ABSTRACT
Tissue engineering constructs to treat intervertebral disc degeneration must adapt to the hypoxic and inflammatory degenerative disc microenvironment. The objective of this study was to determine the effects of two key design factors, cell type and cell configuration, on the regenerative potential of nucleus pulposus cell (NPC) and mesenchymal stem cell (MSC) constructs. Anabolic and catabolic activity was quantified in constructs of varying cell type (NPCs, MSCs, and a 50:50 co‐culture) and varying configuration (individual cells and micropellets). Anabolic and catabolic outcomes were both dependent on cell type. Gene expression of Agg and Col2A1, glycosaminoglycan (GAG) content, and aggrecan immunohistochemistry (IHC), were significantly higher in NPC‐only and co‐culture groups than in MSC‐only groups, with NPC‐only groups exhibiting the highest anabolic gene expression levels. However, NPC‐only constructs also responded to inflammation and hypoxia with significant upregulation of catabolic genes (MMP‐1, MMP‐9, MMP‐13, and ADAMTS‐5). MSC‐only groups were unaffected by degenerative media conditions, and co‐culture with MSCs modulated catabolic induction of the NPCs. Culturing cells in a micropellet configuration dramatically reduced catabolic induction in co‐culture and NPC‐only groups. Co‐culture micropellets, which take advantage of both cell type and configuration effects, had the most immunomodulatory response, with a significant decrease in MMP‐13 and ADAMTS‐5 expression in hypoxic and inflammatory media conditions. Co‐culture micropellets were also found to self‐organize into bilaminar formations with an MSC core and NPC outer layer. Further understanding of these cell type and configuration effects can improve tissue engineering designs. © 2016 The Authors. Journal of Orthopaedic Research published by Wiley Periodicals, Inc. on behalf of the Orthopaedic Research Society. J Orthop Res 35:61–73, 2017.
Keywords: micropellet, spheroid, intervertebral disc degeneration, inflammation, co‐culture
Low back pain is a leading cause of disability worldwide and is most often associated with intervertebral disc (IVD) degeneration.1 Degeneration is irreversible due to the low cellularity and low regenerative capacity of nucleus pulposus (NP) tissue. This has raised interest in cell‐based tissue engineering therapies that could be injected into the NP space to restore mechanical and biochemical properties via synthesis of new NP tissue. However, degenerative IVDs have a hypoxic and inflammatory microenvironment, which inhibits cell survival and proliferation. Inflammatory cytokines also upregulate catabolic factors such as matrix metalloproteinases, which further exacerbate degeneration.2, 3, 4 Therefore, a better understanding of how tissue‐engineering constructs respond to a degenerative microenvironment is needed to evaluate their regeneration potential.
Tissue‐engineering constructs can be designed with many different parameters in mind, including choices of biomaterial, cell types, and exogenous growth factors. Furthermore, the 3D cell configuration within a construct is an inherent design factor that controls the availability and intensity of mechanical and biochemical signals.5 In this current study, we measure the effects of cell type and 3D configuration in IVD tissue engineering constructs. We focus on both anabolic and catabolic gene expression to more comprehensively assess regenerative potential in a degenerative context.
Many cell types are potential candidates for IVD tissue engineering, including autologous nucleus pulposus cells (NPCs), progenitor cells (such as mesenchymal stem cells, or MSCs), or combinations of multiple cell types.6 NPCs, which are native to the disc space, are able to produce disc matrix components even after being expanded and cultivated in vitro. When injected into the IVD in animal studies, NPCs in conjunction with carrier materials or scaffolds improve disc height and material properties.6, 7 However, these methods pose some clinical challenges due to the dilemma of donor cell availability.8
An alternative cell source, MSCs, resolves some of these clinical issues because autologous MSCs can be isolated from bone marrow without causing disc damage. MSCs have the potential to differentiate into chondrogenic lineages, and are able to modulate inflammatory responses in some environments.9, 10 However, chondrogenic differentiation of MSCs requires specific inductive cues (such as addition of exogenous growth factors), and is difficult to regulate due to hypertrophy and calcification in later differentiation stages.8, 11 Combining NPCs and MSCs in co‐culture may be a way to simultaneously address the challenges of NPCs’ limited availability (fewer NPCs are needed) and MSCs’ dependence on an instructive microenvironment (NPCs can provide differentiation cues).12 Previously observed benefits of co‐culturing MSCs with articular chondrocytes or NPCs include increased cell proliferation, upregulated proteoglycan (PG) synthesis, and reduced hypertrophy.13, 14
Another potentially important design consideration for tissue‐engineering constructs is 3D cell configuration, a structural variable that alters the physical, spatial, and biochemical cues that cells perceive.15 Structural factors such as cell density, homotypic and heterotypic cell type proximity, and presence of direct contact affect cell proliferation and chondrogenic potential, but the relative importance of these factors is controversial and unclear.14, 16, 17, 18 One particular 3D configuration that externally controls cell proximity and contact is cell micropellets or microaggregates.19 Pellet culture is commonly used to promote chondrogenic differentiation in vitro, possibly because contact between cells mimics a mesenchymal condensation structure.20, 21, 22 Direct contact in smaller cell clusters can also improve GAG deposition by articular chondrocytes, and chondrogenic differentiation of MSCs,19, 23 so micropellets most likely provide similar contact‐associated benefits. Furthermore, micropellets have fewer diffusion limitations than traditional, larger cell pellets, and can be injectable for clinical applications.24
We hypothesized that both cell type and 3D configuration affect synthetic and catabolic activity, and investigated the extent of these effects by comparing individual‐cell and micropellet constructs. We fabricated nucleus pulposus tissue‐engineering constructs consisting of MSCs and NPCs encapsulated in alginate hydrogels, evaluating both anabolic and catabolic performance to assess the ability of implanted constructs to synthesize matrix while surviving in degenerative environments (simulated by hypoxic and inflammatory media). We first compared anabolic and catabolic performance of co‐cultured constructs with that of single‐cell‐type constructs to more clearly define the benefits of co‐culture in our system. Next, we formed micropellets with single cell types and co‐cultured cells to determine the effect of cell configuration on anabolic and catabolic gene expression. We observed that the co‐cultured micropellets self‐organized into a bilayered structure, and investigated whether this self‐organization might be attributed to differences in intracellular cohesivity in MSCs and NPCs.
METHODS
Experimental Design
To determine the effects of cell type and cell configuration, we varied these parameters through two sets of experiments.
Cell‐Type Effects
To more clearly define the effect of cell type in nucleus pulposus tissue engineering constructs, we compared a 50:50 co‐culture of MSC and NPC with single‐cell‐type cultures (Fig. 1A–C). MSCs, NPCs, and co‐culture groups were all encapsulated in alginate beads as individual cells and cultured as described in the Cell Culture section, with n = 3 for each experimental condition.
Figure 1.
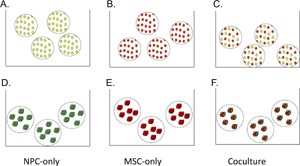
Individual cell (A–C) and micropellet (D–F) groups of NPCs (A and D), MSCs (B and E), and co‐cultured (C and F) cells allow us to compare the effects of cell type and cell configuration. NPCs are shown in green and MSCs are shown in red.
Configuration Effects
We also wanted to study the effect of 3D configuration, particularly in the context of direct cell–cell contact, on cell behavior. Therefore, we formed micropellets of MSCs, NPCs, and a 50:50 co‐culture, which we also encapsulated in alginate beads for long‐term culture (Fig. 1D–F) (n = 3).
Cell Culture
Human MSCs (passage 5, purchased from EMD Millipore, Hayward, CA) were cultured in MSC growth media (low‐glucose DMEM with 10% FBS and 1% antibiotic/antimycotic). Bovine NPCs (passage 4, harvested from caudal discs from a local slaughterhouse as previously described25) were cultured in standard disc media (low‐glucose DMEM with 5% FBS, 1% antibiotic/antimycotic, 1% non‐essential amino acids, and 1.5% osmolarity salt solution containing 5 M NaCl and 0.4 M KCl). Human MSCs were from one donor, and bovine NPCs were from a pooled expansion of three simultaneously harvested tails.
All experimental groups were suspended in alginate beads, which were formed by resuspending cells in 1.2% sodium alginate (FMC BioPolymer, San Jose, CA) in D‐PBS at a concentration of 1 × 106 cells/ml. We expelled the alginate‐cell mixture dropwise through a 22‐gauge needle into a 102 mmol/L CaCl2 solution and allowed the beads to crosslink for 10 min before washing with PBS and media. The groups were cultured in basal MSC growth media or inflammatory and hypoxic media (MSC growth media with 10 ng/ml of IL‐1β and 10 ng/ml of TNF‐α, 2% O2) to simulate normal and degenerative disc environments. All groups were cultured for 21 days, with media changes three times per week.
MSC growth media was used instead of differentiation media because the addition of TGF‐β in differentiation media often leads to hypertrophic differentiation and expression of collagen X,26 and previous studies suggest that co‐cultured MSCs and ACs in growth media induce chondrogenic gene and protein expression.27 Cells were encapsulated in alginate beads to provide a non‐fouling 3D environment with rounded cell morphology, simulating native conditions in the NP.28 We suspended cells with a density of 1 M cells per ml to minimize accidental direct contact between neighboring individual cells, as shown in the individual‐cell group histology sections in Figure 6C.
Figure 6.
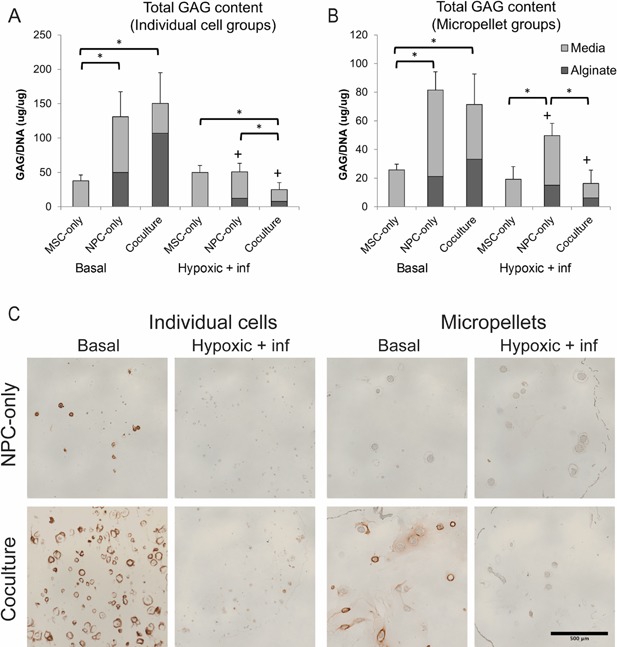
Glycosaminoglycan content and distribution varies with cell type and configuration. We calculated total GAG content of individual cell (A) and micropellet (B) groups as the sum of GAG concentration (normalized by DNA content) in alginate bead and media samples. The + symbol indicates a significant difference (p < 0.05) within a cell type group for different media conditions, while the * symbol indicates a significant difference between different cell type groups under the same media conditions. Error bars represent standard deviation of the total GAG, and statistics were performed on the total as described in the methods section. Panel (C) shows distribution of aggrecan in NPC‐only and co‐culture individual cell, and micropellet constructs after 21 days in basal or inflammatory and hypoxic media conditions. Aggrecan staining (with a hematoxylin counterstain) was visible in co‐culture basal media groups as well as the NPC‐only individual cell baal media group, and was most concentrated in regions surrounding cells and micropellets. Scale bar: 500 μm.
NPC Phenotype Stability
To assess the phenotype stability of NPCs in co‐culture, we measured the gene expression of bovine CD24, a marker of healthy NPCs,29 in NPC‐only and co‐culture groups at 21 days. Gene expression analysis was performed with bovine GAPDH as the housekeeping gene (primer sequences are shown in Table 1). There were no significant differences in CD24 gene expression associated with co‐culture conditions, cell configurations, or media conditions (Fig. S1).
Table 1.
Primer Sequences for Gene Expression Analysis
| Human ref | Bovine ref | Forward | Reverse | |
|---|---|---|---|---|
| GAPDH | NM_002046 | NM_001034034 | AGC TCA CTG GCA TGG CCT TC | CGC CTG CTT CAC CAC CTT CT |
| Aggrecan | NM_001135 | NM_173981 | AGG AGC AGG AGT TTG TCA AC | AGT TGT CAG GCT GGT TGG |
| Collagen 2A1 | NM_001844 | NM_001001135 | AGG AAT TCG GTG TGG ACA TAG | TCA GGT CAG CCA TTC AGT G |
| MMP‐1 | NM_002421 and NM_001145938 | NM_174112 | CTT GCT CAT GCT TTT CAA CCA GG | GCT GAA CAT CAC CAC TGA AGG T |
| MMP‐9 | NM_004994 | NM_174744 | CTA CAC CCA GGA CGG CAA TG | GTC GTA GTT GGC GGT GGT |
| MMP‐13 | NM_002427 | NM_174389 | TGA AAC CTG GAC AAG TAG TTC C | ATG AGT GCT CCT GGG TCC TT |
| ADAMTS5 | NM_007038 | NM_001166515 | GCG CTT AAT GTC TTC CAT CCT | CGT GGT AGG TCC AGC AAA CA |
| Bovine GAPDH | Sequences from ref.49 | GCC ATC ACT GCC ACC CAG AA | GCG GCA GGT CAG ATC CAC AA | |
| Bovine CD24 | N/A | XM_002690126 and XM_015464783 | TCT GGC GCT GCT CTT ACC TA | GCA GGT GAG GTA GTC TGG GA |
All amplify homologous regions of the human and bovine genes of interest, except the species‐specific bovine GAPDH, and CD24 primers, which were used to measure NPC phenotype markers.
Micropellet Formation
Micropellets with 100 μm diameters were formed in molds generated by soft lithography (Fig. 2). Molds were formed by creating a silicon master with 100 μm diameter wells. The master was made with SU‐8 2035 photoresist (MicroCheM, Westborough, MA) and crosslinked under a photo‐mask.30 A polydimethylsiloxane (PDMS) “stamp” was molded from these masters with the negative image: Posts with diameter 100 μm. Finally, these PDMS stamps were inserted into a 3.5% agarose solution (Low gelling agarose type VII‐A, Sigma–Aldrich, St. Louis, MO), which, after gelling, formed 100 μm diameter wells. MSCs and NPCs were loaded into the wells at a 50:50 ratio and total concentration of 1 M cells/ml. They condensed into micropellets over 12 h in MSC growth media. The micropellets, which contain approximately 100 cells each, were released from wells with a D‐PBS wash before alginate encapsulation at a density of 10,000 micropellets per ml, or 1 M cells/ml.
Figure 2.
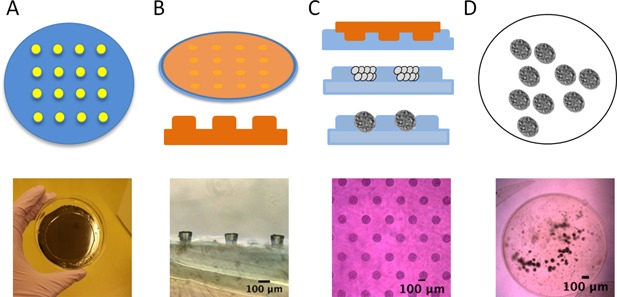
Methods for forming cell micropellets. A. Create 100‐μm wells in a silicon wafer with photolithography. B. Cover the wafer with PDMS, and peel off to form posts. C. Use the PDMS as a “stamp” to form agarose wells. Load the wells with cells, which condense in 12–24 h. D. Release condensed pellets from the wells by washing with D‐PBS, and resuspend in alginate.
Gene Expression Analysis
After 21 days of culture, the alginate beads were dissolved in 55 mM sodium citrate, and cell pellets were isolated by centrifugation. Total RNA was extracted from all groups using QIAShredder and RNeasy mini kits (Qiagen, Valencia, CA), and was converted to cDNA using iScript reverse transcriptase (BioRad, Hercules, CA). Gene expression was measured using quantitative reverse transcription PCR with SYBR green master mix (BioRad, Hercules, CA) using the BioRad CFX96 RealTime Thermal Cycler.
Gene expression was measured for aggrecan and collagen 2A1 (anabolic chondrogenic markers), and MMP‐1, MMP‐9, MMP‐13, and ADAMTS5 (catabolic factors present in disc degeneration31) using primers that amplified homologous regions in human and bovine transcripts (Table 1). Fold changes were calculated using the ΔΔCt method32 and normalized to GAPDH. Results were compared using a one‐way ANOVA test (among cell‐type groups) and multiple t tests (between groups in basal and inflammatory media conditions) with a Tukey HSD correction for multiple hypotheses. P values <0.05 were considered significant.
DNA and Dimethylmethylene Blue Assays for Glycosaminoglycan Quantification
After dissolving the alginate beads in 55 mM sodium citrate, we digested the supernatant in 0.56 U/ml papain (Sigma–Aldrich, St. Louis, MO) at 60°C overnight. Media samples of 1 ml volume were collected at the time of harvest, but did not go through the digest step. DNA content was assayed with a QuantiTPicoGreen kit (Thermo Fisher, Waltham, MA) and measured on a microplate reader (Molecular Devices, Sunnyvale, CA) with excitation at 488 nm and absorption at 525 nm. GAG content was analyzed using a dimethylmethylene blue (DMMB) assay with modifications for alginate33 and media34 measurements, and normalized by DNA content. Statistics on normalized total GAG content were calculated using a one‐way ANOVA test and multiple t tests as described in the Gene Expression Analysis section.
Histological Analysis
Alginate beads were fixed in 10% formalin for 20 min, dehydrated with ethanol washes, embedded in paraffin, and sectioned at seven micron thickness. Immunohistochemistry was performed following manufacturer instructions for the DAB substrate kit (Vector Laboratories, Inc., Burlingame, CA) with a 1:100 dilution of the primary mouse anti‐aggrecan antibody (12/21/1‐C‐6, Developmental Studies Hybridoma Bank, University of Iowa). The slides were counterstained with hematoxylin. The figures show representative images of n = 3 replicates.
Observation of Micropellet Structure and Intracellular Cohesivity Assay
To visualize micropellet organization, we labeled cell populations with Vybrant DiI and DiO cell membrane dyes (5 μl/1*106 million cells) (Life Technologies, Carlsbad, CA). The micropellets were imaged using inverted epifluoresent microscopes (Zeiss Axiovert 200M running SlideBook software and Leica DMi8 running LAS X).
The co‐culture micropellets contain two different cell types that might vary in cohesivity, which could affect their adhesion‐forming behavior. To quantify the intracellular cohesivity, we allowed 100% NPC and 100% MSC populations to interact overnight in agarose microwells and analyzed the contours of the resulting 100% NPC or 100% MSC micropellets. We measured circularity of the contours using FIJI's built‐in circularity measurement tool as previously described.30 Briefly, circularity is a measure of the ratio of a micropellet's area to the square of its perimeter, where C = 4π*area/perimeter2. Higher circularity scores are correlated with smoother micropellet contours, which result from higher intracellular cohesivity.
RESULTS
Cell Type Effects
To determine the role of cell type in synthetic activity and responses to inflammation, we compared NPC‐only and MSC‐only seeded alginate beads with beads containing a 50:50 mix of both cell types (Fig. 1A–C in Methods).
Anabolic Performance
To analyze the anabolic performance of the different cell types, we measured aggrecan and collagen 2A1 gene expression. Under basal media conditions, the MSC‐only group exhibited very low anabolic gene expression: For both aggrecan and collagen 2A1, MSC‐only levels were significantly lower than those of NPC‐only and co‐culture groups (Fig. 3A and B). Although the NPC‐only and co‐culture groups did not show a significant difference in aggrecan or collagen 2A1 gene expression, the NPC‐only group had a trend of higher anabolic gene expression levels.
Figure 3.
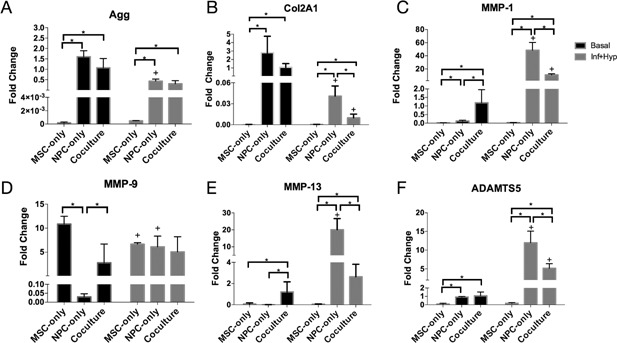
Effect of cell type in individually encapsulated cell constructs: NPCs maximize ECM synthesis while co‐culture provides protection against hypoxic and inflammatory conditions. Aggrecan (A), collagen 2A1 (B), MMP‐1 (C), MMP‐9 (D), MMP‐13 (E), and ADAMTS5 (F) gene expression in MSC‐only, NPC‐only, and co‐culture groups after 21 days in basal or hypoxic and inflammatory media conditions. Fold changes were normalized to the co‐culture basal media group. The + symbol indicates a significant difference (p < 0.05) within a cell type group for different media conditions, while the * symbol indicates a significant difference between different cell type groups under the same media conditions. Error bars represent standard deviation.
We also evaluated changes in anabolic gene expression when cells were cultured in an inflammatory and hypoxic microenvironment (Fig. 3A and B). In this environment, anabolic gene expression remained low for MSCs and decreased for the NPC‐only and co‐culture groups. Similar to the basal media condition, the aggrecan and collagen 2A1 gene expression of the MSC‐only group was significantly lower than that of the NPC‐only and co‐culture groups. The NPC‐only and co‐culture groups did not have a significant difference in aggrecan expression, but the NPC‐only group had significantly higher collagen 2A1 expression than the co‐culture group. Collagen 2A1 expression decreased significantly in NPC‐only and co‐culture groups (p < 0.05), while aggrecan expression decreased significantly in the NPC‐only group and showed a decreasing trend in the co‐culture group (p < 0.05 prior to a multiple hypothesis correction).
Catabolic Performance
To measure the groups’ catabolic response to a hypoxic and inflammatory microenvironment, we measured gene expression of MMP‐1, MMP‐9, MMP‐13, and ADAMTS5 (Fig. 3C–F). The basal expression levels of these genes varied: The MSC‐only group exhibited significantly lower levels of MMP‐1, MMP‐13, and ADAMTS5, but had a high basal expression level of MMP‐9. The co‐culture group exhibited significantly higher basal levels of MMP‐1 and MMP‐13 compared with the single‐cell‐type groups. In inflammatory and hypoxic conditions, the NPC‐only group consistently exhibited a significant upregulation of all four catabolic factors, and the subsequent expression levels of MMP‐1, MMP‐13, and ADAMTS5 were higher in the NPC‐only groups than in MSC‐only and co‐culture groups. The MSC‐only group did not show significant catabolic upregulation, and in fact exhibited a significant decrease in MMP‐9 expression in the inflammatory and hypoxic condition. The co‐culture group exhibited significant catabolic upregulation in two of the genes, MMP‐1 and ADAMTS5. However, in both these cases, the upregulated gene expression levels were still significantly lower than those of the NPC‐only group.
Configuration Effects
To explore the effects of 3D cell configuration, we formed NPC, MSC, and co‐culture micropellets, and compared them to each other and to the previous patterns we observed in individual cell groups (Fig. 1D–F).
Anabolic Performance
The micropellet configuration did not significantly affect MSC anabolic gene expression patterns. Similar to the MSC‐only individual cells, the MSC‐only micropellet group also produced very low expression of aggrecan and collagen 2A1 mRNAs in both basal and hypoxic/inflammatory media (Fig. 4A andB). The MSC micropellet aggrecan and collagen 2A1 expression was significantly lower than NPC and co‐culture micropellet gene expression in almost all the conditions. In hypoxic and inflammatory media, the co‐culture micropellets experienced a significant downregulation of aggrecan gene expression, and their resulting aggrecan level was similar to that of the MSC micropellets (Fig. 4A).
Figure 4.
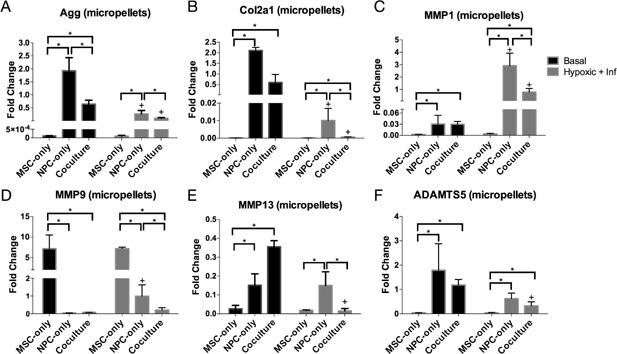
Anabolic and catabolic gene expression in a micropellet configuration: Configuration and co‐culture reduce catabolic upregulation in inflammatory and hypoxic conditions. Aggrecan (A), collagen 2A1 (B), MMP‐1 (C), MMP‐9 (D), MMP‐13 (E), and ADAMTS5 (F) gene expression in MSC‐only, NPC‐only, and co‐culture micropellet groups after 21 days in basal or hypoxic and inflammatory media conditions. Fold changes for each gene were normalized to the co‐culture individual cell basal media group, which is shown in Figure 3. The + symbol indicates a significant difference (p < 0.05) within a cell type group for different media conditions, while the * symbol indicates a significant difference between different cell type groups under the same media conditions. Error bars represent standard deviation.
The micropellet configuration also did not affect the NPC and co‐culture groups’ response to hypoxic and inflammatory media. Both groups had significantly lower aggrecan and collagen 2A1 gene expression in hypoxic and inflammatory media than in basal media.
In the individual‐cells configuration (Fig. 3A and B), the NPC‐only group generally exhibited higher anabolic expression than the co‐culture group. In the micropellet configuration (Fig. 4A and B), the NPC‐only group maintained higher aggrecan expression in both media conditions, but the co‐culture micropellets exhibited a trend of higher collagen 2A1 expression, with a significantly higher expression of Col2A1 than NPCs in hypoxic and inflammatory media.
Catabolic Performance
Similar to the pattern observed in the individual cell constructs, the MSC‐only micropellets exhibited a significantly higher basal level of MMP‐9 and a significantly lower basal level of the other catabolic genes compared with NPC‐only and co‐culture micropellets (Fig. 4C–F). NPC‐only and co‐culture micropellets had similar basal expression levels for all four genes. In inflammatory and hypoxic media conditions, the MSC‐only micropellets did not display any significant changes in catabolic gene expression. NPC‐only micropellets showed a significant upregulation of MMP‐1 and MMP‐9, but unlike in the NPC‐only individual cell group, MMP‐13 and ADAMTS5 expression did not significantly change. In the co‐culture micropellets, MMP‐1 expression significantly increased in inflammatory and hypoxic media, but the increased levels were still significantly lower than those in NPC‐only micropellets (Fig. 4C). MMP‐9 expression did not significantly change in the co‐culture micropellets, and MMP‐13 and ADAMTS5 expression actually showed a significant decrease in degenerative media conditions.
To illustrate how both cell type and configuration influence response to inflammation, we calculated fold induction for the four catabolic markers (fold change difference between basal and hypoxic/inflammatory levels) (Fig. 5). All MSC groups exhibited no significant catabolic induction, while the NPC‐only individual cell group consistently exhibited the highest fold induction, with a greater‐than‐100‐fold induction in all the MMPs that we tested (Fig. 5A–C). Co‐culture groups generally exhibited lower induction levels, and even experienced a significant decrease in MMP‐13 and ADAMTS5 (fold induction was <1) (Fig. 5C and D). Finally, micropellet configurations are associated with lower induction levels in both NPC‐only and co‐culture groups.
Figure 5.
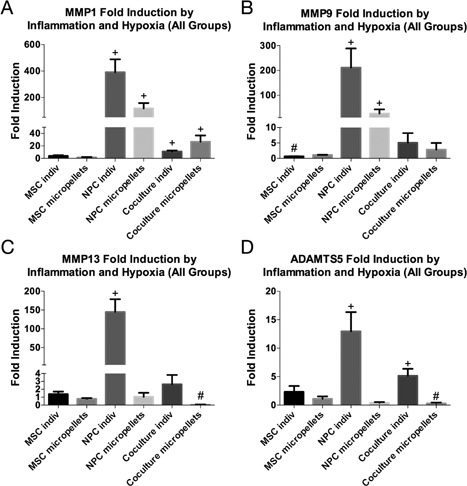
Co‐culture and micropellet configurations exhibit reduced catabolic induction. Fold induction of catabolic markers MMP‐1 (A), MMP‐9 (B), MMP‐13 (C), and ADAMTS5 (D) represents the fold change between basal and hypoxic/inflammatory media conditions (normalized separately to the basal media condition for each group, which is not shown in the graph). The + and # symbols indicate a significant (p < 0.05) increase and decrease, respectively in expression in degenerative media conditions. Error bars represent standard deviation.
Extracellular Matrix Deposition
To evaluate the effects of cell type and configuration on proteoglycan and glycosaminoglycan synthesis, we measured glycosaminoglycan (GAG) content in the alginate beads and media, and performed immunohistochemical staining for aggrecan.
GAG Content
MSC‐only groups exhibited the lowest GAG content in both individual cell and micropellet configurations (Fig. 6A and B), but their GAG secretion was not significantly affected by degenerative media conditions. On the other hand, both NPC‐only and co‐culture groups exhibited a significant decrease in total GAG content in inflammatory and hypoxic media, regardless of cell configuration. In basal media, NPC‐only and co‐culture groups had similar GAG content levels, but in degenerative conditions the co‐culture group had significantly lower GAG content, also regardless of cell configuration. Overall, micropellet groups had lower total GAG content than individual cell groups.
Histological Detection of Aggrecan
Aggrecan staining was clearly present in co‐culture basal groups, in both individual cell and micropellet configurations (Fig. 6C). There was a smaller amount of aggrecan staining visible in the NPC‐only individual cell basal media group, but the staining followed a similar pattern as the co‐culture groups, in which staining was most concentrated in the regions directly surrounding cells or pellets. Staining was very faint in inflammatory and hypoxic media condition groups and NPC‐only micropellet groups, and was negative in all MSC‐only groups (data not shown).
Self‐Organization of Co‐Cultured Cells in Micropellets
We investigated the nature of heterotypic cell–cell contact between MSCs and NPCs in co‐culture micropellets by labeling the MSCs and NPCs with membrane dyes. We observed that the two cell types spontaneously self‐organized into a bilayered structure during pellet condensation.
The differential adhesion hypothesis suggests that differences in intercellular cohesivity can lead to changes in surface tension and subsequent cell segregation, which influences tissue morphologies in development.11 We hypothesized that the two cell types (MSCs and NPCs) in our co‐culture construct might have different adhesivity, leading to changes in the organization of the co‐cultured micropellets. Although two cell types were isolated from different species (human and bovine), previous work suggested that cross‐species interactions did not significantly alter cell behavior or phenotypes.35 To more closely observe changes in micropellet organization over time, we used fluorescent membrane dyes to track MSCs and NPCs before and after the micropellets formed, and condensed. Although MSCs and NPCs were randomly seeded in agarose microwells (Fig. 7A), after condensation, the co‐cultured micropellets exhibited a unique bilayered structure with an MSC core and NPC outer layer (Fig. 7B). This self‐organization was evident across large batches of micropellets (Fig. 7C).
Figure 7.
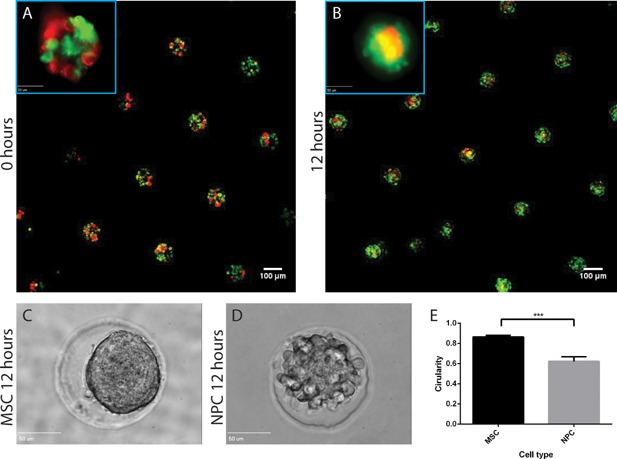
Differential adhesion of MSCs and NPCs leads to a bilaminar self‐organization in co‐culture micropellets. MSCs (red) and NPCs (green) were labeled with DiI and DiO membrane dyes. Micropellets were seeded with a random 50:50 cell mixture (A), but condensed after 12 h to form a bilaminar structure with an MSC core and NPC outer layer (B). To investigate whether the bilaminar self‐organization was caused by differences in intracellular cohesivity, we assessed the circularity of MSCs and NPCs in single‐cell micropellets. After a 12‐h condensation, MSCs (C) had a significantly higher circularity than NPCs (D and E). Scale bar for A and B = 100 μm (50 μm for the 40× inserts). Scale bar for C and D = 50 μm.
To determine if MSCs and NPCs exhibit differences in intracellular cohesivity, we measured the circularity of single‐cell‐type micropellets after 24 h and determined that MSC micropellets had a significantly higher circularity, and therefore, higher intercellular cohesivity (Fig. 6D–F). This concurs with previous work predicting that due to minimization of free surface energy, cells with greater cohesivity tend to migrate toward the core of self‐organized micropellets.30, 36 The observed bilaminar structure of the co‐culture micropellets may be related to the anabolic and catabolic differences we observed between co‐culture micropellets and co‐cultured individual cells.
DISCUSSION
In this study, we varied cell type and configuration to elucidate the roles of these factors in synthesizing new matrix and adapting to a degenerative disc microenvironment. We measured anabolic and catabolic outcomes of the constructs in basal and degenerative culture conditions, and found that both cell type and configuration influence cell behavior.
When comparing the anabolic activity of different cell types, we found that MSC‐only groups had the lowest anabolic gene expression and proteoglycan content, which is consistent with previous literature suggesting that non‐induced MSC monocultures have low synthetic activity.37 NPC‐only groups had the highest anabolic gene expression, but at the protein level, co‐culture and NPC‐only groups had similar total GAG content in basal media conditions. Co‐culture groups also exhibited more positive aggrecan staining, so their synthetic activity may be comparable to that NPC‐only groups despite their lower anabolic gene expression.
Micropellet configurations did not alter the anabolic gene expression trends we observed in individual cell groups. However, NPC‐only and co‐culture micropellet groups exhibited lower overall GAG content and less aggrecan staining than the corresponding individual cell groups, which may indicate that micropellets were less effective at synthesizing matrix.
Overall, we only observed dense aggrecan staining in three of the histological samples, which suggests that immunohistochemistry was less sensitive than the DMMB assay, which was able to measure GAG content in all groups. Low aggrecan staining may be related to the low starting cell density of 1 M cells/ml: In 2010, Watanabe et al. noted that even after 6 months of implantation in a mouse skin nodule, NPC cells that were sparsely arranged did not show much safranin‐O staining.14 Furthermore, cartilage and NP tissue engineering studies use a wide variety of non‐standardized cell densities, media compositions, and co‐culture ratios, which all can affect GAG content, chondrogenic gene expression, and cell proliferation.11, 13, 38, 39, 40 Differences in these experimental culture conditions between our study and previous works may also account for differences in histological outcomes.
Inflammatory and hypoxic media conditions also influenced anabolic activity: Regardless of configuration, NPC‐only and co‐culture groups experienced significant decreases in anabolic gene expression, total GAG content, and aggrecan staining. Previous studies have suggested that co‐culture with MSCs increases NPCs’ resilience to degenerative conditions,41 but we did not observe this phenomenon. As mentioned in the previous paragraph, differences in experimental culture conditions might account for this discrepancy.
In addition to matrix synthesis, matrix degradation is another important process that influences biochemical and mechanical properties of the IVD. Therefore, we investigated the effect of cell type and configuration on catabolic gene expression.
Catabolic activity in hypoxic and inflammatory media conditions was strongly associated with cell type: NPC‐only groups had much higher upregulation of catabolic gene expression than MSC‐only and co‐culture groups. Other studies have also found that co‐cultures containing MSCs are able to modulate inflammatory responses.42, 43 This effect is attributed to immunomodulatory properties of MSCs, which may involve trophic factors and/or contact‐mediated signaling.44
3D configuration also significantly affected cells’ response to inflammatory and hypoxic stimuli. Arranging cells in a micropellet configuration reduced catabolic induction in both NPC‐only and co‐culture groups. Strikingly, MMP‐13 and ADAMTS5 expression significantly decreased in co‐culture micropellets when subjected to the degenerative media conditions, possibly because a micropellet configuration enhances immunomodulatory properties of MSCs.45
Our results motivate further investigation into the mechanisms behind benefits of co‐culture and micropellet configurations. The roles of the different cell types in co‐culture systems is still unclear. Some studies have found evidence of MSC differentiation,11, 12, 46 while others indicate that MSCs play a trophic role.47, 48, 49 To isolate and analyze MSC and NPC or chondrocyte contributions in co‐culture systems, previous studies have labeled MSCs with fluorescent markers and separated them with high‐speed sorting,12 or used species‐specific markers to analyze gene expression contributions of cells isolated from different species.11 Our xenogenic system is well suited to this second approach.
The observed differences in micropellet and individual cell cultures may be related to cell–cell communication mechanisms such as soluble factors and contact‐mediated signaling. Cells in micropellets are in close contact with each other, allowing them to experience higher soluble factor concentrations. Trophic factors secreted by MSCs such as IL‐10 and PGE2 may be involved in immunomodulatory mechanisms, and a decreased diffusion distance could increase their potency in the co‐cultured micropellets.42, 43, 50 Micropellets also allow cells to form direct contacts. Contact‐mediated signaling, including formation of gap junctions51 or exchange of membrane components,52 has been shown to influence anabolic and catabolic gene expression. In particular, previous studies have found that direct cell–cell contact regulates synthetic activity, growth factor secretion, and responses to inflammation in MSCs and co‐culture systems.12, 53, 54 In our system, we measured levels of the trophic factors FGF‐1 and PGE2 and the contact‐associated gene N‐cadherin, but preliminary data did not show strong associations with cell type or cell configuration changes (data not shown), possibly because these factors have time‐dependent expression changes, or because other factors are more influential in the system.
Applying a micropellet configuration to co‐cultured MSCs and NPCs altered structural factors beyond cell proximity and contact. Upon closer observation of the co‐cultured micropellets, we found that they self‐organized during the condensation process to form a bilayered structure with an MSC core and NPC outer layer. The tendency of MSCs to gravitate toward the core in pellet culture with chondrocytes or NPCs has previously been observed in “satellite” pellets that budded from large co‐cultured pellets over time25 and larger pellets that were initially formed without any specific structure.48 The differences we measured in intracellular cohesivity of MSCs and NPCs may also explain these earlier observations. This self‐organization behavior has been observed in both bovine‐human and human–human NPC/MSC combinations, so the effects are not due to xenogenic differences.25, 55
The observed bilayered organization (with an MSC core and NPC or chondrocyte outer layer) has also been fabricated in larger cell pellets using multiple centrifugation steps instead of self‐assembly.25, 27, 56 These Bilaminar Cell Pellets (BCPs) exhibited increased proliferation and chondrogenic gene expression, reduced hypertrophy, and modulated responses in inflammatory environments.11, 27, 41 Although the bilaminar organization minimizes heterotypic cell–cell contact, the MSC‐core structure may be advantageous because it mimics condensations present in cartilage and IVD development.52, 57 In this study, the micropellets did not sustain anabolic gene expression in inflammatory conditions to the extent observed in larger pellets,27 but they showed similar reductions in catabolic responses, and their smaller diameter reduces transport barriers that are associated with larger pellets.24
One limitation of the current study is the use of pooled bovine NPCs from three donor animals and MSCs from a single human donor. This study design prevented us from testing for significant donor‐to‐donor variations in our key findings, and thereby may impact the generalizability of our results. However, related experiments from our group suggest that individual variation among bovine NPC donors has only a minor effect (∼3%) on catabolic induction by inflammatory cytokines, while human MSC donors showed a larger variation. Yet, when comparing human donors in co‐culture systems with the same NPC source, all MSC‐only and co‐culture groups maintained significantly lower catabolic induction than NPC‐only groups. Individual cells also exhibited increased catabolic induction compared with micropellets regardless of donor variability. While these results are consistent with the cell type and configuration effects we report here, future studies with additional biological replicates are needed to identify whether desirable co‐culture behaviors are maintained across a broader donor pool.
Additional factors such as donor age, sex, and location of the MSC source may also influence cell phenotype of both NPCs and MSCs.58, 59, 60 In previous studies, bovine NPCs showed less individual–individual catabolic variation than human NPCs.61 MSCs also showed some individual variation in immunomodulatory activity,59 but induction with pro‐inflammatory cytokines may reduce this variability.62 Additional studies of protein expression and ECM production are needed to ensure that inter‐donor variability does not hinder the applicability of tissue engineering strategies. In addition, co‐cultures containing human NPCs harvested from surgical samples and human MSCs could also be used to test observations from our xenogenic model in a more clinically relevant system. Both cell types could be isolated from the same patients, which would illuminate the extent to which individual variability affects cell behavior in co‐cultured constructs.
Another limitation of our study that hindered matrix deposition was the use of growth culture conditions that were not optimized for chondrogenic synthesis. Therefore, a future step is to determine how our constructs perform in more optimized experimental conditions. For example, overall cell density, ratios of co‐cultured cell types, and addition of growth factors affect chondrogenesis and cell survival both in vitro and in vivo.39, 63, 64 Encapsulation materials also affect matrix synthesis and whether cells benefit from direct cell–cell contact.37 The cell type and configuration results from our initial study provide a template to which additional elements can be added to design a more effective NP tissue engineering construct.
Overall, our results indicate that both cell type and configuration influence cell performance in a degenerative microenvironment. Co‐culture and micropellet configurations had especially strong immunomodulatory effects on catabolic gene expression in inflammatory and hypoxic environments. Further investigations are needed to elucidate mechanisms behind these effects, and to optimize their benefits in an IVD tissue‐engineering context.
AUTHORS’ CONTRIBUTIONS
AO, AEC, and XT designed and performed experiments, and AO analyzed data and wrote the paper. EL performed the histology experiments. AEC also analyzed imaging data. All authors discussed the results and implications and commented on the manuscript at all stages.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. NPC phenotype remains stable in co‐culture.
ACKNOWLEDGMENTS
This work was supported by NIH R21AR063357‐02 to JCL. AO was supported by an NSF Graduate Research Fellowship and AEC was supported by the US Department of Defense through a National Defense Science and Engineering Graduate Fellowship. We received some statistical consulting support from the UCSF Core Center for Musculoskeletal Biology and Medicine (P30‐AR066262‐02). We would also like to thank Kaitlyn Gary for her assistance with histology processing.
Conflicts of interest: None.
REFERENCES
- 1. Aung A, Gupta G, Majid G, et al. 2011. Osteoarthritic chondrocyte‐secreted morphogens induce chondrogenic differentiation of human mesenchymal stem cells. Arthritis Rheum 63:148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fischer J, Dickhut A, Rickert M, et al. 2010. Human articular chondrocytes secrete parathyroid hormone‐related protein and inhibit hypertrophy of mesenchymal stem cells in coculture during chondrogenesis. Arthritis Rheum 62:2696–2706. [DOI] [PubMed] [Google Scholar]
- 3. Vincenti MP, Brinckerhoff CE. 2002. Transcriptional regulation of collagenase (MMP‐1, MMP‐13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene‐specific transcription factors. Arthritis Res 4:157–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Millward‐Sadler SJ, Costello PW, Freemont AJ, et al. 2009. Regulation of catabolic gene expression in normal and degenerate human intervertebral disc cells: implications for the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther 11:R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kinney MA, Hookway TA, Wang Y, et al. 2013. Engineering three‐dimensional stem cell morphogenesis for the development of tissue models and scalable regenerative therapeutics. Ann Biomed Eng 42:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Paesold G, Nerlich AG, Boos N. 2007. Biological treatment strategies for disc degeneration: potentials and shortcomings. Eur Spine J 16:447–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mehrkens A, Müller AM, Valderrabano V, et al. 2012. Tissue engineering approaches to degenerative disc disease—a meta‐analysis of controlled animal trials. Osteoarthritis Cartilage 20:1316–1325. [DOI] [PubMed] [Google Scholar]
- 8. Richardson SM, Kalamegam G, Pushparaj PN, et al. 2015. Mesenchymal stem cells in regenerative medicine: focus on articular cartilage and intervertebral disc regeneration. Methods 99:69–80. [DOI] [PubMed] [Google Scholar]
- 9. Yang HN, Park JS, Na K, et al. 2009. The use of green fluorescence gene (GFP)‐modified rabbit mesenchymal stem cells (rMSCs) co‐cultured with chondrocytes in hydrogel constructs to reveal the chondrogenesis of MSCs. Biomaterials 30:6374–6385. [DOI] [PubMed] [Google Scholar]
- 10. Meretoja VV, Dahlin RL, Kasper FK, et al. 2012. Enhanced chondrogenesis in co‐cultures with articular chondrocytes and mesenchymal stem cells. Biomaterials 33:6362–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mo X, Guo S, Xie H, et al. 2009. Variations in the ratios of co‐cultured mesenchymal stem cells and chondrocytes regulate the expression of cartilaginous and osseous phenotype in alginate constructs. Bone 45:42–51. [DOI] [PubMed] [Google Scholar]
- 12. Strassburg S, Richardson SM, Freemont AJ, et al. 2010. Co‐culture induces mesenchymal stem cell differentiation and modulation of the degenerate human nucleus pulposus cell phenotype. Regen Med 5:701–711. [DOI] [PubMed] [Google Scholar]
- 13. Tsuchiya K, Chen G, Ushida T, et al. 2004. The effect of coculture of chondrocytes with mesenchymal stem cells on their cartilaginous phenotype in vitro. Mater Sci Eng C 24:391–396. [Google Scholar]
- 14. Watanabe T, Sakai D, Yamamoto Y, et al. 2010. Human nucleus pulposus cells significantly enhanced biological properties in a coculture system with direct cell‐to‐cell contact with autologous mesenchymal stem cells. J Orthop Res 28:623–630. [DOI] [PubMed] [Google Scholar]
- 15. Shamir ER, Ewald AJ. 2014. Three‐dimensional organotypic culture: experimental models of mammalian biology and disease. Nat Rev Mol Cell Biol 15:647–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Richardson SM, Walker RV, Parker S, et al. 2006. Intervertebral disc cell‐mediated mesenchymal stem cell differentiation. Stem Cells 24:707–716. [DOI] [PubMed] [Google Scholar]
- 17. Svanvik T, Henriksson HB, Karlsson C, et al. 2010. Human disk cells from degenerated disks and mesenchymal stem cells in co‐culture result in increased matrix production. Cells Tissues Organs 191:2–11. [DOI] [PubMed] [Google Scholar]
- 18. de Windt TS, Hendriks JAA, Zhao X, et al. 2014. Concise review: unraveling stem cell cocultures in regenerative medicine: which cell interactions steer cartilage regeneration and how? Stem Cells Transl Med 3:723–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Albrecht DR, Underhill GH, Wassermann TB, et al. 2006. Probing the role of multicellular organization in three‐dimensional microenvironments. Nat Methods 3:369–375. [DOI] [PubMed] [Google Scholar]
- 20. Gao L, McBeath R, Chen CS. 2010. Stem cell shape regulates a chondrogenic versus myogenic fate through Rac1 and N‐cadherin. Stem Cells 28:564–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang X, Fan L, Pei M, et al. 2015. Evolving concepts of chondrogenic differentiation: history, state‐of‐the‐art and future perspectives. Eur Cell Mater 30:12–27. [DOI] [PubMed] [Google Scholar]
- 22. Lu ZF, Zandieh Doulabi B, Wuisman PI, et al. 2007. Differentiation of adipose stem cells by nucleus pulposus cells: configuration effect. Biochem Biophys Res Commun 359:991–996. [DOI] [PubMed] [Google Scholar]
- 23. Cao B, Li Z, Peng R, et al. 2015. Effects of cell‐cell contact and oxygen tension on chondrogenic differentiation of stem cells. Biomaterials 64:21–32. [DOI] [PubMed] [Google Scholar]
- 24. Goude MC, McDevitt TC, Temenoff JS. 2014. Chondroitin sulfate microparticles modulate transforming growth factor‐β1‐induced chondrogenesis of human mesenchymal stem cell spheroids. Cells Tissues Organs 199:117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Allon A, Schneider R, Lotz J. 2009. Co‐culture of adult mesenchymal stem cells and nucleus pulposus cells in bilaminar pellets for intervertebral disc regeneration. SAS J 3:41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mueller MB, Fischer M, Zellner J, et al. 2010. Hypertrophy in mesenchymal stem cell chondrogenesis: effect of TGF‐beta isoforms and chondrogenic conditioning. Cells Tissues Organs 192:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cooke ME, Allon AA, Cheng T, et al. 2011. Structured three‐dimensional co‐culture of mesenchymal stem cells with chondrocytes promotes chondrogenic differentiation without hypertrophy. Osteoarthritis Cartilage 19:1210–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kluba T, Niemeyer T, Gaissmaier C, et al. 2005. Human anulus fibrosis and nucleus pulposus cells of the intervertebral disc: effect of degeneration and culture system on cell phenotype. Spine (Phila Pa 1976) 30:2743–2748. [DOI] [PubMed] [Google Scholar]
- 29. Risbud MV, Schoepflin ZR, Mwale F, et al. 2015. Defining the phenotype of young healthy nucleus pulposus cells: recommendations of the spine research interest group at the 2014 annual ORS meeting. J Orthop Res 33:283–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cerchiari AE, Garbe JC, Jee NY, et al. 2015. A strategy for tissue self‐organization that is robust to cellular heterogeneity and plasticity. PNAS 112:2287–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Huang YC, Leung VYL, Lu WW, et al. 2013. The effects of microenvironment in mesenchymal stem cell‐based regeneration of intervertebral disc. Spine J 13:352–362. [DOI] [PubMed] [Google Scholar]
- 32. Schmittgen TD, Livak KJ. 2008. Analyzing real‐time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. [DOI] [PubMed] [Google Scholar]
- 33. Enobakhare BO, Bader DL, Lee DA. 1996. Quantification of sulfated glycosaminoglycans in chondrocyte/alginate cultures, by use of 1,9‐dimethylmethylene blue. Anal Biochem 243:189–191. [DOI] [PubMed] [Google Scholar]
- 34. Billington CJ. 2000. Cartilage proteoglycan release assay In: Matrix metalloproteinase protocols. New Jersey: Humana Press; p 451–456. [Google Scholar]
- 35. Liu X, Sun H, Yan D, et al. 2010. In vivo ectopic chondrogenesis of BMSCs directed by mature chondrocytes. Biomaterials 31:9406–9414. [DOI] [PubMed] [Google Scholar]
- 36. Steinberg MS. 1963. Reconstruction of tissues by dissociated cells. Some morphogenetic tissue movements and the sorting out of embryonic cells may have a common explanation. Science 141:401–408. [DOI] [PubMed] [Google Scholar]
- 37. de Windt TS, Saris DBF, Slaper‐Cortenbach ICM, et al. 2015. Direct cell‐cell contact with chondrocytes is a key mechanism in multipotent mesenchymal stromal cell‐mediated chondrogenesis. Tissue Eng Part A 21:2536–2547. [DOI] [PubMed] [Google Scholar]
- 38. Bian L, Zhai DY, Mauck RL, et al. 2011. Coculture of human mesenchymal stem cells and articular chondrocytes reduces hypertrophy and enhances functional properties of engineered cartilage. Tissue Eng Part A 17:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang Y‐H, Lee AJ, Barabino GA. 2012. Coculture‐driven mesenchymal stem cell‐differentiated articular chondrocyte‐like cells support neocartilage development. Stem Cells Transl Med 1:843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mauck RL, Yuan X, Tuan RS. 2006. Chondrogenic differentiation and functional maturation of bovine mesenchymal stem cells in long‐term agarose culture. Osteoarthritis Cartilage 14:179–189. [DOI] [PubMed] [Google Scholar]
- 41. Allon AA, Butcher K, Schneider RA, et al. 2012. Structured bilaminar coculture outperforms stem cells and disc cells in a simulated degenerate disc environment. Spine (Phila Pa 1976) 37:813–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Manferdini C, Maumus M, Gabusi E, et al. 2013. Adipose‐derived mesenchymal stem cells exert antiinflammatory effects on chondrocytes and synoviocytes from osteoarthritis patients through prostaglandin E2. Arthritis Rheum 65:1271–1281. [DOI] [PubMed] [Google Scholar]
- 43. Bertolo A, Thiede T, Aebli N, et al. 2011. Human mesenchymal stem cell co‐culture modulates the immunological properties of human intervertebral disc tissue fragments in vitro. Eur Spine J 20:592–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Caplan AI, Correa D. 2011. The MSC: an injury drugstore. Cell Stem Cell 9:11–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zimmermann JA, McDevitt TC. 2014. Pre‐conditioning mesenchymal stromal cell spheroids for immunomodulatory paracrine factor secretion. Cytotherapy 16:331–345. [DOI] [PubMed] [Google Scholar]
- 46. Wei A, Chung SA, Tao H, et al. 2009. Differentiation of rodent bone marrow mesenchymal stem cells into intervertebral disc‐like cells following coculture with rat disc tissue. Tissue Eng Part A 15:2581–2595. [DOI] [PubMed] [Google Scholar]
- 47. Prockop DJ. 2007. “ Stemness” does not explain the repair of many tissues by mesenchymal stem/multipotent stromal cells (MSCs). Clin Pharmacol Ther 82:241–243. [DOI] [PubMed] [Google Scholar]
- 48. Wu L, Leijten JCH, Georgi N, et al. 2011. Trophic effects of mesenchymal stem cells increase chondrocyte proliferation and matrix formation. Tissue Eng Part A 17:1425–1436. [DOI] [PubMed] [Google Scholar]
- 49. Wu L, Prins H‐J, Helder MN, et al. 2012. Trophic effects of mesenchymal stem cells in chondrocyte co‐cultures are independent of culture conditions and cell sources. Tissue Eng Part A 18:1542–1551. [DOI] [PubMed] [Google Scholar]
- 50. Yañez R, Oviedo A, Aldea M, et al. 2010. Prostaglandin E2 plays a key role in the immunosuppressive properties of adipose and bone marrow tissue‐derived mesenchymal stromal cells. Exp Cell Res 316:3109–3123. [DOI] [PubMed] [Google Scholar]
- 51. Zhang W, Green C, Stott NS. 2002. Bone morphogenetic protein‐2 modulation of chondrogenic differentiation in vitro involves gap junction‐mediated intercellular communication. J Cell Physiol 193:233–243. [DOI] [PubMed] [Google Scholar]
- 52. Strassburg S, Hodson NW, Hill PI, et al. 2012. Bi‐directional exchange of membrane components occurs during co‐culture of mesenchymal stem cells and nucleus pulposus cells. PLoS ONE 7:e33739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Acharya C, Adesida A, Zajac P, et al. 2012. Enhanced chondrocyte proliferation and mesenchymal stromal cells chondrogenesis in coculture pellets mediate improved cartilage formation. J Cell Physiol 227:88–97. [DOI] [PubMed] [Google Scholar]
- 54. MacFarlane RJ, Graham SM, Davies PSE, et al. 2013. Anti‐inflammatory role and immunomodulation of mesenchymal stem cells in systemic joint diseases: potential for treatment. Expert Opin Ther Targets 17:243–254. [DOI] [PubMed] [Google Scholar]
- 55. Wu L, Leijten J, van Blitterswijk CA, et al. 2013. Fibroblast growth factor‐1 is a mesenchymal stromal cell‐secreted factor stimulating proliferation of osteoarthritic chondrocytes in co‐culture. Stem Cells Dev 22:2356–2367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Allon AA, Butcher K, Schneider RA, et al. 2012. Structured coculture of mesenchymal stem cells and disc cells enhances differentiation and proliferation. Cells Tissues Organs 196:99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Smith LJ, Nerurkar NL, Choi K‐S, et al. 2011. Degeneration and regeneration of the intervertebral disc: lessons from development. Dis Model Mech 4:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Rutges J, Creemers LB, Dhert W, et al. 2010. Variations in gene and protein expression in human nucleus pulposus in comparison with annulus fibrosus and cartilage cells: potential associations with aging and degeneration. Osteoarthritis Cartilage 18:416–423. [DOI] [PubMed] [Google Scholar]
- 59. Siegel G, Kluba T, Hermanutz‐Klein U, et al. 2013. Phenotype, donor age and gender affect function of human bone marrow‐derived mesenchymal stromal cells. BMC Med 11:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reinisch A, Etchart N, Thomas D, et al. 2015. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood 125:249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Neidlinger‐Wilke C, Würtz K, Urban JPG, et al. 2006. Regulation of gene expression in intervertebral disc cells by low and high hydrostatic pressure. Eur Spine J 15:S372–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Szabó E, Fajka‐Boja R, Kriston‐Pál É, et al. 2015. Licensing by inflammatory cytokines abolishes heterogeneity of immunosuppressive function of mesenchymal stem cell population. Stem Cells Dev 24:2171–2180. [DOI] [PubMed] [Google Scholar]
- 63. Bernstein P, Dong M, Graupher S, et al. 2009. Sox9 expression of alginate‐encapsulated chondrocytes is stimulated by low cell density. J Biomed Mater Res A 91:910–918. [DOI] [PubMed] [Google Scholar]
- 64. Serigano K, Sakai D, Hiyama A, et al. 2010. Effect of cell number on mesenchymal stem cell transplantation in a canine disc degeneration model. J Orthop Res 28:1267–1275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional supporting information may be found in the online version of this article at the publisher's web‐site.
Figure S1. NPC phenotype remains stable in co‐culture.


