-1, which remain constant throughout adipogenesis (Fig. 3A), suggesting instead that they may be due to fluctuations in the activity of PARP-1. Interestingly, the inflection point for the changes in nuclear PAR levels at 48 hours post-differentiation corresponds with two well-characterized cellular states during adipogenesis: proliferation and differentiation. These cellular states are driven by two distinct waves of transcription factor activation (i.e., C/EBPβ and C/EBPδ in the first wave, and C/EBPα and PPARγ2 in the second wave) (Farmer, 2006; Siersbaek et al., 2012).
Figure 3. Nuclear PAR levels fluctuate and demarcate the transition between two distinct waves of transcription during adipogenesis.
(A) (Top) Western blots showing the levels of PAR, PPARγ1, PPARγ2, and PARP-1 in nuclear extracts from 3T3-L1 cells during a time course of differentiation with MDI. TBP was used as a loading control. (Bottom) The arrows indicate two distinct phases of adipogenesis: (1) mitotic clonal expansion (0 to 48 hours post MDI; elevated C/EBPβ and C/EBPδ; blue) and (2) terminal differentiation (>48 hours post MDI; elevated PPARγ and C/EBPα; green).
(B) Immunofluorescent staining of 3T3-L1 cells for PAR (green) and DNA (red) during a time course of differentiation with MDI.
(C) RT-qPCR analysis showing the expression of early transcription factors (2 hours post MDI; Cebpb, Cebpd), and late transcription factors (4 days post MDI; Cebpa, Pparg2) in 3T3-L1 cells ± knockdown using shRNAs targeting PARP-1 or luciferase (Luc, as a control). Each bar represents the mean + SEM for three replicates. Bars marked with an asterisk are statistically different from the control (Student’s t-test; p-value < 0.05).
(D) Western blots showing the expression of PARP-1, C/EBPβ, and PPARγ in 3T3-L1 cells ± knockdown using shRNAs targeting PARP-1 or luciferase (Luc, as a control) during a time course of differentiation with MDI. snRNP70 was used as a loading control.
[See also Fig. S3]
With this in mind, we determined the effects of PARP-1 knockdown on the expression of mRNAs encoding two early adipogenic transcription factors (i.e., C/EBPβ and C/EBPδ) in the first wave of transcription (at 2 hours), and two late adipogenic transcription factors (i.e., C/EBPα and PPARγ2) in the second wave of transcription (at 4 days). Changes in expression were observed for Cebpa and Pparg2 (encoding late transcription factors), but not for Cebpb and Cebpd (encoding early transcription factors) (Fig. 3C). Similar results were observed in SVF cells and NIH/3T3 cells upon PARP-1 depletion (Fig. 1, C–F and Fig. S3, A and B). Although the levels of PPARγ2 increased upon PARP-1 knockdown, the overall levels and distribution of the three isoforms of C/EBPβ (LIP, LAP, and *LAP) were not affected by PARP-1 knockdown during the adipogenesis (Fig. 3D), consistent with the mRNA expression. Together, these results indicate that hormonal signals may promote the differential regulation of PARP-1 activity at different stages of adipogenesis, leading to sequential regulation of adipogenic transcription factors and the adipogenic transcription program.
PARP-1 and PARylation regulate C/EBPβ-dependent expression of genes encoding late transcription factors
In the sequential model of adipogenic gene expression, early transcription factors, such as C/EBPβ, control the expression of genes encoding late transcription factors, such as Cebpa and Pparg2 (Farmer, 2006; Siersbaek et al., 2012). Thus, we hypothesized that the effects of PARP-1 depletion or inhibition on the expression of Cebpa and Pparg2 that we observed in 3T3-L1 cells might be mediated through C/EBPβ. In this regard, we observed an enrichment of both PARP-1 and C/EBPβ at the promoters of the Cebpa and Pparg2 genes in 3T3-L1 cells 24 hours after MDI-induced differentiation (Fig. 4, A and B). Furthermore, we observed a significant increase in C/EBPβ at both promoters upon knockdown of PARP-1 (Fig. 4A) or inhibition of PARP-1 catalytic activity (Fig. 4B). The latter occurred without a loss of PARP-1 binding at the promoters (Fig. 4B). Importantly, the effects of PARP-1 knockdown on the expression of Cebpa and Pparg2, as well as the expression of Fabp4, were abrogated by shRNA-mediated knockdown C/EBPβ (Fig. 4C; the results for Parp1 and Cebpb are shown to confirm knockdown). Together, these results connect the localization and function of PARP-1 and C/EBPβ to the expression of genes encoding late transcription factors. Similar results were observed in NIH/3T3 fibroblasts (Fig. 3), supporting our conclusions in a non-lineage-committed cell-based model of adipogenesis.
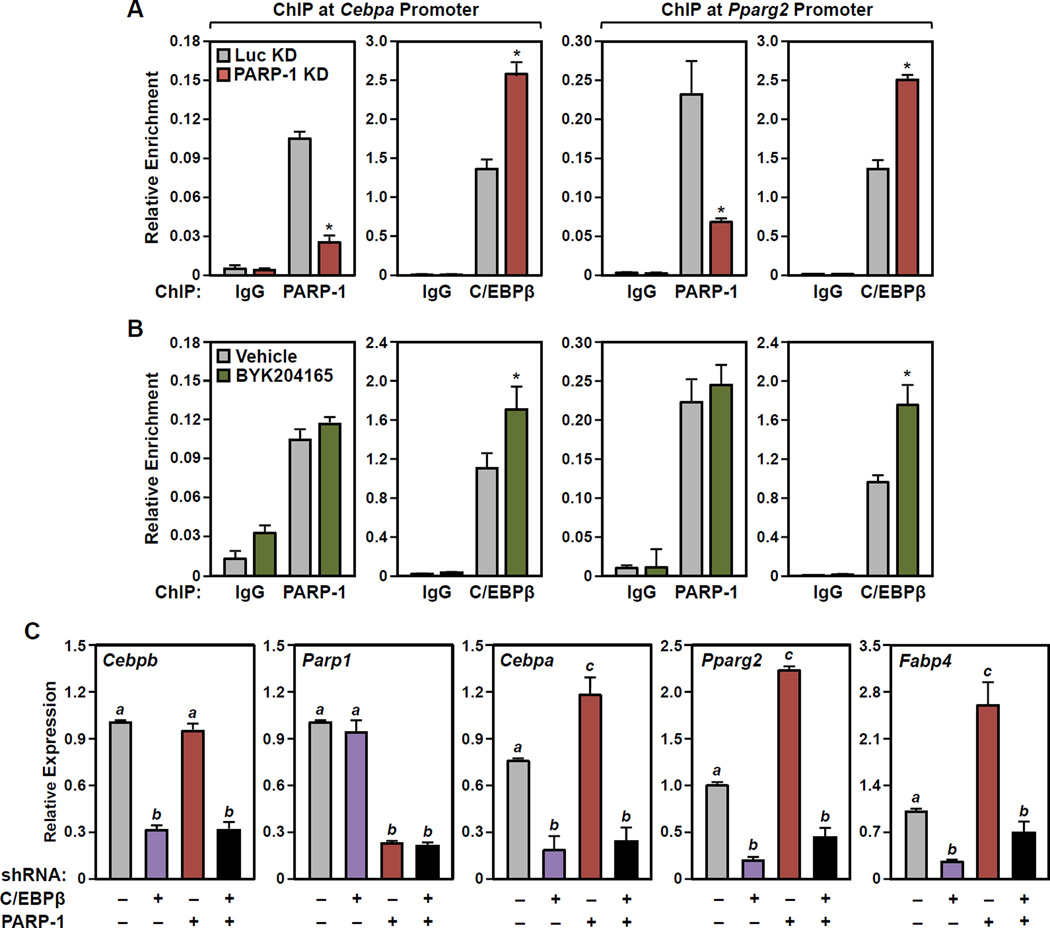
Figure 4. PARP-1 and PARylation modulate the binding of C/EBPβ to the promoters of C/EBPβ target genes encoding late transcription factors in 3T3-L1 cells.
(A and B) Results from ChIP-qPCR assays for PARP-1 or C/EBPβ (as indicated) binding at the Cebpa (left) and Pparg2 (right) gene promoters performed in 3T3-L1 cells 24 hours after differentiation with MDI. The assays were performed (A) with knockdown using shRNAs targeting PARP-1 or luciferase (Luc, as a control), or (B) ± 20 µM BYK204165 (1 hour pretreatment, followed by 24 hours of treatment after adding MDI). Each bar represents the mean + SEM for three replicates. Bars marked with an asterisk are statistically different from the control (Student’s t-test; p-value < 0.05).
(C) Results from RT-qPCR assays showing that the effects of PARP-1 knockdown on the expression of Cebpa, Pparg2, Fabp4 are abrogated by C/EBPβ knockdown in 3T3-L1 cells.
Results for Parp1 and Cebpb are shown to confirm shRNA-mediated knockdown. Each bar represents the mean + SEM for three replicates. Bars marked with different letters are statistically different from each other (ANOVA; p-value < 0.05).
[See also Fig. S3]
C/EBPβ is PARylated by PARP-1 in Preadipocytes
Given the observed requirement of PARP-1 catalytic activity for the attenuation of adipogenesis in preadipocytes (primary, NIH/3T3, and 3T3-L1) (Fig. 2A), as well as the inverse relationship between PARP-1 activity and C/EBPβ enrichment at the promoters of genes encoding late transcription factors (Figs. 4 and S3), we surmised that PARP-1 may directly antagonize the activity of C/EBPβ at those promoters. More specifically, we hypothesized that PARP-1 might PARylate C/EBPβ to inhibit its DNA binding and transcriptional activity. To test this hypothesis, we first determined if C/EBPβ is PARylated by PARP-1. To do so, we prepared nuclear extracts from 3T3-L1 cells after shRNA-mediated knockdown of PARP-1 or C/EBPβ, followed by Western blotting for PAR, C/EBPβ, and PARP-1. These experiments revealed a distinct band of PARylated C/EBPβ, which was dramatically reduced by knockdown of PARP-1 (identifying the PARylated protein as a target of PARP-1) and C/EBPβ (confirming that the PARylated protein is a C/EBPβ) (Figs. 5A and S4A). The PARylation of C/EBPβ was confirmed in PAR-containing immunoprecipitates from 293T cells ectopically expressing HA-tagged C/EBPβ (Fig. 5B), as well as C/EBPβ-containing immunoprecipitates from 3T3-L1 cells ectopically expressing doxycycline (Dox)-inducible HA-tagged C/EBPβ (Fig. 5C and S4, B – D). In the latter, we observed a considerable reduction in C/EBPβ PARylation after 24 hours of MDI-induced differentiation (Fig. 5C), corresponding to a similar reduction in total cellular PAR levels in early differentiation (Fig. 3A). Together, these results demonstrate that (1) C/EBPβ is PARylated by PARP-1 and (2) PARylation of C/EBPβ decreases in the early phases of adipogenesis.
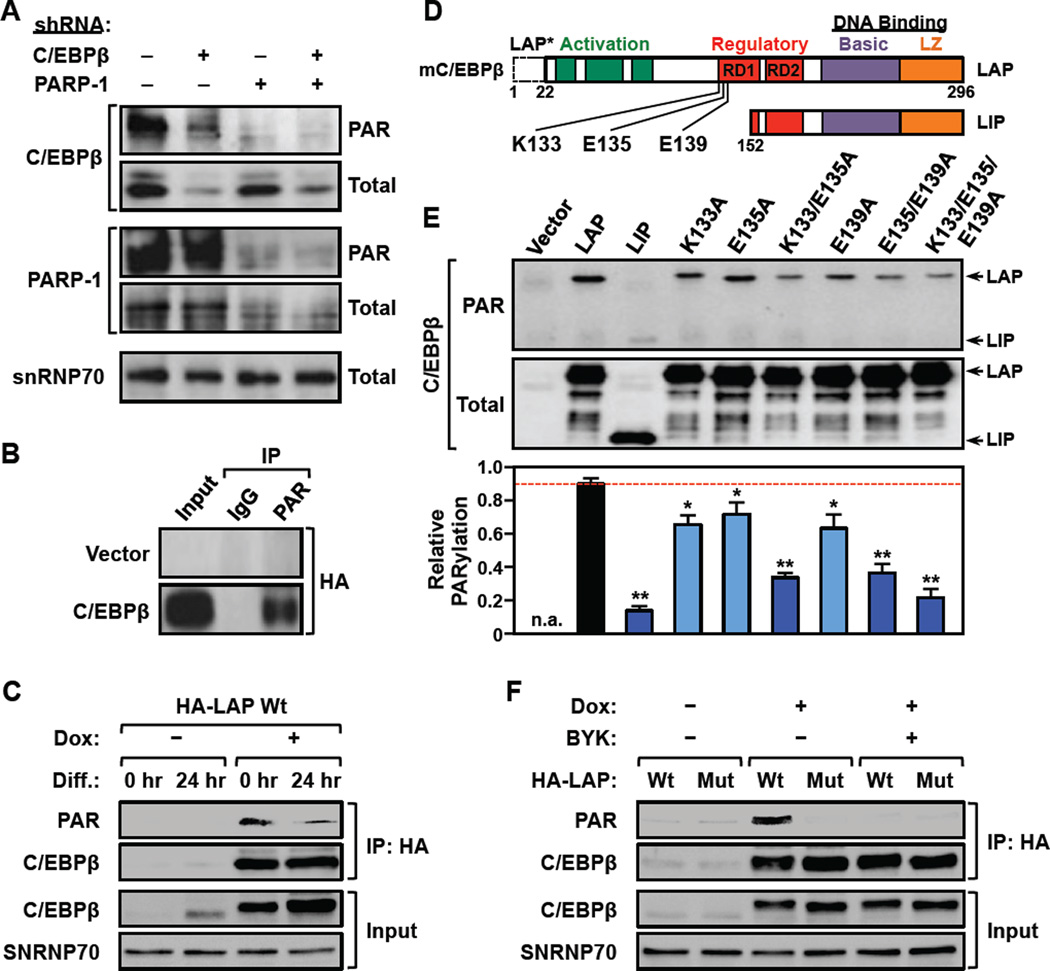
Figure 5. PARP-1 PARylates C/EBPβ at specific sites in 3T3-L1 cells.
(A) PARP-1-dependent PARylation of C/EBPβ. Western blots showing the levels of PARylated and total C/EBPβ and PARP-1 in control, C/EBPβ-, or PARP-1-depleted 3T3-L1 cells. Knockdown was achieved by using shRNAs targeting C/EBPβ, PARP-1, or luciferase (Luc, as a control). snRNP70 was used as a loading control. The full blot of PARylated C/EBPβ is shown in Fig. S4A.
(B) PARylated proteins were immunoprecipitated from 293T cells ectopically expressing HA-tagged C/EBPβ and analyzed by Western blotting as indicated.
(C) Reduced PARylation of C/EBPβ in 3T3-L1 cells upon differentiation. The cells were differentiated with MDI for 24 hours and the expression of HA-tagged mouse C/EBPβ was induced from a transgene by treatment with Dox for 8 hours. The cells were then subjected to immunoprecipitation with an anti-HA antibody. The input and the immunoprecipitate (IP) were analyzed by Western blotting for C/EBPβ and PAR as indicated. snRNP70 was used as a loading control.
(D) Schematic diagram of mouse C/EBPβ showing the ADP-ribosylation site determined by mass spectrometry (E135), as well as two adjacent sites determined by mutagenesis (K133, E139). C/EBPβ isoforms: LAP, common full-length, LAP*, 22 amino acid N-terminal extension; LIP, N-terminally truncated.
(E)Top, Extracts from 293T cells ectopically expressing wild-type or mutant HA-tagged C/EBPβ proteins were analyzed by Western blotting for PARylated and total C/EBPβ. Bottom, Quantification of the relative PARylation of C/EBPβ proteins in Western blots like the one shown in the panel above. Each bar represents the mean + SEM for three independent experiments. Bars marked with asterisks are statistically different from the control (Student’s t-test; ** p-value < 0.01 or * p-value < 0.05).
(F) A PARylation site mutant of C/EBPβ is not PARylated in 3T3-L1 cells. The expression of HA-tagged wild-type (Wt) or PARylation site mutant (K133A/E135A/E139A; Mut) mouse C/EBPβ was induced from a transgene by treatment with Dox. The cells were induced to differentiate with MDI for 24 hours ± 20 µM BYK204165 (BYK) and then subjected to immunoprecipitation (IP) with an anti-HA antibody. The input and the immunoprecipitated material were analyzed by Western blotting for C/EBPβ and PAR. snRNP70 was used as a loading control.
[See also Fig. S4]
To explore the sites at which C/EBPβ is PARylated by PARP-1, we mined an existing mass spectrometry database of experimentally-determined site-specific ADP-ribosylation, which identified glutamate 175 (Glu175 or E175) in human C/EBPβ as a major site of ADP-ribosylation (Zhang et al., 2013). This amino acid, which is homologous to glutamate 135 (Glu135 or E135) in mouse C/EBPβ, is located within a conserved regulatory domain (RD1) (Abdou et al., 2013; Williams et al., 1995) (Fig. 5D). Thus, we focused our subsequent functional analyses on E135 to explore the function of C/EBPβ PARylation in adipogenesis. In addition, since previous analyses have shown that PARP-1 is promiscuous in its site selection, particularly when primary target sites are altered by mutagenesis (Altmeyer et al., 2009), we also considered potential secondary PARylation sites located adjacent to E135, including a nearby lysine (lysine 133; K133) and glutamate (glutamate 139; E139) residues (Fig. 5D).
To verify that these residues are PARylated in cells, we generated vectors to express the following versions of mouse C/EBPβ: (1) wild-type (LAP), (2) a N-terminally truncated form (LIP) lacking K133, E135, and E139, and (3) single, double, or triple mutants with alanine substitutions at K133, E135, and E139. We then expressed these forms of C/EBPβ in 293T cells, and monitored their expression and PARylation by Western blotting. As expected, wild-type C/EBPβ was robustly PARylated, but LIP exhibited little detectable PARylation (Fig. 5E). Furthermore, mutation of the individual sites (K133, E135, or E139) caused a modest (~20%), but significant, reduction of PARylation, whereas mutation of combinations of two or three sites (K133/E135, E135/E139, or K133/E135/E139) caused a dramatic (as much as 70%) reduction of PARylation (Fig. 5E). In 3T3-L1 cells, the triple point mutant (K133/E135/E139) showed no detectable PARylation (Fig. 5F). Furthermore, the PARylation of wild-type C/EBPβ was completed inhibited by treating the cells with the PARP-1-selective inhibitor BYK204165 (Fig. 5F). These results indicate that E135 and adjacent amino acids (K133 and E139) in C/EBPβ can be PARylated by PARP-1 in preadipocytes.
PARylation of C/EBPβ by PARP-1 inhibits its DNA binding activity in vitro and in vivo
Next, we sought to determine the mechanisms by which PARylation of C/EBPβ inhibits its activity. The ChIP-qPCR results shown in Figs. 4 and S3 demonstrate that PARP-1 can attenuate the binding of C/EBPβ to the promoters of its target genes, suggesting that PARylation of C/EBPβ may inhibit its DNA binding activity. To determine how the PARylation by PARP-1 might affect C/EBPβ DNA binding activity, we established an electrophoretic mobility shift assay (EMSA) in which ectopically expressed C/EBPβ in 293T cell nuclear extracts bound to a 32P-labeled double stranded oligonucleotide probe corresponding to the C/EBP regulatory element in the Cebpa promoter (“probe”) (Christy et al., 1991). The ectopically expressed C/EBPβ in the 293T cell nuclear extracts was PARylated, and the PARylation was inhibited by treatment of the cells with PJ34 or BYK204165 (Fig. S5A). In EMSAs, the binding of minimally PARylated C/EBPβ (in the PARP inhibitor-treated extracts) exhibited considerably more binding per unit of protein than more highly PARylated C/EBPβ (Fig. S5, B and C).
We performed a similar experiment comparing wild-type C/EBPβ with the K133A/E135A/E139A triple point mutant, in this case enhancing the PARylation of C/EBPβ by incubating the 293T cell nuclei with 100 µM NAD+ for 30 min. prior to the preparation of nuclear extracts. As expected, wild-type C/EBPβ showed a dramatic increase in PARylation in response to incubation of the nuclei with NAD+, whereas the triple point mutant showed only a very modest increase in PARylation under the same conditions (Fig. 6A). In EMSAs, the binding of wild-type C/EBPβ to the probe was significantly reduced upon NAD+-dependent PARylation (Fig. 6, B and C). In contrast, binding of the triple point mutant was unaffected by incubation with NAD+ (Fig. 6, B and C. Similar results were observed with purified recombinant C/EBPβ PARylated with purified recombinant PARP-1 + NAD+ in vitro (Fig. S5, D – G).
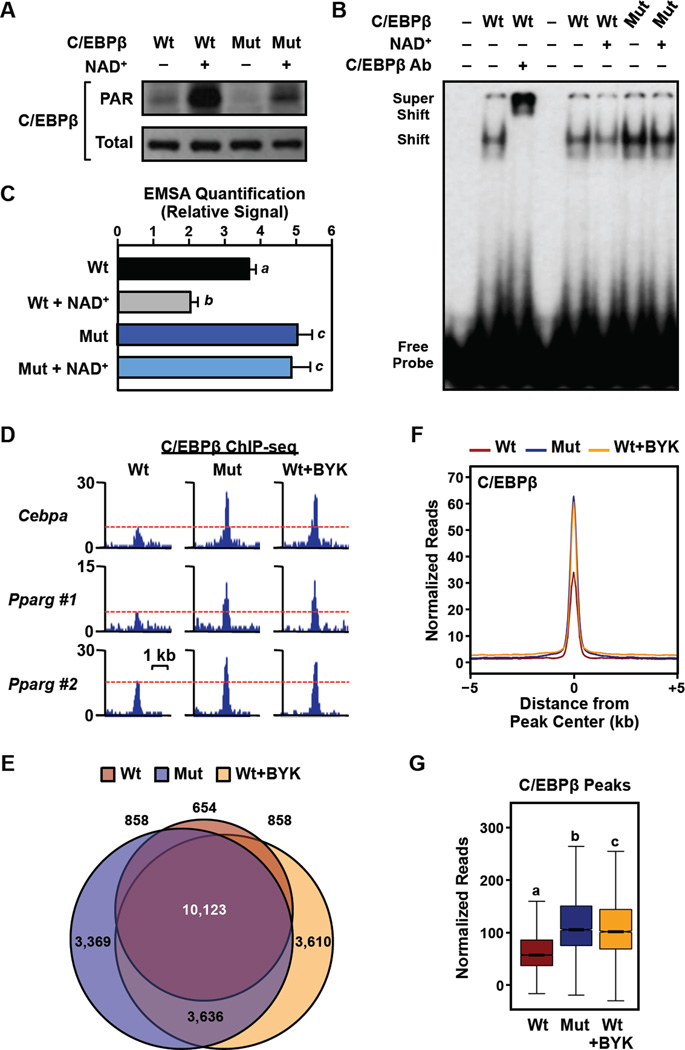
Figure 6. PARylation of C/EBPβ by PARP-1 at specific sites inhibits C/EBPβ binding to DNA in vitro and in cells.
(A through C) Mutation of PARP-1-dependent sites of PARylation render C/EBPβ resistant to PARylation-induced decreases in DNA binding. Wild-type or PARylation site point mutant (K133A/E135A/E139A; Mut) C/EBPβ proteins were expressed in 293T cells. Nuclei were collected and then treated ± 100 µM NAD+ for 30 min. to stimulate PARylation prior to preparation of nuclear extracts. (A) Western blots showing the levels of PARylated (top) and total (bottom) C/EBPβ in nuclear extracts prepared from the transfected 293T cells. (B) Representative EMSA experiment performed using nuclear extract and a labeled double stranded oligonucleotide probe corresponding to the C/EBP regulatory element in the Cebpa promoter (probe) ± supershifting using a C/EBPβ antibody. (C) Quantification of the results from experiments like those shown in (B). Each bar represents the mean + SEM for three independent experiments. Bars marked with different letters are statistically different from each other (ANOVA; p-value < 0.05).
(D through F) Mutation of PARP-1-dependent sites of PARylation render C/EBPβ resistant to PARylation-induced decreases in chromatin binding in 3T3-L1 cells. Expression of HA-tagged wild-type (Wt) or PARylation site mutant (K133A/E135A/E139A; Mut) mouse C/EBPβ was induced from a transgene by treatment with Dox for 8 hours ± 20 µM BYK204165 (BYK). The cells were crosslinked and subjected to ChIP-seq for C/EBPβ. (D) ChIP-seq browser tracks for C/EBPβ in the three conditions tested (Wt, Mut, and Wt+BYK) at genomic loci with C/EBPβ enhancers near the Cebpa and Pparg genes. (E) Venn diagram showing the overlap of statistically significant peaks of C/EBPβ in the three conditions tested. (F) Metaplot and (G) box plot analyses of C/EBPβ binding in 3T3-L1 cells by ChIP-seq in the three conditions tested.
[See also Fig. S5]
To determine the effects of PARylation on C/EBPβ binding to its cognate enhancers in cells, we performed ChIP-seq for C/EBPβ in 3T3-L1 cells ectopically expressing doxycycline (Dox)-inducible wild-type C/EBPβ or the K133A/E135A/E139A triple point mutant. We observed a dramatic increase in C/EBPβ binding at enhancers near the Cebpa and Pparg genes with the triple point mutant compared to wild-type C/EBPβ (Fig. 6D). Furthermore, treatment of the cells with BYK204165 enhanced the binding of wild-type C/EBPβ to the same enhancers (Fig. 6D). Globally, the significant peaks of C/EBPβ binding in the three conditions (Wt C/EBPβ, Mut C/EBPβ, and Wt C/EBPβ + BYK204165) showed considerable overlap (~2/3 of the sites), although unique sites were observed in all conditions (Fig. 6E). For the common sites, Mut C/EBPβ and Wt C/EBPβ + BYK204165 showed ~2-fold more binding than Wt C/EBPβ (Fig. 6, F and G). Together, the results from our in vitro and cell-based assays demonstrate that (1) PARylation inhibits the binding of C/EBPβ to its cognate DNA response elements and (2) the DNA binding activity of the PARylation site point mutant C/EBPβ protein is resistant to PARP-1-mediated PARylation.
PARylation of C/EBPβ inhibits its transcriptional activity
Finally, we sought to determine if PARylation of C/EBPβ and the resulting loss of DNA binding activity inhibits C/EBPβ’s transcriptional activity. To test this, we ectopically expressed wild-type C/EBPβ, or single, double, or triple PARylation site point mutants of C/EBPβ (E135A, K133A/E135A, or K133A/E135A/E139A) in 3T3-L1 cells. RT-qPCR analysis showed equal expression of the mRNAs encoding all four versions of C/EBPβ, as expected (Fig. 7A, left panel). Previous studies have shown that ectopic expression of C/EBPβ in 3T3-L1 cells promotes spontaneous differentiation of the cells in the absence of the inducer (i.e., MDI cocktail) (Yeh et al., 1995). Indeed, we observed an ~5-fold increase in Pparg2 and Fabp4 mRNA after ectopic expression of wild-type C/EBPβ in 3T3-L1 cells (Fig. 7A, middle and right panels). Furthermore, we observed significantly more Pparg2 and Fabp4 mRNA after ectopic expression of the double or triple PARylation site point mutants of C/EBPβ compared to wild-type C/EBPβ (Fig. 7A, middle and right panels). These results demonstrate that blocking PARP-1-dependent PARylation of C/EBPβ enhances the transcription of native C/EBPβ target genes.
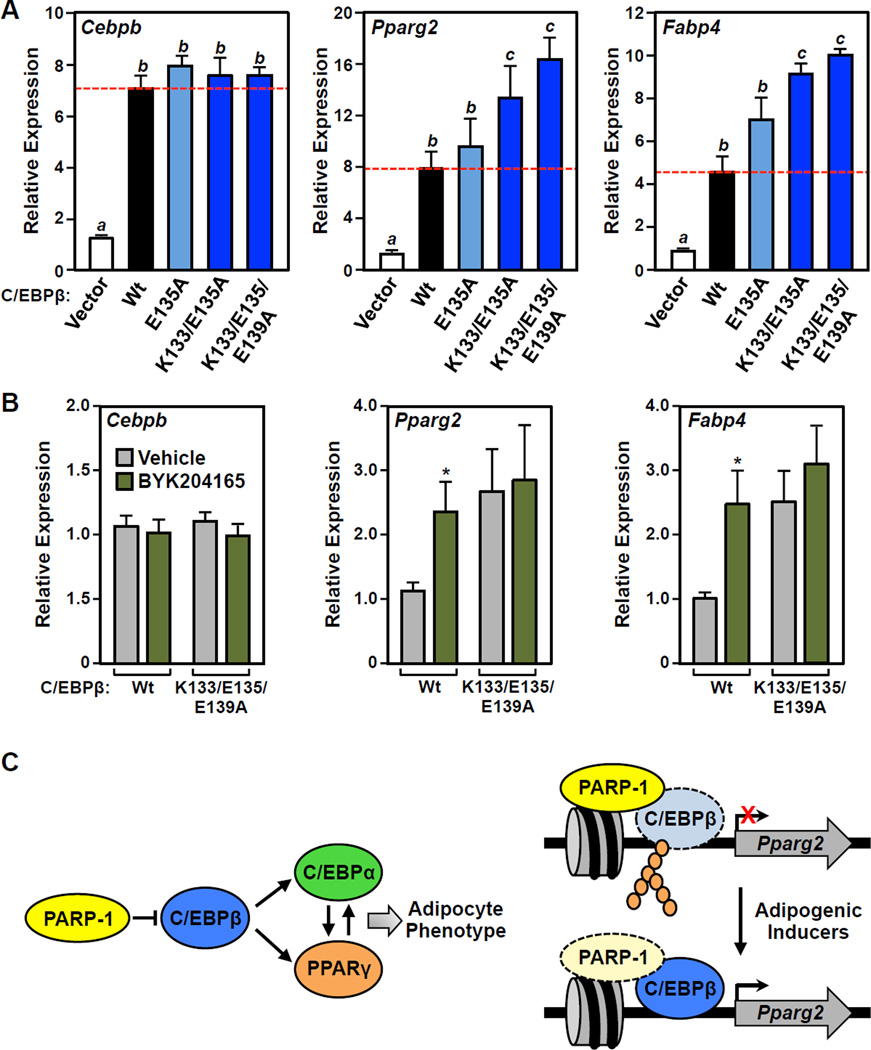
Figure 7. Mutation of PARP-1-dependent of sites PARylation in C/EBPβ enhances adipogenesis and renders C/EBPβ resistant to the effects of PARP inhibitors.
(A and B) Wild-type or mutant C/EBPβ were ectopically expressed in 3T3-L1 cells, as indicated, which induces spontaneous differentiation of the cells in the absence of MDI. The expression of Pparg and Fabp4 was determined by RT-qPCR performed 4 days post-transfection. Results for Cebpb are shown to confirm equal ectopic expression of the mutants.
(A) Mutation of PARP-1-dependent sites of PARylation in C/EBPβ enhances adipogenesis. Bars marked with different letters are statistically different from each other (ANOVA; p-value < 0.05). (B) PARylation site C/EBPβ mutants are resistant to the effects of PARP inhibitor. The cells were treated ± 20 µM BYK204165 (1 hour pretreatment, followed by 4 days of treatment after adding MDI). Bars marked with an asterisk are statistically different from the corresponding control (Student’s t-test; p-value < 0.05).
(C) Model for the role of PARP-1 in attenuating adipogenesis, as described in the text.
To link the enhanced activity of the C/EBPβ PARylation site point mutants more directly to impaired modification by PARP-1, we performed assays similar to those shown in Fig. 7A using the K133A/E135A/E139A mutant in the presence or absence of BYK204165. Again, we observed no differences in the expression of the mRNAs encoding wild-type or mutant C/EBPβ under the different treatment conditions (Fig. 7B, left panel). Treatment with BYK204165 significantly enhanced the expression of Pparg2 and Fabp4 in the presence of ectopically expressed C/EBPβ, as expected based on the results shown in Fig. 2, A and B, and S2B (Fig. 7B, middle and right panels). In contrast, BYK204165 had no effect on the expression of Pparg2 and Fabp4 with the K133A/E135A/E139A C/EBPβ mutant (Fig. 7B, middle and right panels). These results provide evidence for a direct link between PARP-1 activity, PARylation of C/EBPβ, and C/EBPβ gene regulatory activity.
DISCUSSION
In the studies described herein, we identified amino acid residues in C/EBPβ, a pro-adipogenic transcription factor, that are PARylated by the NAD+-dependent catalytic activity of PARP-1. In addition, we connected PARP-1-dependent PARylation of C/EBPβ at those residues to the regulation of adipogenesis in various cell-based models. Few examples of definitive biological roles for site-specific PARylation of proteins exist in the literature. In this regard, our results provide a clear example of how PARylation of specific amino acids in a key transcriptional regulatory protein can affect the molecular and biochemical functions of a protein, as well as the biological outcomes that it controls. More broadly, our results provide new insights into the NAD+-dependent regulation of the transcriptional program controlling the differentiation of adipocyte precursors (preadipocytes) during adipogenesis.
Site-specific PARylation by PARP-1 modulates the molecular functions of C/EBPβ
Our understanding of the biological roles of ADP-ribosylation has been hampered by the limited information available on the specific residues targeted by the catalytic activity of PARP family members in various biological contexts. Recent advances in mass spectrometry and proteomics have begun to address this issue, leading to the identification of PARP target proteins and the amino acids at which they are modified (reviewed in (Daniels et al., 2015)). In this regard, a previous unbiased mass spectrometry-based screen identified E175 in human C/EBPβ (homologous to E135 in mouse C/EBPβ) as a major site of ADP-ribosylation (Zhang et al., 2013). We confirmed E135 in mouse C/EBPβ as a functional site of PARP-1-mediated PARylation. In addition, we identified two nearby residues (K133 and E139) that can also be PARylated by PARP-1. Together, these three amino acids account for the vast majority of PARP-1-dependent PARylation of C/EBPβ in cells (see Fig. 5F) and mediate PARP-1-dependent effects on C/EBPβ function (see Figs. 6 and 7).
Previous studies have demonstrated clear effects of PARylation of target proteins, although the sites of PARylation were not mapped in most cases. For example, we have shown that PARylation of KDM5B, a histone H3 lysine 4 demethylase, by PARP-1 prevents its binding to chromatin and blocks its enzymatic activity (Krishnakumar and Kraus, 2010b). Additional studies have shown that a number of DNA-binding transcription factors (e.g., Sox, YY1, and NFAT) are PARylated by PARP-1 (Gao et al., 2009; Oei and Shi, 2001; Olabisi et al., 2008). The few examples in the literature where the sites of PARylation on target proteins have been identified are NFAT (increases DNA binding; (Olabisi et al., 2008)), p53 (inhibits nuclear export; (Kanai et al., 2007)), and NELF-E (inhibits RNA binding and NELF-dependent promoter-proximal pausing by RNA polymerase II; (Gibson et al., 2016)). Many of the molecular and biochemical effects of PARylation on these target proteins are likely due to the nature of PAR itself, namely a large charged polymer that can elicit both steric and charge effects on target protein functions (Gibson and Kraus, 2012).
Our results indicate that PARylation by PARP-1 can inhibit the sequence-specific binding of C/EBPβ to DNA and chromatin and, conversely, that inhibition of PARylation can enhance the binding of C/EBPβ to DNA and chromatin. Reduced binding of PARylated C/EBPβ to promoters is likely to account for the relatively low C/EBPβ-dependent transcription of target genes. We cannot, however, exclude additional effects on C/EBPβ transcriptional activity. C/EBPβ possesses an amino-terminal activation domain, a carboxyl-terminal basic-leucine zipper (bZIP) DNA binding domain, and a central regulatory domain (Fig. 5D). The latter, which contains the sites of PARylation, functions as a bipartite regulatory domain that controls DNA binding by C/EBPβ and supports its transcriptional activity under some cell types and promoter contexts (Abdou et al., 2013; Williams et al., 1995). Previous studies have shown that signal-induced post-translational modification (e.g., phosphorylation, sumoylation, or acetylation) of the regulatory domain regulate C/EBPβ DNA binding activity and context specific transcriptional activity by: (1) changing the conformation of the protein, (2) modulating its exposure to target DNA elements, or (3) altering other sites of post-translational modification (Kowenz-Leutz et al., 1994; Lynch et al., 2011; Williams et al., 1995). PARylation of this domain may directly inhibit or block intra- or intermolecular interactions required for the function of the regulatory domain through allosteric effects, thus affecting DNA binding by C/EBPβ. This possibility will require additional analysis in future studies.
A role for site-specific PARylation of C/EBPβ in adipogenesis
As noted above, adipogenesis is controlled by two sequential waves of transcription driven by two distinct waves of transcription factor activation (Farmer, 2006; Siersbaek et al., 2012). Identifying the regulators that mediate the transition between these the two waves is key to understanding the process. Interestingly, the transition between the two waves of transcription, which occurs 24–48 hour post-differentiation, corresponds to a nadir of nuclear PAR levels, as well as the following effects on C/EBPβ: reduced PARylation, increased DNA binding, and enhanced transcriptional activity. Together, these results support a role for PARP-1 in the transcriptional switch during early adipogenesis (i.e., the transition of preadipocytes into adipocytes) through the modulation C/EBPβ transcriptional activity. How PARP-1 catalytic activity is regulated to control the timing and cessation of C/EBPβ PARylation during adipogenesis is currently unknown, but may involve cellular signaling directly to PARP-1 (Krishnakumar and Kraus, 2010a; Luo and Kraus, 2012) or possibly the availability of nuclear NAD+ (Zhang et al., 2012). Also, since C/EBPβ levels fluctuate during adipogenesis, target availability for PARP-1 may also play a role.
Our results on the role of PARP-1 in adipogenesis different from those reported previously by Hottiger and colleagues using whole body Parp1 null mice, as well as cell-based models of adipogenesis (Erener et al., 2012a; Erener et al., 2012b; Lehmann et al., 2015). While we observed an inhibitory effect of PARP-1 on adipogenesis through PARylation of C/EBPβ at early stages (i.e., differentiation of precursors) that persists throughout the differentiation process, Hottiger et al. observed a stimulatory effect of PARP-1 on adipogenesis that occurs by enhancing the transcriptional activity of PPARγ, which functions in adipocyte maturation and mature adipocyte function (Erener et al., 2012a; Erener et al., 2012b; Lehmann et al., 2015). Thus, the timing of PARP-1 activity (or PARP-1 knockdown or inhibition) may be critical in interpreting the results of these experiments. Our results, which (1) were generated using three different cell-based model systems (i.e., SVF, 3T3-L1, and NIH/3T3 cells) and four different approaches for modulating PARP-1 levels and activity (inducible knockout, knockdown, PARP inhibitors, ectopic expression), (2) were replicated in two independent labs (W.L.K. and R.E.S.) using independent batches of 3T3-L1 cells purchased directly from the ATCC, and (3) included the identification of specific sites of PARylation on C/EBPβ that, when mutated, yielded predictable results that are consistent with the rest of our observations, fully support the model that we have proposed.
We also note that conflicting results about the role of PARP-1 in adipogenesis and other metabolic processes have been reported by at least three different labs using whole body Parp1 null mice (Asher et al., 2010; Bai et al., 2011; Devalaraja-Narashimha and Padanilam, 2010; Erener et al., 2012b; Luo and Kraus, 2011). As described above, PARP-1 contributes to overall metabolic outcomes by playing distinct (and sometimes opposing) roles in different metabolism-related tissues, including muscle, liver, pancreas, brain, and adipose, with actions in one tissue presumably impacting actions in the other tissues (Luo and Kraus, 2011). As such, tissue-specific conditional Parp1 knockout mice, like those we describe herein, are needed to unambiguously address questions about the role of PARP-1 in tissue-specific physiological outcomes. In particular, genetic models that allow lineage tracing to track the differentiation of adipocyte precursors, as opposed to mature adipocyte biology, will be especially relevant to test the model that we have proposed herein. In general, studies with such genetic models should be coupled with assays using PARylation site point mutants of the target protein, the results of which should correspond logically with the results of assays using PARP-1 knockout, knockdown, or chemical inhibition.
EXPERIMENTAL PROCEDURES
Additional details on the experimental procedures can be found in the Supplemental Materials.
Generation of Parp1 conditional knockout mice
Parp1tm1a(EUCOMM)Hmgu ES cells on a C57BL/6N background were obtained from the International Mouse Phenotyping Consortium and used to generate chimeric mice. A multiple step breeding scheme (see Fig. S1A) was used to produce homozygous Parp1loxP/loxP mice on a C57BL/6N background, as well as Parp1loxP/loxP;Pdgfra-cre/ERT mice, which have a Tamoxifen-inducible conditional allele of Parp1.
Isolation and differentiation of stromal vascular fraction (SVF) cells
Isolation and culture of SVF cells from 6-week-old male wild-type C57BL/6 mice, or the transgenic mice described above, were performed as described previously (Gupta et al., 2012). Cre-mediated deletion of the floxed cassette in SVF cells from Parp1loxP/loxP or Parp1loxP/loxP;Pdgfra-cre/ERT mice was achieved by treating the cells in culture with adenovirus-Cre or 4-hydroxytamoxifen (4-OHT), respectively, prior to experiments. Adipogenesis was induced in contact-inhibited cells by the addition of a cocktail of differentiation agents (MDI: IBMX, dexamethasone, and insulin). In some cases, the cells were treated with vehicle, 20 µM BYK204165, or 5 µM PJ34, as specified.
Cell culture and treatments
3T3-L1 cells (Green and Kehinde, 1975) were obtained from the American Type Cell Culture (ATCC, CL-173TM) and NIH/3T3 cells (Todaro and Green, 1963) were obtained from Dr. Rana Gupta at UT Southwestern Medical Center. The cells were maintained in DMEM supplemented with 10% fetal bovine serum. Adipogenesis was induced in contact-inhibited cells by the addition of MDI cocktail. For induction of adipogenesis in NIH-3T3 cells, the cells were with treated MDI cocktail supplemented with 5 µM Rosiglitazone. In some cases, the cells were treated with vehicle, 20 µM BYK204165, or 5 µM PJ34, as specified. Adipogenesis was monitored by assessing the expression of adipogenic marker genes by RT-qPCR and staining oil droplets in intact cells with 5% Oil Red O or BODIPY 493/503 NHS Ester. 293T cells were purchased from the ATCC and maintained in DMEM supplemented with 10% fetal bovine serum.
Antibodies
The rabbit polyclonal antiserum against PARP-1 was generated using an antigen comprising the amino-terminal half of PARP-1 (Kim et al., 2004) (now available from Active Motif; cat. no. 39559). The custom recombinant anti-poly-ADP-ribose binding reagent (anti-PAR) was generated and purified in-house (now available from EMD Millipore; cat. no. MABE1031). Other antibodies are listed in the Supplemental Materials.
Cloning, mutagenesis, and generation of retroviral and lentiviral vectors for expression and knockdown
Standard recombinant DNA cloning and mutagenesis strategies were used to make (1) retroviral vectors for constitutive expression (pQCXIP) or shRNA-mediated knockdown (pSUPER.retro) of C/EBPβ, or PARP-1 or (2) lentiviral vectors (modified pINDUCER20) for doxycycline (Dox)-inducible expression of C/EBPβ in 3T3-L1 or 293T cells. pcDNA 3.1(−) mouse C/EBPβ (LAP isoform) and pcDNA 3.1(−) mouse C/EBPβ (LIP isoform) were purchased from Addgene (plasmids 12557 and 12561). A sequence encoding an N-terminal HA tag was added to the C/EBPβ cDNA by PCR. Mutant C/EBPβ (LAP) cDNAs encoding single, double, and triple point mutants of C/EBPβ were generated by site-directed mutagenesis.
Generation of stable knockdown and overexpression cell lines
Retroviruses and lentiviruses were generated from the vectors described above and used to infect 3T3-L1 or NIH/3T3 cells under selection with puromycin or G418 sulfate. Dox-inducible expression of C/EBPβ was achieved by treating the cells with 1 µM Dox for 8 or 24 hours, as specified.
Immunoprecipitation and detection of PARylated proteins and HA-tagged C/EBPβ
293T cells were plated and transfected with the pcDNA-based CEBPβ expression vectors described above using GeneJuice transfection reagent (Novagen). 3T3-L1 cells expressing inducible HA-tagged wild-type or triple mutant (K133A, E135A, E139A) C/EBPβ were cultured, differentiated, and treated as described above. The expression of HA-tagged C/EBPβ expression was induced with 1 µg/ml of doxycycline (Dox) for 8 hours before collection. Nuclear extracts from the cells were subjected to immunoprecipitation using anti-PAR binding reagent, anti-HA antibody, or rabbit IgG (as a control). The immunoprecipitated material was subjected to Western blotting for C/EBPβ ,the HA tag, or PAR.
RNA isolation and RT-qPCR
Total RNA was isolated from 3T3-L1 cells, NIH/3T3 cells, or SVF cells using Trizol Reagent (Invitrogen) according to the manufacturer’s protocol. RNA isolation and RT-qPCR were performed as described previously (Luo et al., 2014) using the gene-specific primers described in the Supplemental Materials. All target gene expression was normalized to the expression of the gene encoding TBP.
Chromatin immunoprecipitation-qPCR (ChIP-qPCR)
ChIP on 3T3-L1 cells was performed as described previously (Kininis et al., 2007; Krishnakumar et al., 2008), with a few modifications. The ChIPed genomic DNA was subjected to qPCR as described previously (Luo et al., 2014) using the gene-specific primers described in the Supplemental Materials
C/EBPβ ChIP-seq
The expression of HA-tagged wild-type or triple mutant (K133A, E135A, E139A) C/EBPβ was induced in 3T3-L1 cells by treatment with 1 µg/ml of Dox for 8 hours, with or without the addition of BYK204165. ChIP-seq libraries were generated and analyzed largely as described previously (Franco et al., 2015). The ChIP-seq data can be accessed from the NCBI’s Gene Expression Omnibus (GEO) repository using the series accession number GSE85100.
In nuclei PARylation reactions
293T cells were plated and transfected with the pcDNA-based CEBPβ expression vectors described above using GeneJuice transfection reagent (Novagen). Luc, PARP-1 or C/EBPβ knockdown 3T3-L1 cells were plated and differentiated for 4 hours as described above. Nuclei were isolated from the cells as described previously (Dignam et al., 1983) and then incubated with an equal volume of 2x reaction buffer (10 mM Tris-HCl pH 8.5, 20 mM KCl, 2 mM DTT, protease inhibitors) with (3T3-L1 nuclei) or without (293T cells nuclei) 200 µM NAD+ for 30 minutes at room temperature. Stopped reactions were subjected to Western blotting for PAR and C/EBPβ.
Electrophoretic mobility shift assays (EMSAs)
The effects of PARylation on C/EBPβ binding to DNA was performed using C/EBPβ PARylated in nuclei or in vitro. For in nuclei PARylation assays, nuclei were isolated from 293T cells expressing wild-type or mutant mouse C/EBPβ and PARylation reactions were performed as described above. For in vitro PARylation assays, recombinant wild-type or mutant mouse C/EBPβ was expressed in 293T cells as described above and then purified using HA affinity chromatography. PARylation reactions were performed in the EMSA reactions using purified recombinant PARP-1 and NAD+. The EMSA reactions contained 2.5 pmol of a 32P-end-labeled double-stranded oligonucleotide probe corresponding to the C/EBP regulatory element in the Cebpa promoter. The reactions were run on acrylamide gels in 0.5x TBE and analyzed by autoradiography or phosphorimager.
Supplementary Material
Highlights.
Inhibition or depletion of PARP-1 promotes the adipogenic differentiation program
PARP-1 PARylates C/EBPβ on three residues in a conserved regulatory domain
PARylation of C/EBPβ inhibits DNA binding, transcriptional activity, and adipogenesis
Mutation of the C/EBPβ PARylation sites produces a superactive transcription factor
Acknowledgments
The authors thank Hector Franco and Ziying Liu for critical comments on this manuscript. This work was supported by a predoctoral fellowship from the DOD Breast Cancer Research Program (BC093731) to X.L., a predoctoral fellowship from the American Heart Association to B.A.G., a grant from the a the NIH/NIDDK (DK094968), a grant from the Welch Foundation (I-1800) to Y.Y., and a grant from the NIH/NIDDK (DK058110) and support from the Cecil H. and Ida Green Center for Reproductive Biology Sciences Endowment to W.L.K.,
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURE
W.L.K. is a founder and consultant for Ribon Therapeutics, Inc.
AUTHOR CONTRIBUTIONS
X.L. and W.L.K. conceived this project and developed it with input from K.W.R. and R.K.G. X.L. and K.W.R. performed most of the experiments and analyzed all of the data, with assistance as follows: D-S.K. performed the EMSA experiments with recombinant C/EBPβ, T.N. analyzed the ChIP-seq data, C.J.M. and R.E.S. performed the time course experiments with PARP inhibitor in 3T3-L1 cells, R.G. generated the Parp1flox/flox mice and derivatives, B.A.G. made and produced the PAR detection reagent, R.K.G helped with the isolation of the SVF cells and the experiments using them, Y.Y. performed the mass spectrometry-based identification of PARylation sites. X.L. and K.W.R. prepared the initial drafts of the figures and text, which were edited and finalized by W.L.K. with input from the other authors. W.L.K secured funding to support this project and provided intellectual support for all aspects of the work.
REFERENCES
- Abdou HS, Atlas E, Hache RJ. A positive regulatory domain in CCAAT/enhancer binding protein beta (C/EBPBeta) is required for the glucocorticoid-mediated displacement of histone deacetylase 1 (HDAC1) from the C/ebpalpha promoter and maximum adipogenesis. Endocrinology. 2013;154:1454–1464. doi: 10.1210/en.2012-2061. [DOI] [PubMed] [Google Scholar]
- Altmeyer M, Messner S, Hassa PO, Fey M, Hottiger MO. Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic Acids Res. 2009;37:3723–3738. doi: 10.1093/nar/gkp229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Reinke H, Altmeyer M, Gutierrez-Arcelus M, Hottiger MO, Schibler U. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell. 2010;142:943–953. doi: 10.1016/j.cell.2010.08.016. [DOI] [PubMed] [Google Scholar]
- Bai P, Canto C, Oudart H, Brunyanszki A, Cen Y, Thomas C, Yamamoto H, Huber A, Kiss B, Houtkooper RH, et al. PARP-1 inhibition increases mitochondrial metabolism through SIRT1 activation. Cell Metab. 2011;13:461–468. doi: 10.1016/j.cmet.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy RJ, Kaestner KH, Geiman DE, Lane MD. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci U S A. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels CM, Ong SE, Leung AK. The promise of proteomics for the study of ADP-ribosylation. Mol Cell. 2015;58:911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalaraja-Narashimha K, Padanilam BJ. PARP1 deficiency exacerbates diet-induced obesity in mice. J Endocrinol. 2010;205:243–252. doi: 10.1677/JOE-09-0402. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Hesse M, Kostadinova R, Hottiger MO. Poly(ADP-ribose)polymerase-1 (PARP1) controls adipogenic gene expression and adipocyte function. Mol Endocrinol. 2012a;26:79–86. doi: 10.1210/me.2011-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erener S, Mirsaidi A, Hesse M, Tiaden AN, Ellingsgaard H, Kostadinova R, Donath MY, Richards PJ, Hottiger MO. ARTD1 deletion causes increased hepatic lipid accumulation in mice fed a high-fat diet and impairs adipocyte function and differentiation. FASEB J. 2012b;26:2631–2638. doi: 10.1096/fj.11-200212. [DOI] [PubMed] [Google Scholar]
- Farmer SR. Transcriptional control of adipocyte formation. Cell Metab. 2006;4:263–273. doi: 10.1016/j.cmet.2006.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco HL, Nagari A, Kraus WL. TNFalpha signaling exposes latent estrogen receptor binding sites to alter the breast cancer cell transcriptome. Mol Cell. 2015;58:21–34. doi: 10.1016/j.molcel.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Kwon SW, Zhao Y, Jin Y. PARP1 poly(ADP-ribosyl)ates Sox2 to control Sox2 protein levels and FGF4 expression during embryonic stem cell differentiation. J Biol Chem. 2009;284:22263–22273. doi: 10.1074/jbc.M109.033118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol. 2012;13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- Gibson BA, Zhang Y, Jiang H, Hussey KM, Shrimp JH, Lin H, Schwede F, Yu Y, Kraus WL. Chemical genetic discovery of PARP targets reveals a role for PARP-1 in transcription elongation. Science. 2016;353:45–50. doi: 10.1126/science.aaf7865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green H, Kehinde O. An established preadipose cell line and its differentiation in culture. II. Factors affecting the adipose conversion. Cell. 1975;5:19–27. doi: 10.1016/0092-8674(75)90087-2. [DOI] [PubMed] [Google Scholar]
- Gupta RK, Mepani RJ, Kleiner S, Lo JC, Khandekar MJ, Cohen P, Frontini A, Bhowmick DC, Ye L, Cinti S, et al. Zfp423 expression identifies committed preadipocytes and localizes to adipose endothelial and perivascular cells. Cell Metab. 2012;15:230–239. doi: 10.1016/j.cmet.2012.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen OE, Hilz H. Differentiation of 3T3-L1 pre-adipocytes induced by inhibitors of poly(ADP-ribose) polymerase and by related noninhibitory acids. Eur J Biochem. 1989;180:595–602. doi: 10.1111/j.1432-1033.1989.tb14686.x. [DOI] [PubMed] [Google Scholar]
- Kanai M, Hanashiro K, Kim SH, Hanai S, Boulares AH, Miwa M, Fukasawa K. Inhibition of Crm1-p53 interaction and nuclear export of p53 by poly(ADP-ribosyl)ation. Nat Cell Biol. 2007;9:1175–1183. doi: 10.1038/ncb1638. [DOI] [PubMed] [Google Scholar]
- Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL. NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell. 2004;119:803–814. doi: 10.1016/j.cell.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL. Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol. 2007;27:5090–5104. doi: 10.1128/MCB.00083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowenz-Leutz E, Twamley G, Ansieau S, Leutz A. Novel mechanism of C/EBP beta (NF-M) transcriptional control: activation through derepression. Genes Dev. 1994;8:2781–2791. doi: 10.1101/gad.8.22.2781. [DOI] [PubMed] [Google Scholar]
- Kraus WL, Hottiger MO. PARP-1 and gene regulation: progress and puzzles. Mol Aspects Med. 2013;34:1109–1123. doi: 10.1016/j.mam.2013.01.005. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Gamble MJ, Frizzell KM, Berrocal JG, Kininis M, Kraus WL. Reciprocal binding of PARP-1 and histone H1 at promoters specifies transcriptional outcomes. Science. 2008;319:819–821. doi: 10.1126/science.1149250. [DOI] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell. 2010a;39:8–24. doi: 10.1016/j.molcel.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010b;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann M, Pirinen E, Mirsaidi A, Kunze FA, Richards PJ, Auwerx J, Hottiger MO. ARTD1-induced poly-ADP-ribose formation enhances PPARgamma ligand binding and co-factor exchange. Nucleic Acids Res. 2015;43:129–142. doi: 10.1093/nar/gku1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Chae M, Krishnakumar R, Danko CG, Kraus WL. Dynamic reorganization of the AC16 cardiomyocyte transcriptome in response to TNFalpha signaling revealed by integrated genomic analyses. BMC Genomics. 2014;15:155. doi: 10.1186/1471-2164-15-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL. A one and a two … expanding roles for poly(ADP-ribose) polymerases in metabolism. Cell Metab. 2011;13:353–355. doi: 10.1016/j.cmet.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X, Kraus WL. On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev. 2012;26:417–432. doi: 10.1101/gad.183509.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch VJ, May G, Wagner GP. Regulatory evolution through divergence of a phosphoswitch in the transcription factor CEBPB. Nature. 2011;480:383–386. doi: 10.1038/nature10595. [DOI] [PubMed] [Google Scholar]
- Oei SL, Shi Y. Poly(ADP-ribosyl)ation of transcription factor Yin Yang 1 under conditions of DNA damage. Biochem Biophys Res Commun. 2001;285:27–31. doi: 10.1006/bbrc.2001.5115. [DOI] [PubMed] [Google Scholar]
- Olabisi OA, Soto-Nieves N, Nieves E, Yang TT, Yang X, Yu RY, Suk HY, Macian F, Chow CW. Regulation of transcription factor NFAT by ADP-ribosylation. Mol Cell Biol. 2008;28:2860–2871. doi: 10.1128/MCB.01746-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pekala PH, Lane MD, Watkins PA, Moss J. On the mechanism of preadipocyte differentiation. Masking of poly(ADP-ribose) synthetase activity during differentiation of 3T3-L1 preadipocytes. J Biol Chem. 1981;256:4871–4876. [PubMed] [Google Scholar]
- Rodeheffer MS, Birsoy K, Friedman JM. Identification of white adipocyte progenitor cells in vivo. Cell. 2008;135:240–249. doi: 10.1016/j.cell.2008.09.036. [DOI] [PubMed] [Google Scholar]
- Ryu KW, Kim DS, Kraus WL. New facets in the regulation of gene expression by ADP-ribosylation and poly(ADP-ribose) polymerases. Chem Rev. 2015;115:2453–2481. doi: 10.1021/cr5004248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siersbaek R, Nielsen R, Mandrup S. Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol Metab. 2012;23:56–64. doi: 10.1016/j.tem.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van RL, Bayliss CE, Roncari DA. Cytological and enzymological characterization of adult human adipocyte precursors in culture. J Clin Invest. 1976;58:699–704. doi: 10.1172/JCI108516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vyas S, Chesarone-Cataldo M, Todorova T, Huang YH, Chang P. A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nat Commun. 2013;4:2240. doi: 10.1038/ncomms3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams SC, Baer M, Dillner AJ, Johnson PF. CRP2 (C/EBP beta) contains a bipartite regulatory domain that controls transcriptional activation, DNA binding and cell specificity. EMBO J. 1995;14:3170–3183. doi: 10.1002/j.1460-2075.1995.tb07319.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh WC, Cao Z, Classon M, McKnight SL. Cascade regulation of terminal adipocyte differentiation by three members of the C/EBP family of leucine zipper proteins. Genes Dev. 1995;9:168–181. doi: 10.1101/gad.9.2.168. [DOI] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Yao J, DuMond ME, Krishnakumar R, Ruhl DD, Ryu KW, Gamble MJ, Kraus WL. Regulation of poly(ADP-ribose) polymerase-1-dependent gene expression through promoter-directed recruitment of a nuclear NAD+ synthase. J Biol Chem. 2012;287:12405–12416. doi: 10.1074/jbc.M111.304469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Ding M, Yu Y. Site-specific characterization of the Asp- and Glu-ADP-ribosylated proteome. Nat Methods. 2013;10:981–984. doi: 10.1038/nmeth.2603. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.