Abstract
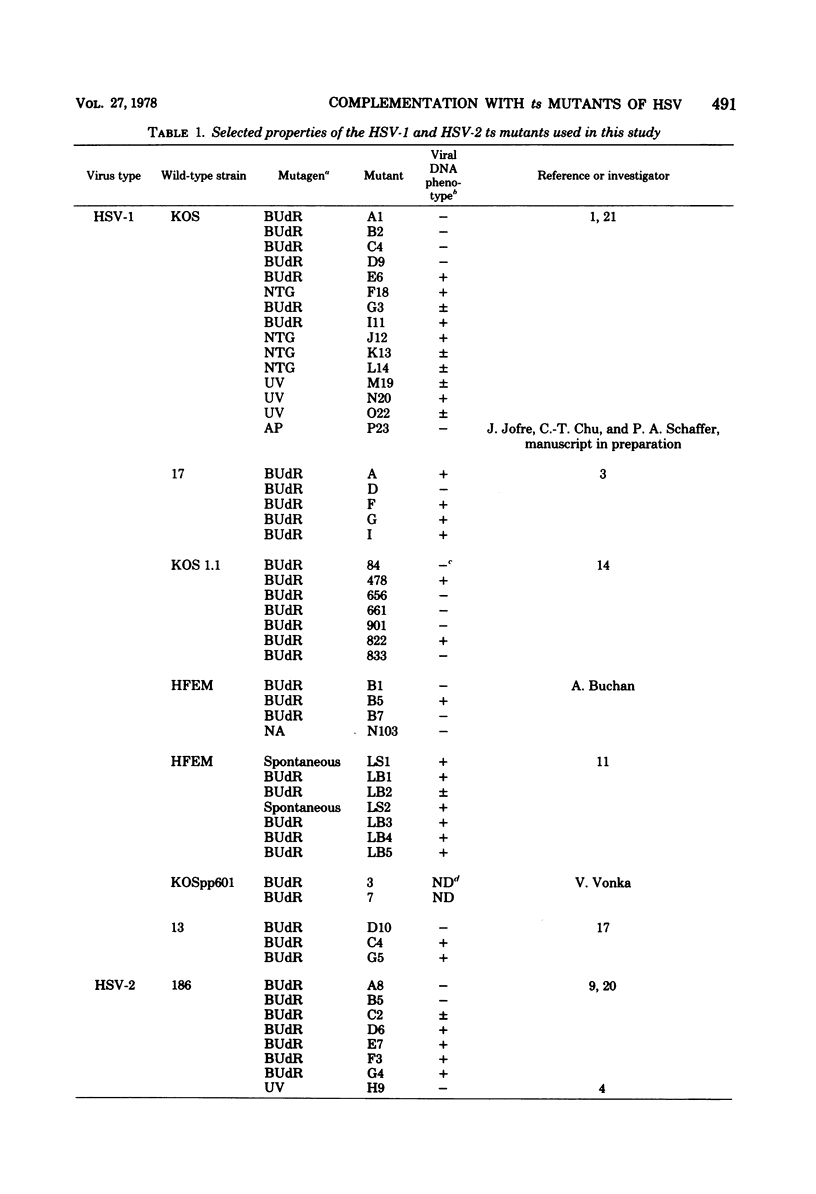
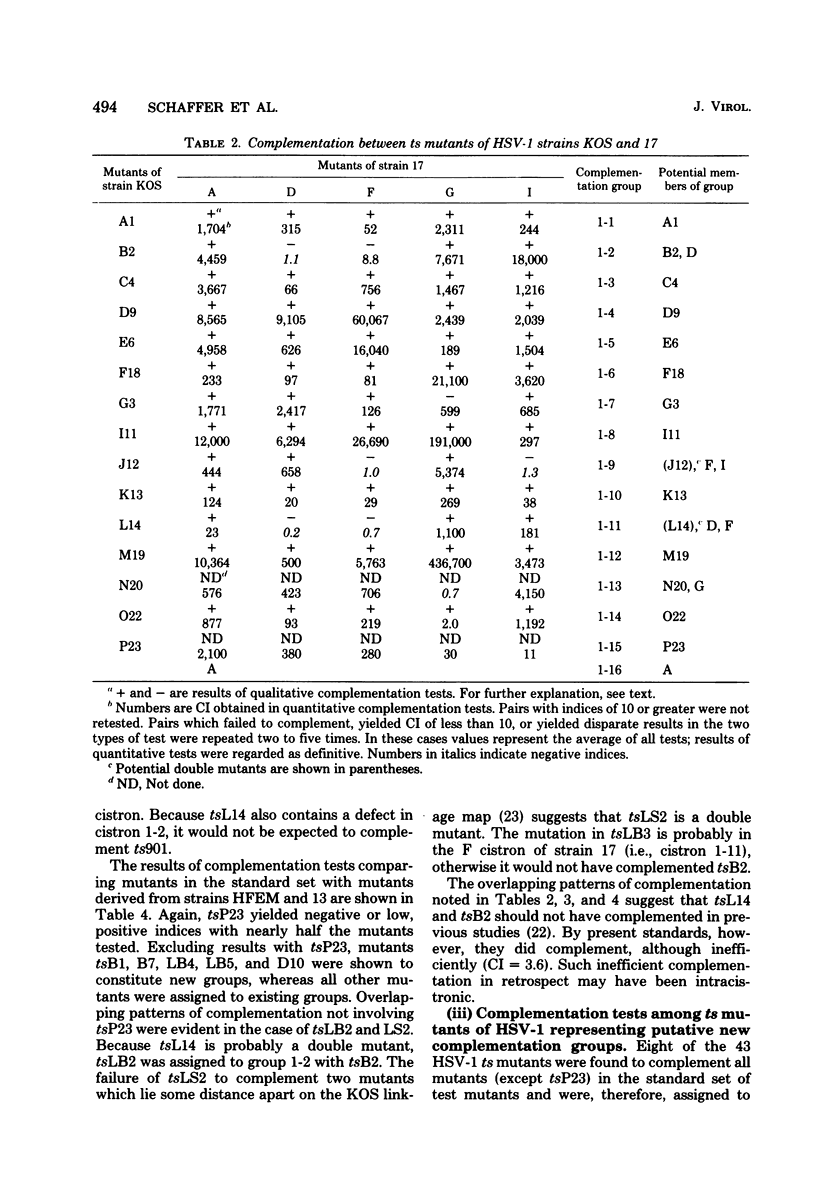
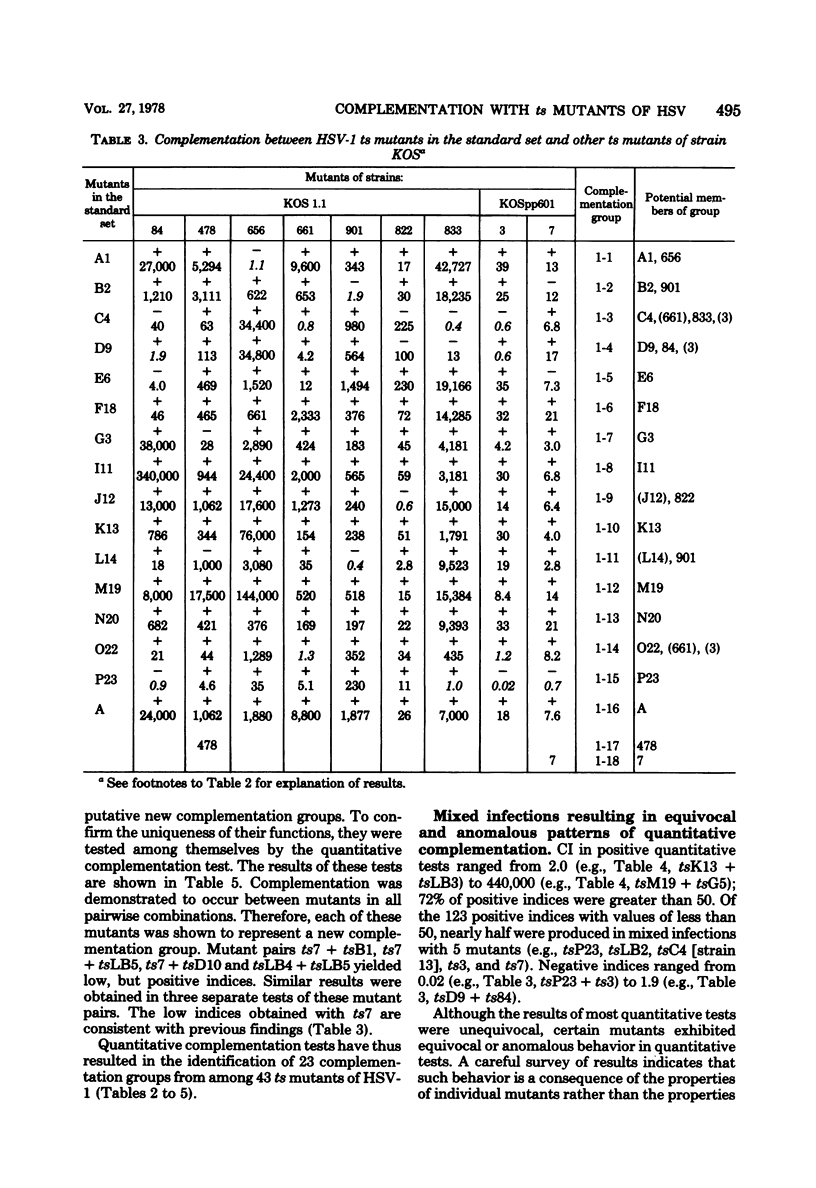
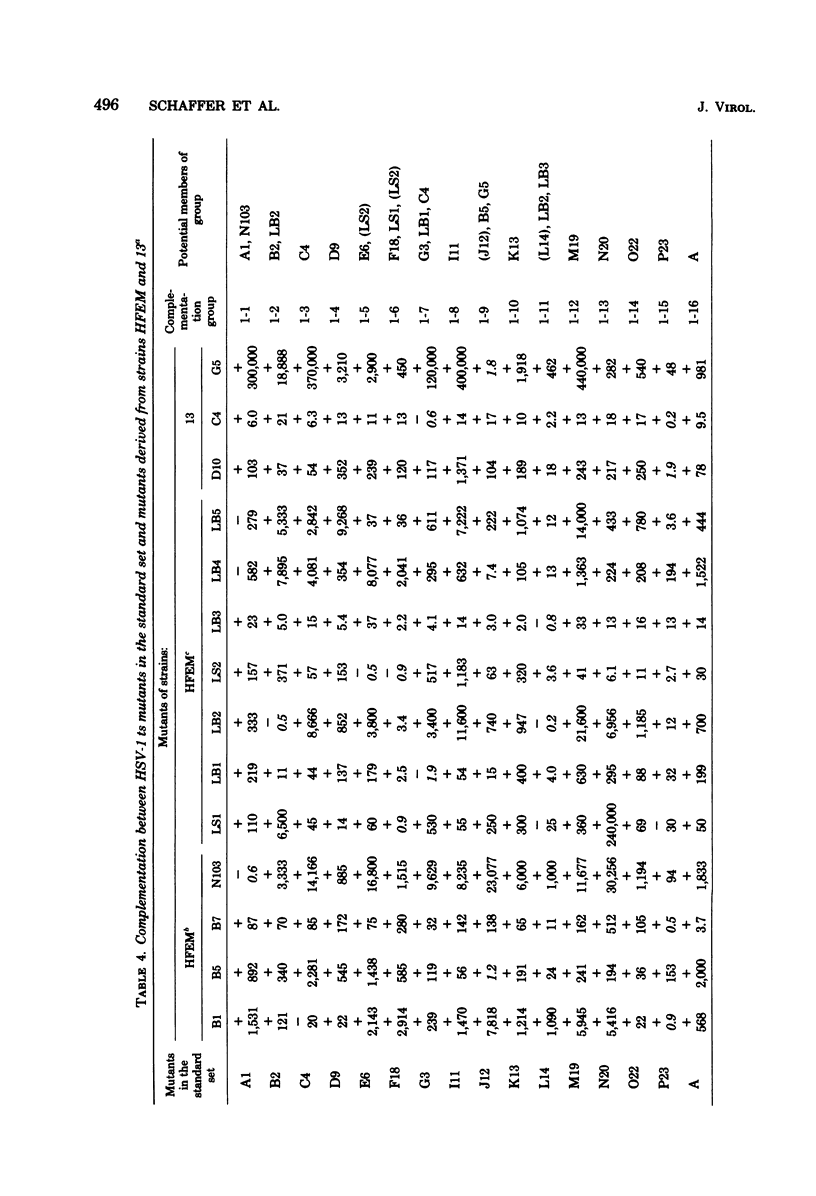
Twenty-three complementation groups of herpes simplex virus type 1 (HSV-1) and 20 of HSV-2 were identified by qualitative and quantitative complementation analysis from among 43 temperature-sensitive (ts) mutants of HSV-1 and 29 ts mutants of HSV-2 which had been isolated independently in 10 laboratories.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aron G. M., Purifoy D. J., Schaffer P. A. DNA synthesis and DNA polymerase activity of herpes simplex virus type 1 temperature-sensitive mutants. J Virol. 1975 Sep;16(3):498–507. doi: 10.1128/jvi.16.3.498-507.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown S. M., Ritchie D. A., Subak-Sharpe J. H. Genetic studies with herpes simplex virus type 1. The isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J Gen Virol. 1973 Mar;18(3):329–346. doi: 10.1099/0022-1317-18-3-329. [DOI] [PubMed] [Google Scholar]
- Chu C. T., Schaffer P. A. Qualitative complementation test for temperature-sensitive mutants of herpes simplex virus. J Virol. 1975 Nov;16(5):1131–1136. doi: 10.1128/jvi.16.5.1131-1136.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delius H., Clements J. B. A partial denaturation map of herpes simplex virus type 1 DNA: evidence for inversions of the unique DNA regions. J Gen Virol. 1976 Oct;33(1):125–133. doi: 10.1099/0022-1317-33-1-125. [DOI] [PubMed] [Google Scholar]
- Dreesman G. R., Benyesh-Melnick M. Spectrum of human cytomegalovirus complement-fixing antigens. J Immunol. 1967 Dec;99(6):1106–1114. [PubMed] [Google Scholar]
- EDGAR R. S., DENHARDT G. H., EPSTEIN R. H. A COMPARATIVE GENETIC STUDY OF CONDITIONAL LETHAL MUTATIONS OF BACTERIOPHAGE T4D. Genetics. 1964 Apr;49:635–648. doi: 10.1093/genetics/49.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esparza J., Purifoy D. J., Schaffer P. A., Benyesh-Melnick M. Isolation, complementation and preliminary phenotypic characterization of temperature-sensitive mutants of herpes simplex virus type 2. Virology. 1974 Feb;57(2):554–565. doi: 10.1016/0042-6822(74)90194-9. [DOI] [PubMed] [Google Scholar]
- Guerola N., Ingraham J. L., Cerdá-Olmedo E. Induction of closely linked multiple mutations by nitrosoguanidine. Nat New Biol. 1971 Mar 24;230(12):122–125. doi: 10.1038/newbio230122a0. [DOI] [PubMed] [Google Scholar]
- Halliburton I. W., Randall R. E., Killington R. A., Watson D. H. Some properties of recombinants between type 1 and type 2 herpes simplex viruses. J Gen Virol. 1977 Sep;36(3):471–484. doi: 10.1099/0022-1317-36-3-471. [DOI] [PubMed] [Google Scholar]
- Halliburton I. W., Timbury M. C. Characterisation of temperature-sensitive mutants of herpes simplex virus type 2. Growth and DNA synthesis. Virology. 1973 Jul;54(1):60–68. doi: 10.1016/0042-6822(73)90114-1. [DOI] [PubMed] [Google Scholar]
- Halliburton I. W., Timbury M. C. Temperature-sensitive mutants of herpes simplex virus type 2: description of three new complementation groups and studies on the inhibition of host cell DNA synthesis. J Gen Virol. 1976 Feb;30(2):207–221. doi: 10.1099/0022-1317-30-2-207. [DOI] [PubMed] [Google Scholar]
- Hughes R. G., Jr, Munyon W. H. Temperature-sensitive mutants of herpes simplex virus type 1 defective in lysis but not in transformation. J Virol. 1975 Aug;16(2):275–283. doi: 10.1128/jvi.16.2.275-283.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koment R. W., Rapp F. Temperature-sensitive host range mutants of herpes simplex virus type 2. J Virol. 1975 Apr;15(4):812–819. doi: 10.1128/jvi.15.4.812-819.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manservigi R. Method for isolation and selection of temperature-sensitive mutants of herpes simplex virus. Appl Microbiol. 1974 Jun;27(6):1034–1040. doi: 10.1128/am.27.6.1034-1040.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Buchman T. G., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus DNA. IX. Apparent exclusion of some parental DNA arrangements in the generation of intertypic (HSV-1 X HSV-2) recombinants. J Virol. 1977 Oct;24(1):231–248. doi: 10.1128/jvi.24.1.231-248.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse L. S., Pereira L., Roizman B., Schaffer P. A. Anatomy of herpes simplex virus (HSV) DNA. X. Mapping of viral genes by analysis of polypeptides and functions specified by HSV-1 X HSV-2 recombinants. J Virol. 1978 May;26(2):389–410. doi: 10.1128/jvi.26.2.389-410.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purifoy D. J., Benyesh-Melnick M. DNA polymerase induction by DNA-negative temperature-sensitive mutants of herpes simplex virus type 2. Virology. 1975 Dec;68(2):374–386. doi: 10.1016/0042-6822(75)90280-9. [DOI] [PubMed] [Google Scholar]
- Schaffer P. A., Aron G. M., Biswal N., Benyesh-Melnick M. Temperature-sensitive mutants of herpes simplex virus type 1: isolation, complementation and partial characterization. Virology. 1973 Mar;52(1):57–71. doi: 10.1016/0042-6822(73)90398-x. [DOI] [PubMed] [Google Scholar]
- Schaffer P. A. Temperature-sensitive mutants of herpesviruses. Curr Top Microbiol Immunol. 1975;70:51–100. doi: 10.1007/978-3-642-66101-3_3. [DOI] [PubMed] [Google Scholar]
- Schaffer P. A., Tevethia M. J., Benyesh-Melnick M. Recombination between temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1974 Mar;58(1):219–228. doi: 10.1016/0042-6822(74)90156-1. [DOI] [PubMed] [Google Scholar]
- Scotti P. D. A new class of temperature conditional lethal mutants of bacteriophage T4D. Mutat Res. 1968 Jul-Aug;6(1):1–14. doi: 10.1016/0027-5107(68)90098-5. [DOI] [PubMed] [Google Scholar]
- Sheldrick P., Berthelot N. Inverted repetitions in the chromosome of herpes simplex virus. Cold Spring Harb Symp Quant Biol. 1975;39(Pt 2):667–678. doi: 10.1101/sqb.1974.039.01.080. [DOI] [PubMed] [Google Scholar]
- Siccardi A. G., Ferrari F. A., Mazza G., Galizzi A. Identification of coreplicating chromosomal sectors in Bacillus subtilis by nitrosoguanidine-induced comutation. J Bacteriol. 1976 Mar;125(3):755–761. doi: 10.1128/jb.125.3.755-761.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M., Yamanishi K. Transformation of hamster embryo and human embryo cells by temperature sensitive mutants of herpes simplex virus type 2. Virology. 1974 Sep;61(1):306–311. doi: 10.1016/0042-6822(74)90267-0. [DOI] [PubMed] [Google Scholar]
- Timbury M. C., Hendricks M. L., Schaffer P. A. Collaborative study of temperature-sensitive mutants of herpes simplex virus type 2. J Virol. 1976 Jun;18(3):1139–1142. doi: 10.1128/jvi.18.3.1139-1142.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timbury M. C. Temperature-sensitive mutants of herpes simplex virus type 2. J Gen Virol. 1971 Nov;13(2):373–376. doi: 10.1099/0022-1317-13-2-373. [DOI] [PubMed] [Google Scholar]
- Wadsworth S., Jacob R. J., Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975 Jun;15(6):1487–1497. doi: 10.1128/jvi.15.6.1487-1497.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmoreland D., Rapp F. Host range temperature-sensitive mutants of herpes simplex virus type 2. J Virol. 1976 Apr;18(1):92–102. doi: 10.1128/jvi.18.1.92-102.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zygraich N., Huygelen C. In vivo behaviour of a temperature-sensitive (ts) mutant of herpesvirus hominis type 2. Arch Gesamte Virusforsch. 1973;43(1):103–111. doi: 10.1007/BF01249353. [DOI] [PubMed] [Google Scholar]



