Abstract
Vitiligo is an autoimmune disease of the skin that results in the destruction of melanocytes and the clinical appearance of white spots. Disease pathogenesis depends on IFN-γ and IFN-γ-induced chemokines to promote T cell recruitment to the epidermis where melanocytes reside. The skin is a complex organ, with a variety of resident cell types. We sought to better define the microenvironment and distinct cellular contributions during autoimmunity in vitiligo, and found that the epidermis is a chemokine-high niche in both a mouse model and human vitiligo. Analysis of chemokine expression in mouse skin revealed that CXCL9 and CXCL10 expression strongly correlate with disease activity, whereas CXCL10 alone correlates with severity, supporting them as potential biomarkers for following disease progression. Further studies in both our mouse model and human patients revealed that keratinocytes were the major chemokine-producers throughout the course of disease, and functional studies using a conditional STAT1 knockout mouse revealed that IFN-γ signaling in keratinocytes was critical for disease progression and proper autoreactive T cell homing to the epidermis. In contrast, epidermal immune cell populations including endogenous T cells, Langerhans cells, and γδ T cells were not required. These results have important clinical implications, as topical therapies that target IFN-γ signaling in keratinocytes could be safe and effective new treatments, and skin expression of these chemokines could be used to monitor disease activity and treatment responses.
Keywords: Vitiligo, Chemokine, Keratinocyte, CXCL9, CXCL10
Introduction
Vitiligo is an organ-specific autoimmune disease of the skin mediated by autoreactive CD8+ T cells that destroy melanocytes, the pigment-producing cells, resulting in the appearance of white spots. There are no Food and Drug Administration (FDA)-approved treatments, and current off-label treatments are time-consuming and only moderately effective (Dell’Anna ML, 2015; Ezzedine et al., 2015). We reported that IFN-γ is central to disease pathogenesis, and helps to promote autoreactive CD8+ T cell recruitment into the skin (Harris et al., 2012). T cells respond to CXCL9 for bulk recruitment to the skin, while CXCL10 appears to fine-tune T cell localization to the epidermis where melanocytes reside, and promote their effector function (Rashighi et al., 2014). These non-redundant roles for CXCL9 and CXCL10 are surprising, because both chemokines share a single receptor, CXCR3. A similar pattern has been reported in other inflammatory models, and one possible explanation for this is different temporal and spatial patterns of expression of each chemokine, where the timing and location of expression influences their function within the tissue (Groom and Luster, 2011). Such a pattern would implicate distinct contributions of individual cell types during disease progression. For example, melanocyte-specific T cells in vitiligo are first recruited into the vascularized dermis, but then must efficiently find their targets that reside in the basal layer of the epidermis. The timing and expression of chemokines within different layers of the skin could create microenvironments that coordinate the movement of T cells, playing an important role during disease pathogenesis.
The skin is a complex organ made up of various cell types, including both non-immune and immune cells (reviewed in (Pasparakis et al., 2014)). This complexity is analogous to the gut, another epithelial tissue that interfaces with the environment and in which multiple inflammatory diseases are found, including chemokine-dependent autoimmunity (Kunkel et al., 2003; Zimmerman et al., 2008). Studies to address the source of chemokines within peripheral tissues and the functional consequences in vivo are limited, as chemokines are difficult to detect using conventional methods and robust mouse models of autoimmune diseases for mechanistic studies are not widely available. Understanding the contributions of individual cell types to autoimmunity within complex tissues would provide mechanistic understanding of disease pathogenesis that is clinically relevant, as it may dictate therapeutic strategies, including the route of delivery.
We developed a mouse model of vitiligo that closely reflects the effector phase, or progression, of human disease (Agarwal et al., 2015; Harris et al., 2012; Rashighi et al., 2014). Here we use this model, a novel chemokine reporter mouse, and human tissues from vitiligo patients to answer these important mechanistic and clinical questions. We found that CXCL9 and CXCL10 are indeed produced by different epidermal cell types with different timing of expression during vitiligo progression. Keratinocytes outnumber other epidermal cell types by 25:1 in mice and 10:1 in humans, making them excellent candidates to amplify inflammatory signals initiated by a small number of autoreactive T cells through widespread chemokine production. Immunohistochemistry on lesional skin from vitiligo patients confirmed chemokine production by keratinocytes in the epidermis. Functional experiments in our mouse model revealed that eliminating IFN-γ signaling in keratinocytes protected mice from disease, while immune populations in the epidermis were dispensable. Therefore, our data identify keratinocytes as major contributors to vitiligo pathogenesis through chemokine production, and suggest that targeting IFN-γ signaling with topical therapy may be a safe and effective treatment strategy in vitiligo.
Results
CXCL9 and CXCL10 are expressed individually in the skin during vitiligo and form gradients within the tissue
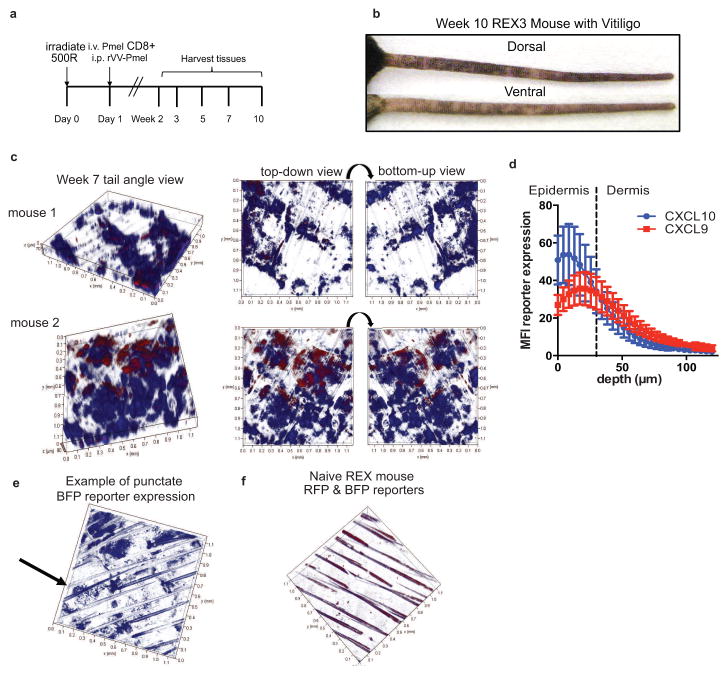
Our previous studies using CXCL9 and CXCL10-deficient animals indicated that CXCL9 recruits a large number of T cells to the skin during vitiligo, while CXCL10 was important for their epidermal localization and function (Rashighi et al., 2014). Therefore, we wanted to determine how these chemokines were functioning differently in the skin during vitiligo. We first determined the location and pattern of CXCL9 and CXCL10 expression using the REX3 mouse strain, which expresses a transgene containing RFP under the control of the CXCL9 promoter and BFP under the control of the CXCL10 promoter (Groom et al., 2012). We bred the REX3 mice to Krt14-Kitl* mice, which have epidermal melanocytes like human skin, to use as hosts in an established vitiligo model (Harris et al., 2012). Vitiligo was induced by transfer of melanocyte-specific T cells that recognize pre-melanosome protein (PMEL, also called gp100) followed by in vivo activation with a recombinant vaccinia virus expressing their cognate antigen (Figure 1A). Importantly, the PMEL T cells recognize both human and mouse gp100 and target mouse melanocytes, thereby providing a clinically-relevant, antigen-specific disease model. Five to seven weeks post-induction, mice develop white spots (depigmentation) on the skin where PMEL T cells have killed epidermal melanocytes (Figure 1B).
Fig. 1. CXCL9 and CXCL10 are expressed in the skin during vitiligo and form gradients within the tissue.
a. REX3 mice were used as hosts in the vitiligo model as shown. b. Representative image depicting a REX3 mouse with vitiligo on the tail. c. Sample images from week 7 mice depicting chemokine clouds (10x z stack). d. Quantification of reporter expression versus depth in skin. CXCL10 is expressed more superficially than CXCL9 (3 representative mice with similar tail scores at week 7, mean ± SEM; dashed line indicates dermal-epidermal junction; 2-way ANOVA significant for depth only). e. Example of punctate CXCL10 reporter expression in a vitiligo mouse. f. Representative z stack from a naïve mouse shows very little reporter expression (10x, auto-fluorescent hairs visible; each tick on axes = 0.1mm).
Confocal microscopy revealed clouds of chemokine expression in the tail skin between weeks 5–10, with some mice expressing chemokine earlier at week 3. Interestingly, most cells were single chemokine producers, making either CXCL9 or CXCL10, with very few foci of dual producers (Figure 1C). We found chemokine expression in lesional, perilesional, and uninvolved skin sites, indicating that chemokine expression precedes depigmentation and is maintained after the process is initiated. Perilesional skin sites often exhibited only CXCL9 expression, suggesting that this chemokine turns on first when a new spot is forming or extending. Quantification of chemokine reporter median fluorescence intensity (MFI) in z stacks from mice with similar disease scores revealed gradients of CXCL9 and CXCL10 that differed spatially, with CXCL10 more superficially expressed (Figure 1D). The dermal-epidermal junction is approximately 30μm below the surface of the tail skin (So et al., 1998), indicating that most chemokine is expressed in the epidermis. In some mice, we found punctate CXCL10 expression (Figure 1E) rather than diffuse expression, indicating that some cells may turn on CXCL10 prior to others in the skin. Naïve mice were taken down at all time points to compare expression levels and control laser settings on the microscope (Figure 1F). Taken together, these data demonstrate distinct spatial expression of CXCL9 and CXCL10 during vitiligo.
Both CXCL9 and CXCL10 reflect vitiligo disease activity, whereas only CXCL10 progressively increases in the epidermis and reflects disease severity
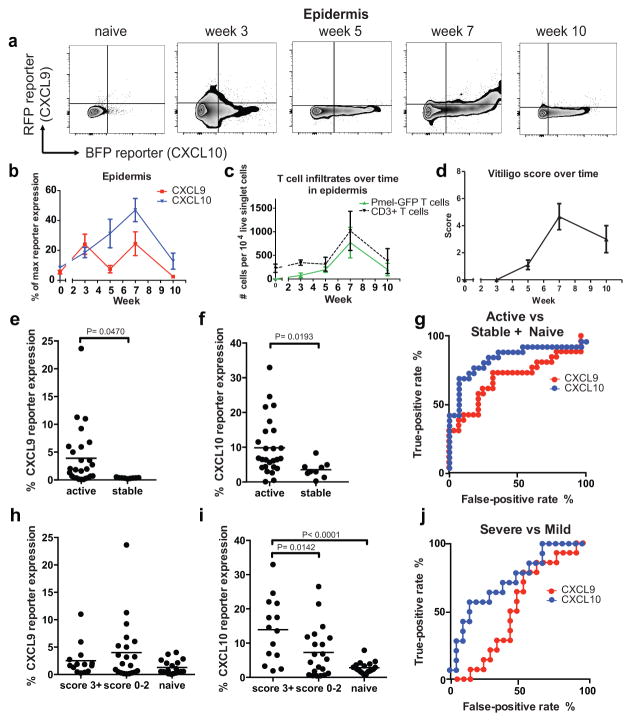
We next quantified and assessed the kinetics of CXCL9 and CXCL10 expression in REX3 mice using flow cytometry (Figure 2A). Chemokine expression in vitiligo mice was assessed in live single cells at 2–3 week intervals after clearance of the virus (Figure S1A). This schedule includes time points before, during, and after onset of clinically evident autoimmunity. Importantly, mice that did not develop clinically-evident vitiligo (i.e. a score of 0) did not exhibit elevated expression of chemokines in the skin, ruling out late effects of viral infection alone (data not shown). Chemokine expression in the lymph node was indistinguishable from naïve control mice by week 3, indicating skin-specific expression during the vitiligo autoimmune response (Figure S1B & C). We found that CXCL9 was highly expressed in the epidermis prior to (3wks) and during (7wks) peak disease (Figure 2B). We suspect this pattern of expression reflects the early formation of new lesions (not yet visible at week 3), and that expression at later time points is due to continued formation of new lesions during active progression, before disease stabilizes. In contrast, epidermal CXCL10 expression peaked with disease and was maintained up to 10 weeks after vitiligo induction when disease stabilized (Figure 2B). Therefore, CXCL9 is maximally upregulated first in a new lesion, followed by peak upregulation of CXCL10. We confirmed that chemokine mRNA expression correlated with reporter expression (Figure S1D). Antigen-specific T cell and endogenous host T cell infiltration in the epidermis increased with chemokine expression during active disease and stabilized between weeks 7 and 10 (Figure 2C).
Fig. 2. CXCL9 and CXCL10 are expressed with different kinetics during the progression of vitiligo in mice.
a. Representative flow plots depicting chemokine reporter expression in the epidermis during vitiligo progression. b. Quantification of chemokine reporter expression in the epidermis of vitiligo mice. c. Melanocyte-specific (PMEL) and endogenous T cell infiltration in the epidermis during vitiligo. d. Scores of all animals used for REX3 analysis. e. CXCL9 and f. CXCL10 reporter expression in active and stable mice. g. Receiver operator characteristics (ROC) curves for CXCL9 and CXCL10 as biomarkers of disease activity. h. CXCL9 and i. CXCL10 reporter expression in severe, mild, or naïve mice. j. ROC curves for CXCL9 and CXCL10 as biomarkers of disease severity. (n=19 naïve mice, 5 mice at week 3, 9 at week 5, 12 at week 7, and 9 at week 10 pooled from 2 to 3 separate experiments; mean ± SEM).
To better understand how CXCL9 and CXCL10 were contributing to disease, we analyzed expression levels based on active disease (weeks 3–7) versus stable disease (week 10, scores stop increasing and T cell infiltrates decrease to a maintenance level, Figure 2D). We found that mice with active disease expressed high levels of CXCL9 and CXCL10 compared to stable mice (Figure 2E & F). To determine the usefulness of CXCL9 and CXCL10 expression as biomarkers of disease activity, we performed Receiver Operator Characteristic (ROC) curve analysis and found that both CXCL9 and CXCL10 had high sensitivity and specificity in mice (CXCL9 AUC = 0.6923, P= 0.0154; CXCL10 AUC = 0.8379, P < 0.0001; Figure 2G). However, CXCL9 levels were lower than CXCL10 levels in mice with stable disease, indicating that CXCL9 may be a more specific biomarker of disease activity. Of note, the entire tail was homogenized for flow cytometry analysis. Therefore, individual lesion activity is difficult to assess by this method, though trends across the tissue indicate that CXCL9 is likely turned on first in a new lesion, followed by CXCL10.
We next assessed whether CXCL9 and CXCL10 could serve as biomarkers of disease severity. Mice with severe disease (score 3+, or >25% depigmentation) had significantly higher levels of CXCL10 but not CXCL9 than mice with mild disease or naïve mice (Figure 2H & I). ROC analysis of chemokine expression for disease severity revealed that only CXCL10 serves as a biomarker of severe disease (CXCL9: AUC = 0.5306, P = 0.7619; CXCL10: AUC = 0.7381, P = 0.01846; Figure 2J). Taken together, these data demonstrate that CXCL9 and CXCL10 can serve as biomarkers of disease activity, whereas CXCL10 serves as a biomarker of disease severity in mice.
Active vitiligo lesions in both mouse and human highly express CXCL9 and CXCL10 in the epidermis
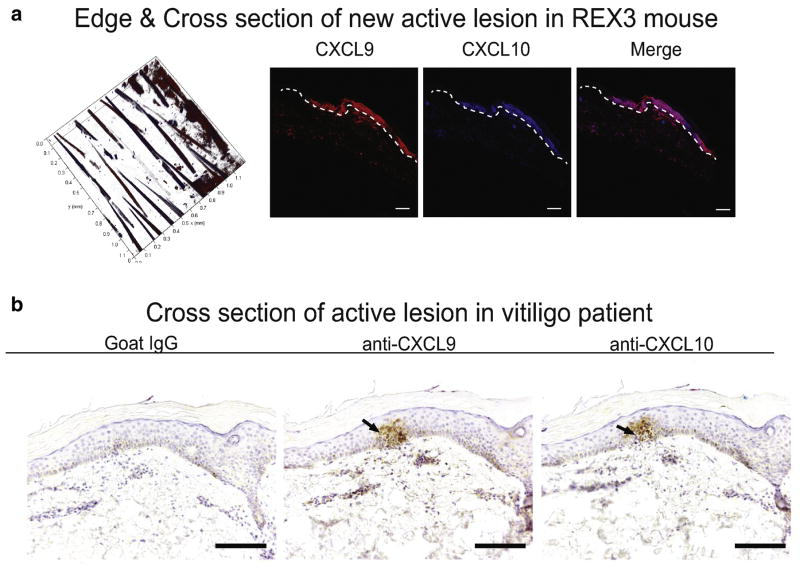
We next determined whether reporter expression in the skin in our mouse model reflected CXCL9 and CXCL10 expression in human disease. We performed confocal microscopy on cross sections of REX3 mouse tail skin during active disease (week 5). In concert, we performed CXCL9 and CXCL10 immunostaining on skin biopsies from vitiligo patients with active disease (T cell infiltrate present). To do this, we acquired elliptical incisional skin biopsies from patients that crossed the lesional border, with one tip in depigmented lesional skin and the other in normally pigmented skin, and sectioned each specimen longitudinally from tip to tip. We found strong epidermal CXCL9 and CXCL10 expression in both mouse and human skin samples (Figure 3).
Fig. 3. Active lesions in both mouse and human express both CXCL9 and CXCL10.
a. Sample lesion from a REX3 vitiligo mouse at week 5 demonstrates expression of both CXCL9 and CXCL10 with high epidermal expression (10x; scale bar = 100μm). b. Cross section of an active lesion from a vitiligo patient also reveals superficial expression of both CXCL9 and CXCL10 (10x; scale bar = 250μm).
Keratinocytes produce the bulk of epidermal CXCL9 and CXCL10 during vitiligo
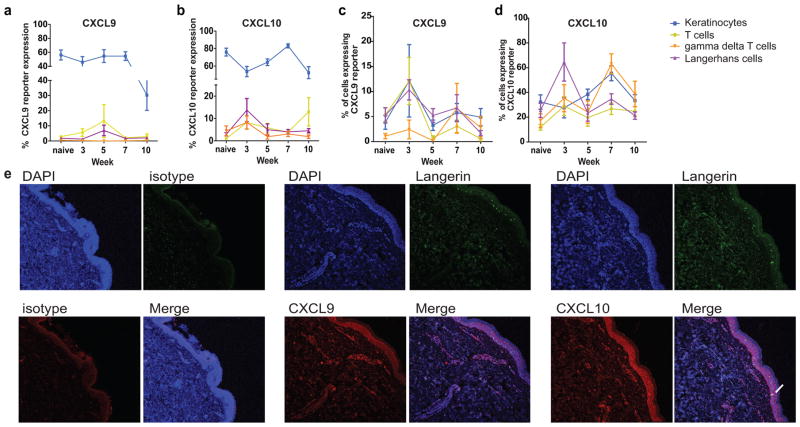
To quantify the contribution of each cell type within the epidermis to the total chemokine produced during the progression of vitiligo, we used flow cytometry and stained for cell-type specific markers (summarized in Table S1). We confirmed that our gating strategy for cell types assessed was appropriate via cell sorting for surface markers followed by qPCR for specific cell transcripts (Figure S2). We found that keratinocytes comprised the largest percentage of total chemokine producers in the skin throughout the course of disease (Figure 4A & B). Keratinocytes also outnumber all other cell types in the epidermis in both mice and humans, and these ratios are presented in Table S2. We next assessed what percentage of each cell population was producing chemokine by first gating on the cell type and then assessing reporter expression (Figure 4C & D). At the peak of disease at week 7, approximately 60% of keratinocytes and γδT cells were expressing CXCL10, compared to just 30% of Langerhans cells and T cells. CXCL9 expression was highest in week 3 in most cell types assessed with variable expression at later time points. Taken together, our data indicate that keratinocytes, which comprise the bulk of the cells within the epidermis, produce the majority of chemokine, while epidermal immune populations represent small foci of chemokine expression.
Fig. 4. CXCL9 and CXCL10 are expressed by different cell types during vitiligo progression. a.
Epidermal cell types represented as a % of total RFP+ gate and b. total BFP+ gate. c. Percent of each cell type producing CXCL9 or d. CXCL10 over time. (n=19 naïve mice, 5 mice at week 3, 9 at week 5, 12 at week 7, and 9 at week 10 pooled from 2 to 3 separate experiments; two-way ANOVAs significant). e. Vitiligo skin ellipse biopsies demonstrate keratinocytes and LHC expressing CXCL9 and CXCL10 in lesions (10x; representative images from 1 of 7 patients evaluated; Langerin, CXCL9, or CXCL10 antibodies were used to stain sections as indicated to identify Langerhans cells and chemokine-producing cells, respectively, and keratinocytes were identified based on their morphology, epidermal location, and negative Langerin staining; white arrows indicate dual staining of Langerin and chemokines; scale bar = 250μm).
To examine whether these results reflected similar characteristics in human tissue, we stained human ellipse biopsies from patients with varied VDAS scores with CXCL9 or CXCL10 antibody and Langerin antibody. As in our mouse model, we found that human keratinocytes in the basal epidermis as well as Langerhans cells express CXCL9 and CXCL10 (Figure 4E). These patterns in vitiligo patients are consistent with reporter mice that reveal chemokine expression in both cell types at distinct stages of disease.
Keratinocyte-derived chemokine production drives vitiligo, whereas other epidermal cell types are dispensable for disease
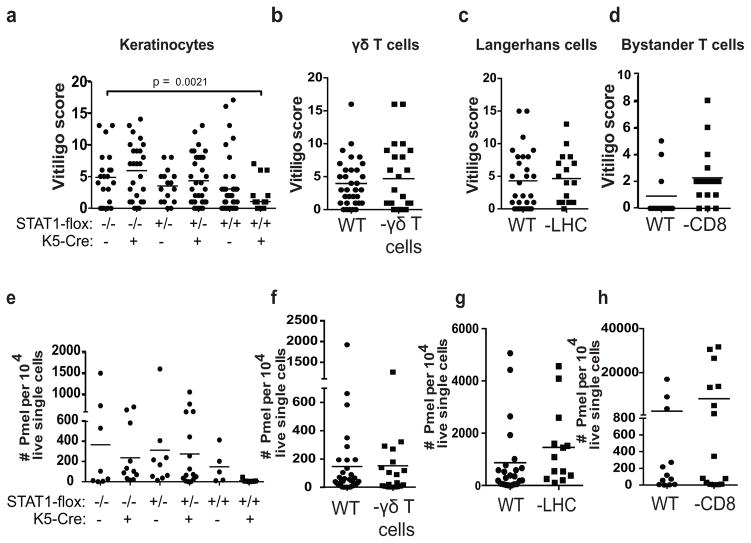
Due to the complex expression patterns of each chemokine within the skin during vitiligo, we sought to determine how individual chemokine-producers functionally contributed to the progression of disease. Therefore, we tested genetically modified hosts in the model to eliminate cell type-specific contributions to vitiligo based on our observations in reporter mice. Keratinocytes are required for viability, so in order to eliminate their contribution we bred STAT1-floxed mice to keratin-5-Cre mice to render them incapable of responding to IFN-γ and making IFN-inducible chemokines. K5-Cre expression was previously reported using LacZ expression, and is limited to the epidermis (Gannon et al., 2011). In vitro treatment of skin from K5-Cre+ STAT1flox/flox mice with TNF-α/IFN-γ revealed no chemokine induction compared to control mice (Figure S3). We found that STAT1 signaling in keratinocytes was required for maximal clinical depigmentation and epidermal PMEL infiltration, revealing that keratinocytes promote vitiligo through recruitment of autoreactive T cells (Figure 5A & E). In contrast, TCR-delta−/− mice (lack γδ T cells (Itohara et al., 1993)), hu-Lang-DTA mice (lack Langerhans cells (Kaplan et al., 2005)), and CD8−/− mice (lack endogenous CD8+ T cells) all developed vitiligo, indicating that these cells are dispensable for disease (Figure 5). These data demonstrate that keratinocytes and autoreactive T cells collaborate to drive autoimmunity in vitiligo.
Fig. 5. Keratinocyte responses to IFN-γ are required for vitiligo induction in mice, whereas other epidermal cell types are dispensable.
a. Mice with keratinocytes unable to respond to IFN-γ (STAT1-flox +/+ K5-cre+) were significantly protected from vitiligo compared to littermate control mice. b. Mice lacking gamma-delta T cells (TCRδ−/−), c. Langerhans cells (hu-Langerin DTA), or d. bystander CD8+ T cells (CD8−/−) did not have significantly different average disease scores as compared to WT controls. e–h. Number of PMEL T cells in the epidermis of STAT1-flox +/+ K5-cre+, TCRδ −/−, hu-Langerin DTA, or CD8−/− mice compared to controls. Each symbol represents data from an individual animal, pooled from 3–4 separate experiments.
Discussion
Evidence suggests that IFN-γ is expressed by autoreactive T cells in vitiligo lesions at very low levels that are difficult to detect. Low-level IFN-γ production appears to be amplified through expression of its target chemokines CXCL9 and CXCL10 (Rashighi et al., 2014), and this is predominantly via expression by keratinocytes. This phenomenon has also been hypothesized to play a role in contact dermatitis (Tokuriki et al., 2002), atopic dermatitis (Klunker et al., 2003), and psoriasis (Lowes et al., 2014), where keratinocytes are major producers of inflammatory cytokines and chemokines, however functional studies in these diseases are lacking. Our functional studies using STAT1-floxed mice in vitiligo support a mechanistic role of keratinocytes rather than immune cells in propagating T cell-derived IFN-γ signals through chemokine expression.
Our data reveal that CXCL9 and CXCL10 are produced in distinct spatial and temporal patterns during the progression of vitiligo, which may explain their non-redundant roles during inflammation within peripheral tissues (Groom and Luster, 2011). This pattern of expression may be responsible for the coordinated movement of T cells through this complex tissue to identify their targets in the epidermis, far from where they enter the skin via blood vessels in the dermis, and may model similar patterns in other complex organs with stratified epithelia, such as the gut. How each cell type is poised to respond to IFN-γ production differently is not currently known, but could involve different mechanisms, such as differential expression of the IFN-γ receptor (Qin and Blankenstein, 2000), unique subcellular localization of the receptor chains (Larkin et al., 2000), different transcription factor expression (Brownell et al., 2014; Tomura and Narumi, 1999), epigenetic modifications of each chemokine promoter (Nancy et al., 2012), cell-specific lncRNA expression (Atianand et al., 2016; Gomez et al., 2013), or variable quantities of both positive and negative components of the downstream signaling pathway (Schneider et al., 2014). Epigenetic modifications would likely be established during development, creating consistent microenvironments within the skin to coordinate migration of T cells. Differences in IFNγR expression or negative signaling components could change over the course of the immune response. Future studies will be required to better understand the detailed control of expression of each chemokine in each cell type.
Despite sharing the CXCR3 receptor, CXCL9 and CXCL10 bind to different regions and mediate slightly different signals to receptor-bearing cells (Colvin et al., 2004). Our reporter mouse data reveal that both CXCL9 and CXCL10 expression reflect disease activity, although CXCL9 may be expressed first in a lesion, which fits with its role as a “recruit” signal. Other models support these observations, including CXCL9 expression in the vaginal mucosa following HSV or LCMV infection that recruits T cells in to tissue in bulk (Iijima and Iwasaki, 2014; Schenkel et al., 2013; Shin and Iwasaki, 2012). Our data also demonstrated that CXCL10 is most reflective of disease severity, which supports its role as a “fine-tune function” signal. This was also observed in a vaccine model that showed CXCL10-dependent tethering of T cells to dendritic cells in the draining lymph node (Groom et al., 2012). Importantly, our mouse data appear to accurately reflect chemokine expression during different phases of human vitiligo. In a recent study that examined a large number of vitiligo patients, serum levels of CXCL9 and CXCL10 correlated with disease activity, while only CXCL10 reflected disease severity (Wang et al., 2016), and another study reported low-level expression of CXCL10 in stable lesions (Regazzetti et al., 2015). Therefore, targeting both CXCL9 and CXCL10 may be important for treating active disease, whereas stable disease may be treated by blocking CXCL10 alone (Rashighi et al., 2014; Regazzetti et al., 2015; Wang et al., 2016). Future clinical studies will need to be conducted to determine the therapeutic potential for targeting CXCR3 ligand expression in keratinocytes as a treatment for vitiligo.
Materials and Methods
Study Design
The objectives of this study were to determine which cells in the skin produce CXCL9 and CXCL10, how they contribute to disease in a mouse model of vitiligo, and to determine the relevance to human vitiligo skin. These objectives were proposed to test the hypothesis that cells in the skin that express high levels of CXCL10 also are functionally required to promote T cell recruitment in vitiligo. This hypothesis was formed based on previous observations reported in our mouse model (Rashighi et al., 2014).
Study subjects
Inclusion/exclusion criteria for human patient samples were as follows: patients with a diagnosis of vitiligo by clinical exam. Patient ellipse skin biopsies were collected after written informed patient consent under IRB-approved protocols at UT Southwestern Medical Center and UMass Medical School by board-certified dermatologists, and all samples were de-identified before use in experiments. The depigmented and pigmented edge of the ellipse biopsy were marked with red and blue ink, respectively, for orientation during sectioning.
Mice
All mice were housed in pathogen-free facilities at UMMS, and procedures were approved by the UMMS Institutional Animal Care and Use Committee and in accordance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals. Mice used for these studies were on the C57BL/6J (B6) background or a mixed 129 × C57BL/6 background that had been backcrossed to B6 for more than 10 generations. Age and sex-matched mice were used, and both male and female mice of all strains were tested to avoid gender bias. Replicate experiments were performed two to five times.
KRT14-Kitl*4XTG2Bjl (Krt14-Kitl*) mice were a gift from B. J. Longley (University of Wisconsin, Madison, WI). The following strains were bred to Krt14-Kitl* mice for use as hosts in the vitiligo model: Tcrd−/− (The Jackson Laboratory, stock no. 002120), Cd8−/− (The Jackson Laboratory, stock no. 002665), hu-Lang-DTA (provided by D. Kaplan, University of Minnesota, Minneapolis, MN), Stat1fl/fl (provided by L. Hennighausen, National Institute of Health, Bethesda, MD), Keratin 5-Cre (provided by S. Jones, University of Massachusetts Medical School, Worcester, MA), and REX3 (provided by A. Luster, Massachusetts General Hospital). Thy1.1+ or GFP PMEL TCR transgenic mice (The Jackson Laboratory, stock no. 005023; or crossed to DPEGFP mice, provided by U. von Andrian, Harvard Medical School, Boston, MA) were used as donors in the vitiligo model.
Vitiligo induction
Vitiligo was induced as previously described (Harris et al., 2012). Briefly, CD8+ T cells were isolated from the spleens of PMEL TCR transgenic mice through negative selection on microbeads (Miltenyi Biotec) according to the manufacturer’s instructions, and 1×106 cells were injected intravenously into sublethally irradiated (500 rads 1 day before transfer) Krt14-Kitl* hosts (8 to 16 weeks of age). On the same day of transfer, recipient mice received intraperitoneal injection of 1×106 plaque-forming units of rVV-hPMEL (N. Restifo, National Cancer Institute, NIH). Vitiligo score was objectively quantified by an observer blinded to the experimental groups, using a point scale based on the extent of depigmentation on the ears, nose, tail and footpads. Points were awarded as follows: no evidence of depigmentation (0%) received a score of 0, >0 to 10% = 1 point, >10 to 25% = 2 points, >25 to 75% = 3 points, >75 to <100% = 4 points, and 100% = 5 points. The “vitiligo score” is the sum of the scores at all four sites, with a maximum score of 20 points.
Flow cytometry
Tissues were harvested at the indicated times. Lymph nodes were mechanically disrupted, and tail skin was incubated with 5U/mL Dispase II (Roche) for 1h at 37°C. Epidermis was removed and mechanically dissociated using 70μm filters and syringe plungers. Dermis was incubated with 1mg/mL collagenase IV and 2mg/mL DNAse I (Sigma Aldrich) for 1h at 37°C before mechanical dissociation. All samples were filtered prior to staining. Naïve REX3 mice were taken down at each time point and used to set gates for chemokine reporter analysis, and UltraComp beads (eBioscience) stained with Alexa Fluor 555 antibodies (Invitrogen) or Pacific Blue antibodies (Biolegend) were used to set up compensation for RFP and BFP. All murine flow cytometry samples were blocked with 2.4G2 and stained with LiveDead Blue (Invitrogen, 1:1000). The following antibodies were used at a 1:200 dilution: CD45 APC-Cy7 or AF700, Thy1.1 FITC, CD3 APC, CD8 PerCP-Cy5.5, CD49f PerCP-Cy5.5, CD31 PE-Cy7, CD11c PE-Cy7, Langerin APC, and CD11b PerCP-Cy5.5 (Biolegend).
Immunohistochemistry and immunofluorescence on human skin samples
Unstained sections of formalin-fixed paraffin-embedded (FFPE) patient skin samples were prepared for immunohistochemistry (IHC) using Bond-Max automated immunohistochemistry system (Leica Biosystems Inc., Buffalo Grove, IL). Following routine dewaxing and rehydration, FFPE sections were subjected to heat-induced epitope retrieval using Bond Epitope Retrieval Solution 2, pH 9 (Leica Biosystems) according to the manufacturer’s instructions, followed by blocking with 10% donkey serum. Sections were incubated with goat polyclonal anti-human CXCL10 antibody (R&D Systems, cat #AF266-NA; 5 μg/ml) or anti-human CXCL9 antibody (R&D Systems, cat #AF392; 5 μg/ml) for one hour at room temperature; washed with Tris-buffered saline (TBS) with 0.5% Tween and subsequently incubated with biotinylated donkey anti-goat IgG (Jackson ImmunoResearch Laboratories, Inc. West Grove, PA, cat# 705-065-147; 1.25ug/mL) for 20 min at room temperature. Color reaction was visualized using Vectastatin Elite avidin-biotin complex kit (Vector Labs, Burlingame, CA, cat #PK-6100). Photomicrographs were obtained using Nikon DS-Ri1 bright-field digital camera.
For dual fluorescence immunohistochemistry, sections were incubated with goat polyclonal anti-human CXCL10 or CXCL9 antibody at 10 μg/ml, along with mouse anti-human Langerin antibody (Leica Biosystems; 1.04 μg/ml) for one hour at room temperature. Secondary incubation was carried out using Alexa Fluor 488 donkey anti-mouse IgG1 (Invitrogen, Carlsbad, CA, 1:600 dilution) and Alexa Fluor 555 donkey anti-goat IgG (Invitrogen, 1:600 dilution) for 30 minutes at room temperature. Sections were coverslipped using ProLong ® Gold Antifade Mountant with DAPI (Life Technologies Grand Island, NY) and sealed with nail polish. Images were taken with a Leica SP8 at 10x magnification, room temperature, with Leica LAS-AF software version 3.3.0.
Confocal microscopy
For analysis of CXCL9 and CXCL10 reporter expression, REX3 mouse tail biopsies were whole-mounted in PBS and kept on ice during tissue processing. All images were taken using a Leica SP8 at 10x magnification, room temperature, with Leica LAS-AF software version 3.3.0. For z stack images, the surface of the skin was determined by using the focal plane of the hair and images were taken through at least 120μm of tissue to pass the dermal-epidermal junction.
Statistics
All statistical analyses were performed with GraphPad Prism software. Dual comparisons were made with unpaired Student’s t test. Groups of three or more were analyzed by ANOVA with Tukey’s or Dunnett’s post-tests. ROC curve analysis was performed to determine the sensitivity and specificity of chemokine biomarkers in mice. P values < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank clinic patients (of J.E.H. and A.G.P.) for donating tissue. We thank L. Hennighausen for STAT1-floxed mice; S. Jones for K5-Cre mice; D. Kaplan for Hu-Lang-DTA mice; B. J. Longley for Krt14-Kitl* mice; U. von Andrian for DPEGFP mice; N. Restifo for recombinant vaccinia virus; and A. Rothstein for insightful comments on the manuscript. We thank members of the Harris Lab including P. Agarwal, M. Damiani, M. Frisoli, M. Rashighi, R. Riding, and J. Strassner for technical assistance.
Funding: Supported by a Research Grant and Calder Research Scholar Award from the American Skin Association (to J.M.R.), the National Institute of Arthritis and Musculoskeletal and Skin Diseases, part of the NIH, under Award Numbers AR061437 and AR069114, and research grants from the Kawaja Vitiligo Research Initiative, Vitiligo Research Foundation, and Dermatology Foundation Stiefel Scholar Award (to J.E.H.). Flow cytometry and confocal microscopy equipment used for this study is maintained by the UMMS Flow Cytometry Core Facility and Morphology Core Facility, and tissue sectioning and pathology services are maintained by the UMMS DERC Morphology Core.
Footnotes
Conflict of Interest: The authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agarwal P, Rashighi M, Essien KI, Richmond JM, Randall L, Pazoki-Toroudi H, et al. Simvastatin prevents and reverses depigmentation in a mouse model of vitiligo. J Invest Dermatol. 2015;135:1080–8. doi: 10.1038/jid.2014.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atianand MK, Hu W, Satpathy AT, Shen Y, Ricci EP, Alvarez-Dominguez JR, et al. A Long Noncoding RNA lincRNA-EPS Acts as a Transcriptional Brake to Restrain Inflammation. Cell. 2016;165:1672–85. doi: 10.1016/j.cell.2016.05.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell J, Bruckner J, Wagoner J, Thomas E, Loo YM, Gale M, Jr, et al. Direct, interferon-independent activation of the CXCL10 promoter by NF-kappaB and interferon regulatory factor 3 during hepatitis C virus infection. J Virol. 2014;88:1582–90. doi: 10.1128/JVI.02007-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin RA, Campanella GS, Sun J, Luster AD. Intracellular domains of CXCR3 that mediate CXCL9, CXCL10, and CXCL11 function. J Biol Chem. 2004;279:30219–27. doi: 10.1074/jbc.M403595200. [DOI] [PubMed] [Google Scholar]
- Dell’Anna MLEK, Hamzavi I, Harris J, Parsad D, Taieb A, Picardo M. Vitiligo. Nature Reviews Disease Primers. 2015;1:1–16. doi: 10.1038/nrdp.2015.11. [DOI] [PubMed] [Google Scholar]
- Ezzedine K, Eleftheriadou V, Whitton M, van Geel N. Vitiligo. Lancet. 2015;386:74–84. doi: 10.1016/S0140-6736(14)60763-7. [DOI] [PubMed] [Google Scholar]
- Gannon HS, Donehower LA, Lyle S, Jones SN. Mdm2-p53 signaling regulates epidermal stem cell senescence and premature aging phenotypes in mouse skin. Developmental biology. 2011;353:1–9. doi: 10.1016/j.ydbio.2011.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez JA, Wapinski OL, Yang YW, Bureau JF, Gopinath S, Monack DM, et al. The NeST long ncRNA controls microbial susceptibility and epigenetic activation of the interferon-gamma locus. Cell. 2013;152:743–54. doi: 10.1016/j.cell.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol. 2011;89:207–15. doi: 10.1038/icb.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groom JR, Richmond J, Murooka TT, Sorensen EW, Sung JH, Bankert K, et al. CXCR3 chemokine receptor-ligand interactions in the lymph node optimize CD4+ T helper 1 cell differentiation. Immunity. 2012;37:1091–103. doi: 10.1016/j.immuni.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris JE, Harris TH, Weninger W, Wherry EJ, Hunter CA, Turka LA. A mouse model of vitiligo with focused epidermal depigmentation requires IFN-gamma for autoreactive CD8(+) T-cell accumulation in the skin. J Invest Dermatol. 2012;132:1869–76. doi: 10.1038/jid.2011.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima N, Iwasaki A. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 2014;346:93–8. doi: 10.1126/science.1257530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itohara S, Mombaerts P, Lafaille J, Iacomini J, Nelson A, Clarke AR, et al. T cell receptor delta gene mutant mice: independent generation of alpha beta T cells and programmed rearrangements of gamma delta TCR genes. Cell. 1993;72:337–48. doi: 10.1016/0092-8674(93)90112-4. [DOI] [PubMed] [Google Scholar]
- Kaplan DH, Jenison MC, Saeland S, Shlomchik WD, Shlomchik MJ. Epidermal langerhans cell-deficient mice develop enhanced contact hypersensitivity. Immunity. 2005;23:611–20. doi: 10.1016/j.immuni.2005.10.008. [DOI] [PubMed] [Google Scholar]
- Klunker S, Trautmann A, Akdis M, Verhagen J, Schmid-Grendelmeier P, Blaser K, et al. A second step of chemotaxis after transendothelial migration: keratinocytes undergoing apoptosis release IFN-gamma-inducible protein 10, monokine induced by IFN-gamma, and IFN-gamma-inducible alpha-chemoattractant for T cell chemotaxis toward epidermis in atopic dermatitis. J Immunol. 2003;171:1078–84. doi: 10.4049/jimmunol.171.2.1078. [DOI] [PubMed] [Google Scholar]
- Kunkel EJ, Campbell DJ, Butcher EC. Chemokines in lymphocyte trafficking and intestinal immunity. Microcirculation. 2003;10:313–23. doi: 10.1038/sj.mn.7800196. [DOI] [PubMed] [Google Scholar]
- Larkin J, 3rd, Johnson HM, Subramaniam PS. Differential nuclear localization of the IFNGR-1 and IFNGR-2 subunits of the IFN-gamma receptor complex following activation by IFN-gamma. J Interferon Cytokine Res. 2000;20:565–76. doi: 10.1089/10799900050044769. [DOI] [PubMed] [Google Scholar]
- Lowes MA, Suarez-Farinas M, Krueger JG. Immunology of psoriasis. Annu Rev Immunol. 2014;32:227–55. doi: 10.1146/annurev-immunol-032713-120225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nancy P, Tagliani E, Tay CS, Asp P, Levy DE, Erlebacher A. Chemokine gene silencing in decidual stromal cells limits T cell access to the maternal-fetal interface. Science. 2012;336:1317–21. doi: 10.1126/science.1220030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M, Haase I, Nestle FO. Mechanisms regulating skin immunity and inflammation. Nat Rev Immunol. 2014;14:289–301. doi: 10.1038/nri3646. [DOI] [PubMed] [Google Scholar]
- Qin Z, Blankenstein T. CD4+ T cell--mediated tumor rejection involves inhibition of angiogenesis that is dependent on IFN gamma receptor expression by nonhematopoietic cells. Immunity. 2000;12:677–86. doi: 10.1016/s1074-7613(00)80218-6. [DOI] [PubMed] [Google Scholar]
- Rashighi M, Agarwal P, Richmond JM, Harris TH, Dresser K, Su MW, et al. CXCL10 Is Critical for the Progression and Maintenance of Depigmentation in a Mouse Model of Vitiligo. Sci Transl Med. 2014;6:223ra23. doi: 10.1126/scitranslmed.3007811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regazzetti C, Joly F, Marty C, Rivier M, Mehul B, Reiniche P, et al. Transcriptional Analysis of Vitiligo Skin Reveals the Alteration of WNT Pathway: A Promising Target for Repigmenting Vitiligo Patients. J Invest Dermatol. 2015;135:3105–14. doi: 10.1038/jid.2015.335. [DOI] [PubMed] [Google Scholar]
- Schenkel JM, Fraser KA, Vezys V, Masopust D. Sensing and alarm function of resident memory CD8(+) T cells. Nat Immunol. 2013;14:509–13. doi: 10.1038/ni.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol. 2014;32:513–45. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Iwasaki A. A vaccine strategy that protects against genital herpes by establishing local memory T cells. Nature. 2012;491:463–7. doi: 10.1038/nature11522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So P, Kim H, Kochevar I. Two-Photon deep tissue ex vivo imaging of mouse dermal and subcutaneous structures. Optics express. 1998;3:339–50. doi: 10.1364/oe.3.000339. [DOI] [PubMed] [Google Scholar]
- Tokuriki A, Seo N, Ito T, Kumakiri M, Takigawa M, Tokura Y. Dominant expression of CXCR3 is associated with induced expression of IP-10 at hapten-challenged sites of murine contact hypersensitivity: a possible role for interferon-gamma-producing CD8(+) T cells in IP-10 expression. Journal of dermatological science. 2002;28:234–41. doi: 10.1016/s0923-1811(01)00172-4. [DOI] [PubMed] [Google Scholar]
- Tomura K, Narumi S. Differential induction of interferon (IFN)-inducible protein 10 following differentiation of a monocyte, macrophage cell lineage is related to the changes of nuclear proteins bound to IFN stimulus response element and kappaB sites. Int J Mol Med. 1999;3:477–84. doi: 10.3892/ijmm.3.5.477. [DOI] [PubMed] [Google Scholar]
- Wang X, Wang Q, Wu J, Jiang M, Chen L, Zhang C, et al. Increased Expression of CXCR3 and its Ligands in Vitiligo Patients and CXCL10 as a Potential Clinical Marker for Vitiligo. Br J Dermatol. 2016 doi: 10.1111/bjd.14416. [DOI] [PubMed] [Google Scholar]
- Zimmerman NP, Vongsa RA, Wendt MK, Dwinell MB. Chemokines and chemokine receptors in mucosal homeostasis at the intestinal epithelial barrier in inflammatory bowel disease. Inflamm Bowel Dis. 2008;14:1000–11. doi: 10.1002/ibd.20480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.