Abstract
CCAR2 is a widely expressed protein involved in the regulation of a variety of transcriptional complexes. High expression of CCAR2 correlates with poor outcomes in a variety of human tumor types such as Squamous Cell Carcinoma (SCC). Paradoxically, loss of Ccar2 in the mouse results in an increased tumor burden, suggesting that CCAR2 may in fact function as a tumor suppressor. Importantly, this tumor suppressor function is dependent on p53, a protein that is inactivated in the vast majority of SCC tumors, leaving the role of CCAR2 in p53-null tumors unclear. We sought to identify p53-independent CCAR2 functions in Squamous Cell Carcinoma, and to examine its role in tumorigenesis. We find that CCAR2 is highly over-expressed in p53-deficient SCC cell lines compared to normal primary keratinocytes due to increased protein stability. We identify a role for CCAR2 in promoting the stability of the transcription factors RFX1 and CREB1, which are both required for proliferation. Finally, we demonstrate that CCAR2 is required for proliferation in vitro and in established SCC tumors in vivo. Our data suggest an important role for CCAR2 in maintaining cell cycle progression and promoting SCC tumorigenesis.
Keywords: Squamous Cell Carcinoma, CCAR2, DBC1, SIRT1, RFX1, CREB
INTODUCTION
Non-melanoma skin cancer is the most common form of cancer, and although the majority of patients exhibit positive outcomes, a subset of patients succumb to metastatic disease (Karia et al., 2013). Clinical studies across a variety of tumor types, including SCC, have suggested that high levels of CCAR2 (Cell Cycle activator and Apoptosis Regulator 2; also known as DBC1) may be an indicator of poor outcomes (Chen et al., 2014; Kim et al., 2013; Kim et al., 2012; Yu et al., 2013), and could potentially have pro-tumorigenic functions. However, studies in Ccar2−/− mice suggest that Ccar2 may function as a tumor-suppressor, although SCC did not develop in these mice (Qin et al., 2015).
CCAR2 has been shown to bind and inhibit the NAD-dependent deacetylase activity of SIRT1 (Kim et al., 2008; Zhao et al., 2008). The CCAR2-SIRT1 complex is dynamically regulated by nutrient levels, as fasting results in dissociation to release highly active SIRT1, while high nutrient levels promote the SIRT1-CCAR2 association, blocking SIRT1 deacetylase activity (Escande et al., 2010; Nin et al., 2014). In addition, the CCAR2-SIRT1 interaction can be inhibited through acetylation of CCAR2 by hMOF (Zheng et al., 2013), or enhanced through phosphorylation of CCAR2 by ATM/ATR kinases in response to DNA damage (Yuan et al., 2012; Zannini et al., 2012). Importantly, inhibition of SIRT1 activity by CCAR2 following DNA damage results in acetylation and activation of p53, which can then activate downstream targets (Kim et al., 2008; Luo et al., 2001; Vaziri et al., 2001; Zhao et al., 2008). In addition, CCAR2 can regulate the function of a variety of transcription factors through inhibition of SIRT1-dependent deacetylation (Chini et al., 2013a; Huan et al., 2015; Kim et al., 2014; Koyama et al., 2010; Park et al., 2013; Sakurabashi et al., 2015; Tanikawa et al., 2013; Yu et al., 2015).
In this study, we sought to clarify the role of CCAR2 in Squamous Cell Carcinoma. We demonstrate that SCC cells stabilize CCAR2 protein, leading to significant overexpression, and that CCAR2 is essential for continued proliferation of SCC tumors, independent of p53 function. We have identified key transcription factors, CREB and RFX1, which bind and are stabilized by CCAR2. Finally, we demonstrate that both CREB and RFX1 are required for the maintenance of proliferation in Squamous Cell Carcinoma.
RESULTS
CCAR2 protein expression is elevated in Squamous Cell Carcinoma
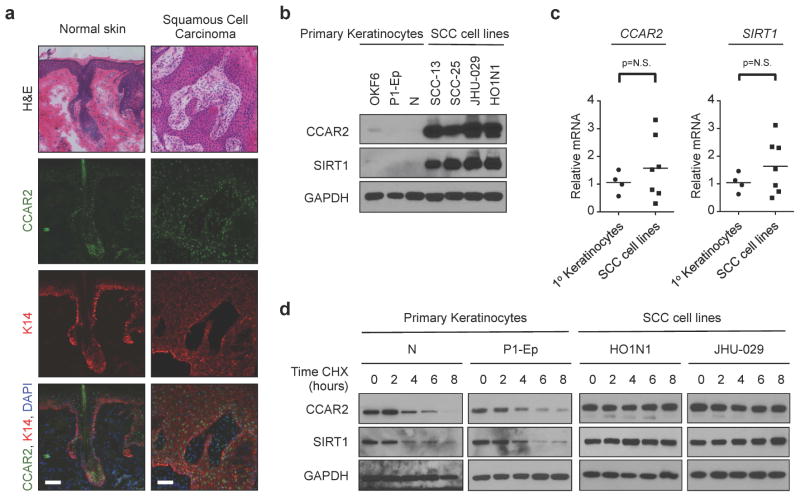
In order to understand the role of CCAR2 in SCC, we examined CCAR2 expression in normal epidermis and cutaneous SCC (Figure 1a). In normal skin, CCAR2 was expressed in both the interfollicular epidermis and the hair follicle. Similar to esophageal SCC (Kim et al., 2012), CCAR2 was also expressed in cutaneous SCC (Figure 1a). Examination of cultured primary human and murine keratinocytes and SCC cell lines from the epidermis (SCC-13), oral cavity (SCC-25, HO1N1), and larynx (JHU-029) by western blot identified a large increase in CCAR2 protein in SCC cells compared keratinocytes (Figures 1b and S1a). One major function of CCAR2 is to inhibit the deacetylase activity of SIRT1 through direct interaction (Kim et al., 2008; Zhao et al., 2008), and high SIRT1 expression correlates with poor prognosis in SCC (Chen et al., 2014). Similar to CCAR2, we observed an increase in SIRT1 protein in SCC cells compared to normal keratinocytes (Figures 1b and S1a). Despite the large increases in protein, there was no significant difference in either CCAR2 or SIRT1 mRNA levels between primary keratinocytes and SCC cell lines (Figure 1c and S1b). We treated primary keratinocytes and SCC cells with cycloheximide, an inhibitor of protein translation, and examined CCAR2 and SIRT1 protein by western blot. In primary keratinocytes, the half-life of both CCAR2 and was between 2 and 4 hours; while in SCC cells no detectable decrease in CCAR2 or SIRT1 protein was observed within the 8-hour experiment (Figure 1d and S1c). These data demonstrate that CCAR2 and SIRT1 protein is more abundant in SCC cells compared to normal keratinocytes due to a significant increase in protein stability.
Figure 1. CCAR2 and SIRT1 proteins are stablized in squamous cell carcinoma.
(a) Histology (H&E) and Immunofluorescence of CCAR2 (green) and Keratin 14 (K14, red) protein expression in normal human skin and Squamous Cell Carcinoma (SCC) frozen sections. DAPI (blue) stains the nucleus. Scale = 50μm. (b) Western blot analysis of CCAR2 and SIRT1 protein in keratinocytes (OKF6, P1-Ep and N) and SCC (SCC-13, SCC-25, JHU-029 and HO1N1) cell lines. GAPDH provides the loading control. (c) Quantitative RT-PCR of CCAR2 (left) and SIRT1 (right) mRNA expression in keratinocytes (Ker) and SCC cell lines. p > 0.05, not significant (N.S.). (d) Western blot of CCAR2 and SIRT1 protein expression in keratinocytes (N, P1-Ep) and SCC (HO1N1 and JHU-029) cell lines treated with Cyclohexamide (CHX, 100 μg/ml) for 0, 2, 4, 6 or 8 hours. GAPDH provides the loading control.
CCAR2 is required for SCC cell proliferation
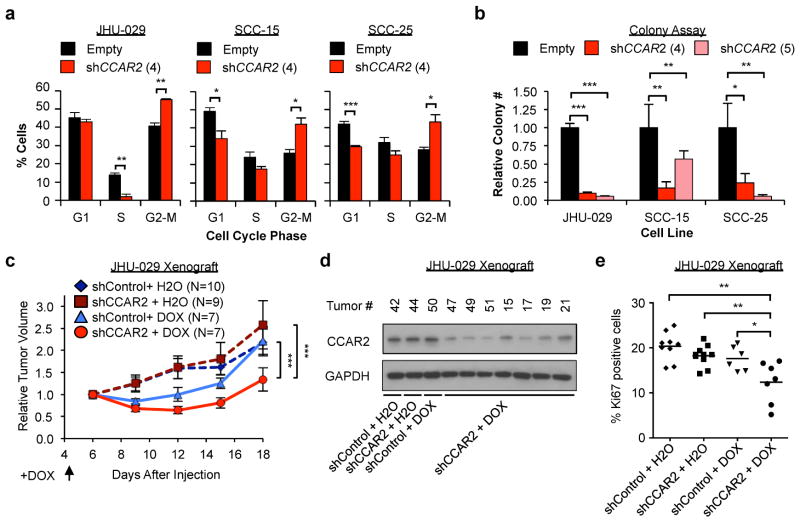
In order to assess the physiological effects of CCAR2 loss, we infected SCC cells with shRNA targeting CCAR2 and examined cell cycle by flow cytometry. Following reduction in CCAR2 levels (Figure S2a), all cell lines examined (JHU-029, SCC-15, and SCC-25) exhibited a significant increase in the percentage of G2 cells, with a coincident reduction in G1 and S-phase cells (Figure 2a). Consistent with these results, we observed a significant reduction in colony-forming ability of SCC cells following reduction of CCAR2 levels (Figure 2b and S2b). Of note, these cells all have functional inactivation of p53 due to frameshift or point mutation (Supplementary Table S1). We then investigated the in vivo requirement for CCAR2 in tumor maintenance. We stably expressed a doxycycline-inducible shRNA directed against CCAR2 or control in tumorigenic JHU-029 cells. Addition of doxycycline results in greater than 50% reduction of CCAR2 protein and mRNA by 48 hours in shCCAR2 cells, but not control cells (Figure S2c and S2d). Cells were injected subcutaneously into Nude mice, and then treated with doxycycline-containing water after four days. Although doxycycline alone had a modest inhibitory effect on tumor growth at early time-points, there was a significant reduction in tumor growth in shCCAR2 tumors treated with doxycycline compared to control and shCCAR2 tumors treated with vehicle (Figure 2c). Importantly, only the shCCAR2 tumors treated with doxycycline showed reduced CCAR2 protein expression (Figure 2d). Tumors showed no change in Cleaved Caspase 3 staining, suggesting that CCAR2 does not regulate apoptosis in vivo (Figure S2e). However, there was a significant reduction in Ki67 staining, a marker of proliferation, in shCCAR2 tumors treated with doxycycline compared to all other groups (Figure 2e). These data point to an essential p53-independent role for CCAR2 in promoting SCC cell proliferation and tumor maintenance.
Figure 2. CCAR2 is essential for proliferation and tumor maintenance.
(a) Flow cytometry analysis of the cell cycle (G1, S or G2/M) in control (Empty) or CCAR2 knockdown (shCCAR2) SCC cell lines. N=3 replicates. Error bars indicate +/− Standard Deviation. Representative from 2 independent repeats. (b) Colony assay in control or CCAR2 knockdown SCC cell lines. Colony number is relative to the control of each cell line. Error bars indicate +/− Standard deviation. N= 4 replicates per cell line. (c) Growth of Xenograft tumors derived from JHU-029 cells with Doxycycline-inducible shRNA, injected subcutaneously in Nude mice. Doxycycline was added to drinking water on day 4. Tumor volume is normalized to average day 6 tumor volume for each group. Error bars +/− SEM. (d) Western blot analysis of CCAR2 protein in JHU-029 Xenograft tumors with indicated treatment as in (c). (e) Analysis of proliferative index of tumors from (c) at day 18 as assessed by Ki67 staining. *p<0.05, **p<0.01, ***p<0.001 for all experiments.
Identification of transcriptional programs regulated by CCAR2
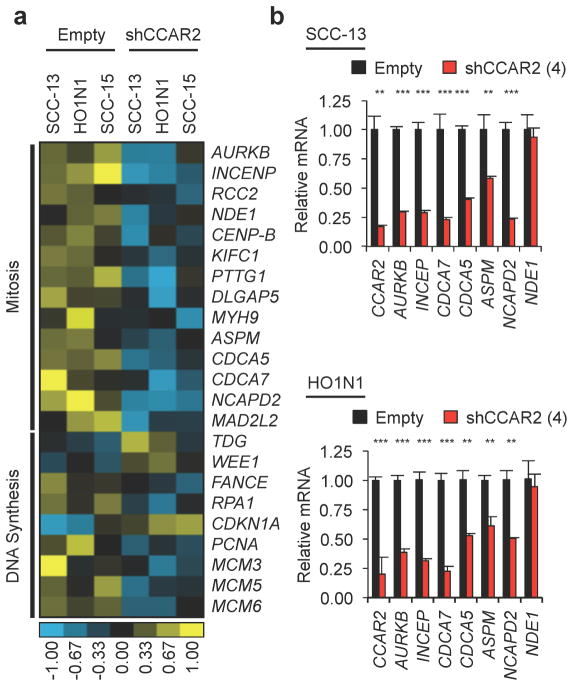
Given the clear requirement for CCAR2 in proliferation, we hypothesized that CCAR2 could regulate one or more transcription factors controlling important cell cycle genes. SCC-13, SCC-15, and HO1N1 cells were infected with shRNA empty vector (Empty) or shRNA directed against CCAR2 (shCCAR2), cells were harvested after 48 hours, RNA was prepared, and global gene expression profiling was performed using Illumina Human HT-12 BeadChip arrays. 164 transcripts had a greater than 0.5log2 fold-change and a p-value of <0.05 following CCAR2 knockdown, with 128 transcripts decreasing expression and 36 transcripts increasing expression (Supplementary Table S2). Pathway and Gene Ontogeny (GO) analysis of this dataset was then performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) tool (Dennis et al., 2003). The large majority of GO terms and pathways centered on the cell cycle, in particular genes involved in DNA synthesis and mitosis (Supplementary Tables S3 and S4). Examination of transcripts identified by DAVID analysis showed that the majority of these genes promoted cell cycle progression, and expression decreased following loss of CCAR2 (Figure 3a). Using Quantitative Real-Time PCR, we validated decreases in mitotic genes (AURKB, INCENP, CDCA7, CDCA5, ASPM, and NCAPD2) 48 hours following shRNA knockdown of CCAR2 in SCC-13 and HO1N1 cells (Figure 3b), confirming the microarray results. These data identify a subset of cell cycle genes controlled by CCAR2, further supporting a key role in promoting proliferation.
Figure 3. CCAR2 regulates genes involved in cell division.
(a) Heat-map depicting gene expression changes from microarray in CCAR2 knockdown (shCCAR2) compared to control cells (Empty) in SCC cell lines (SCC-13, HO1N1, SCC-15). (b) Quantitative RT-PCR validation of genes identified by the microarray in SCC-13 (top panel) and HO1N1 cells (bottom panel) with either control (Empty) or CCAR2 knockdown (shCCAR2). **p<0.01, ***p<0.001, N.S. = Not significant.
CCAR2 regulates stability of RFX1 and CREB to promote proliferation
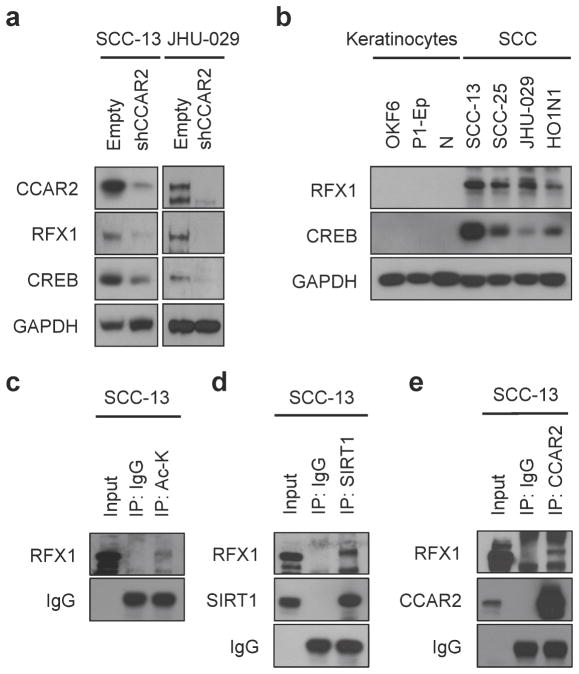
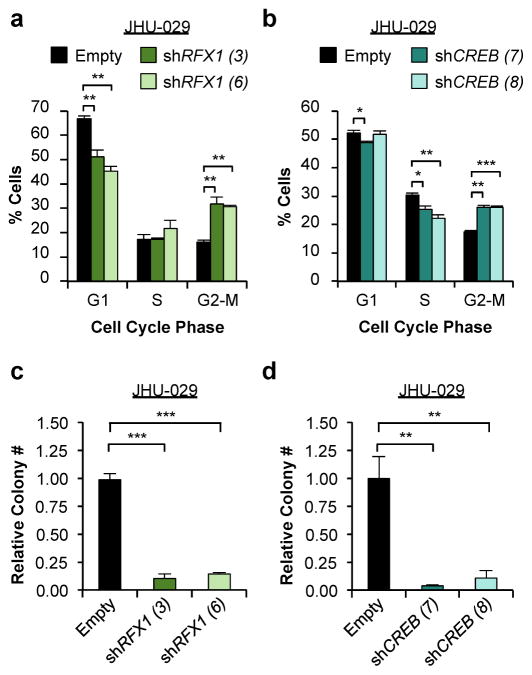
CCAR2 has been found to regulate a variety of transcription factors in a context-specific manner. Using the DAVID tool, we generated a list of transcription factors with canonical binding sites that are significantly enriched in our microarray gene set (Supplementary Table S5). When CCAR2 expression was depleted in SCC cell lines using shRNA, the protein, but not mRNA of a subset of transcription factors, notably RFX1 and CREB, were reduced (Figures 4a and S3). Interestingly, both RFX1 and CREB have been shown to bind in close proximity to the PCNA promoter (Lee and Mathews, 1997; Liu et al., 1999), suggesting they could contribute to cell cycle control. Given our observations that CCAR2 levels are high in SCC cells compared to keratinocytes (Figure 1b), we examined RFX1 and CREB levels in SCC cells and mid-lifespan primary keratinocytes. As with CCAR2, both RFX1 and CREB protein, but not mRNA, were increased in SCC cells (Figure 4b, Figure S3), suggesting that CCAR2 regulates protein levels of RFX1 and CREB. SIRT1 has been shown to decrease protein stability through deacetylase activity (Li et al., 2010). We hypothesized that CCAR2 inhibits SIRT1-mediated deacetylation through binding to CREB and RFX1, increasing protein stability. Immunoprecipitation of RFX1 from the nuclear and cytoplasmic fractions of HO1N1 cells demonstrated binding of RFX1 to CCAR2 and CREB in the nucleus (Figure S3). While SIRT1 has been previously found to regulate acetylation of CREB (Paz et al., 2014), RFX1 has not. In order to determine if RFX1 is also acetylated, we performed immunoprecipitation with α-Acetyl Lysine antibodies followed by western blotting for RFX1 in SCC-13 cells, suggesting that RFX1 is acetylated (Figure 4c). Immunoprecipitation with α-SIRT1 antibodies demonstrated that SIRT1 does interact with RFX1 (Figures 4d and S3). In addition, we also observed binding of CCAR2 to both RFX1 (Figures 4e and S3) and SIRT1 (Figure S3). CCAR2 controls proliferation, and we sought to examine the interaction of CCAR2 and SIRT1 with RFX1 during the cell cycle. Cells were synchronized prior to entering S-phase with the CDK4/6 inhibitor PD0332991, released, and then examined during G1 arrest and S-phase entry. Interestingly, while both CCAR2 and SIRT1 interacted similarly during S-phase entry, there was a shift in the mobility of RFX1 upon S-phase entry. In addition, CCAR2 and SIRT1 appear to physically interact with different forms of RFX1, suggesting that they are in unique complexes (Figure S3). Finally, we sought to examine whether RFX1 and CREB functionally contribute SCC proliferation. Similar to reduction of CCAR2 (Figure 2), shRNA-mediated knockdown of either RFX1 or CREB (Figure S4) resulted in a significant increase in G2 phase cells (Figures 5a and 5b) and a reduction in colony forming ability (Figures 5c and 5d). In total, these data support a role for the CCAR2-RFX1-CREB complex in promoting proliferation in SCC tumors.
Figure 4. CCAR2 regulates stability of acetylated transcription factors in SCC.
(a) Western blot analysis of CCAR2, RFX1, and CREB protein in SCC cell lines with CCAR2 knockdown (shCCAR2) or control (Empty). GAPDH provides the loading control. (b) Western blot analysis of RFX1 and CREB protein in keratinocytes and SCC cells. GAPDH provides the loading control. (c) Immunoprecipitation of Acetylated RFX1 protein from whole cell extracts of SCC-13 cells. IgG serves as a loading control for IP. (d) Co-Immunoprecipitation of RFX1 and SIRT1 proteins from whole cell extracts of SCC-13. IgG serves as a loading control for IP. (e) Co-Immunoprecipitation of RFX1 and CCAR2 proteins from whole cell extracts of SCC-13. IgG serves as a loading control for IP.
Figure 5. RFX1 and CREB are required for proliferation in SCC.
(a) Flow cytometry analysis of the cell cycle (G1, S or G2/M) in control (Empty), RFX1 knockdown (shRFX1) in JHU-029 cells (n=3). (b) Flow cytometry analysis of the cell cycle in control (Empty) or CREB knockdown (shCREB) in JHU-029 cells (n=3). Error bars indicate +/− Standard Deviation. (c) Colony assay in control (Empty), RFX1 knockdown (shRFX1) in JHU-029 cells. Colony number is relative to the control of each cell line. Error bars indicate +/− Standard deviation. N=4 replicates per cell line. Representative from 2 independent repeats. (d) Colony assay in control (Empty) CREB knockdown (shCREB) in JHU-029 cells.
DISCUSSION
Previous studies have reported conflicting results regarding the role of CCAR2 in tumorigenesis (Chini et al., 2013b; Song and Surh, 2012), resulting in uncertainty whether CCAR2 would serve as a useful predictive biomarker. Some reports have suggested that CCAR2 is reduced in tumors (Shim et al., 2013; Won et al., 2015), consistent with data from Ccar2−/− mice exhibiting an increase in tumor formation (Qin et al., 2015). This also fits with the established role for CCAR2 in regulating stability of p53 through acetylation (Kim et al., 2008; Zhao et al., 2008) or inhibition of the p53 ubiquitin ligase Mdm2 (Qin et al., 2015). Indeed, regulation of p53 by CCAR2 appears to be important for tumor initiation in some tissues, as Ccar2−/− mice have increased tumor incidence compared to wild-type controls, dependent on p53 but Sirt-independent (Qin et al., 2015). In contrast to this data, many tumor types have been reported to have high expression of CCAR2, and this correlates with poor clinical outcomes (Cha et al., 2009; Cho et al., 2015; Kim et al., 2013; Kim et al., 2012; Yu et al., 2015; Yu et al., 2013; Zhang et al., 2014). In addition, the large majority of SCC tumors have mutated or lost p53 (Giglia-Mari and Sarasin, 2003), while CCAR2 is rarely altered in human cancer (Cerami et al., 2012), suggesting there may be p53-independent roles for CCAR2 in SCC. In this context, we sought to examine the requirement for CCAR2 in established SCC tumors, where high expression of CCAR2 is strongly correlated with poor clinical outcomes (Kim et al., 2012; Yu et al., 2013). Consistent with these reports, we find high levels of CCAR2 in human and mouse SCC compared to normal keratinocytes. This high expression of CCAR2 is functionally important, as reduction of CCAR2 results in decreased proliferation, colony formation, and in vivo tumor growth, strongly supporting a role for CCAR2 in the maintenance of proliferation in p53-null tumors. Thus, in addition to serving as a negative prognostic indicator, CCAR2 may serve as an attractive therapeutic target in SCC.
CCAR2 has been revealed to affect the activity of a variety of transcription factors in different contexts, serving as a positive regulator of p53 function (Kim et al., 2008; Zhao et al., 2008), but an inhibitor of others such as the Liver X Receptor α (Sakurabashi et al., 2015). We have identified an interaction between CCAR2 and the transcription factors RFX1 and CREB, which our data suggests serves to stabilize these proteins. We demonstrate a requirement for CREB in maintaining proliferation in SCC cells, and consistent with this data, CREB is required for papilloma formation following chemical carcinogenesis (Rozenberg et al., 2009). While SIRT1-dependent acetylation of CREB activity has been reported (Paz et al., 2014), CCAR2 was not previously known to be involved in this regulatory axis. To our knowledge, a role for RFX1 in promoting tumor maintenance has not been previously described, and our data demonstrate that RFX1 makes a functional contribution to SCC proliferation. Our data also demonstrate that RFX1 is an acetylated protein and physically interacts with SIRT1, suggesting that SIRT1 may regulate the RFX1 acetylation state. Regulation of RFX1 stability by SIRT1 fits with a model proposed for the WRN protein, where SIRT1-mediated deacetylation can reduce protein stability (Li et al., 2010). Our data supports a model in which CCAR2 functions to maintain appropriate temporal acetylation of RFX1 and CREB by blocking SIRT1 activity and thus facilitating transcription of key cell cycle genes.
MATERIALS AND METHODS
Cell Lines and Xenografts
The unique identity of SCC-13, SCC-15, SCC-25, JHU-029, and HO1N1 cell lines were verified by STR profiling performed by the ATCC. Individual adult B6 mice were used to generate each primary keratinocyte cell line, Mu-K1 and Mu-K2 (Supplementary Methods). Culture conditions can be found in Supplementary Methods. Xenograft tumors were generated as previously described (Ramsey et al., 2011). 4 days after injections, water was exchanged with 2 % sucrose and 2 mg/mL doxycycline water, or sham water, which was freshly prepared every other day. Tumor volumes were calculated using the formula: tumor volume (mm3) = 4/3π x (length ÷ 2) x (width ÷ 2).
Western blotting and Immunoprecipitation
Cells were lysed in RIPA buffer (10 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 % (w/v) Sodium deoxycholate, 0.1 % (w/v) SDS, 1 % (v/v) NP-40), and western blotting was performed as previously described (Ramsey et al., 2011). Fractionation of nuclear and cytoplasmic lysates and immunoprecipitation was performed as previously described (Ramsey et al., 2011). In all other immunoprecipitation experiments, cells were incubated at 4 °C for 2 hours with NP-40 lysis buffer (50 mM Tris-HCl pH 7.9, 120 mM NaCl, 0.75% NP-40), and then incubated with protein A beads for 30 minutes. Cleared lysates were incubated with antibody and protein A beads for 3 hours at 4 °C, and immunocomplexes were washed with 5 times with NP-40 buffer. For protein stability experiments, cells were treated with 100 μg/ml Cyclohexamide (VWR) for up to 8 hours.
Lentiviral infection with shRNA constructs
Lentivirus was generated by transfecting 293T cells using calcium phosphate transfection (Clontech) with plasmid of interest and packaging plasmids (VSV-G, RGR, RSV, Addgene). Viral supernatant was filter through a 0.45μm filter, then 8 μg/ml polybrene (Santa Cruz) was added and viral supernant was applied to cell lines in 6-well plates. After a 1-hour spin infection at 1000g, the media was replaced and puromycin added to the cells after 24 hours. Following 48-hours incubation with puromycin, cells were harvested for downstream assays. Sequences for hairpins targeting CCAR2, RFX1 (GE Healthcare Biosciences), and CREB1 (Sigma) are listed in Supplementary Table S6.
Cell cycle analysis and colony forming assays
Cells were treated with 10 mM Bromodeoxyuridine (BrdU) 2 hours prior to trypsinization and fixation with cold ethanol. Cell pellets were incubated in 0.08 % (w/v) pepsin (in 0.1 M HCl) for 20 minutes at 37 °C and centrifuged. Nuclei were incubated in 2 M HCl for 20 minutes at 37 °C and 0.1 M Sodium Borate added while vortexing. After centrifugation, pellets were washed in IFA/Tween 20 (10 mM HEPES, 150 mM NaCl, 4 % Fetal Bovine Serum, 0.1 % Sodium Azide, 0.5 % Tween 20) and incubated in primary BrdU antibody (MoBu-1, Invitrogen), washed and incubated in secondary AlexaFluor488 (Invitrogen). Nuclei were resuspended in IFA with 50μg/mL propidium iodide for flow cytometry on BD Canto Analyser using FlowJo software. For colony forming assays, 1000 cells were seeded per well of 12-well tissue culture plates, with media changed every 48 hours. After 8 days, cells were fixed with 10 % buffered formalin, stained with 0.1 % crystal violet for 20 minutes, and plates were scanned electronically for colony counts. Only colonies meeting a minimum size cut-off were counted.
Quantitative Real-Time PCR and Microarray
RNA was extracted from cell lines using Qiagen RNeasy Mini Kit according to manufacturer’s instructions. Quantitative Real-Time PCR analysis was performed using KAPA SYBR FAST ABI Prism qPCR Kit (Kapa Biosystems) according to manufacturer’s instructions. Primers used for qRT-PCR are listed in Supplementary Table S7. For microarray analysis, synthesis of cDNA, hybridization, and reading of signal intensity were performed by the Partners HealthCare Center for Personalized Genetic Medicine. RNA quality was assessed on a 2100 bioanalyzer (Agilent) and total RNA was hybridized to Human HT-12 BeadChip arrays. Chips were scanned with Illumina BeadArray Reader and data were processed using the R statistical software environment version 2.12.0 (Gentleman et al., 2004). Data were background corrected, normalized, and underwent variance stabilizing transformation using the Lumi package (Du et al., 2008). Differential gene expression was determined based on a moderated t-test using the Limma package (Smyth, 2004). Unprocessed raw data is available through the NCBI Gene Expression Omnibus (GSE85966).
Statistical Analysis
For cell cycle and colony forming assays, student’s T-test was used to assess statistical significance. For Xenograft experiments, p-values were determined using multiple measures ANOVA. For mRNA correlations, Pearson’s Product-moment Correlation Coefficient (R2) was calculated and two-tailed P-value was generated from a probability table. p-values for GO, KEGG, and Panther Pathways as analyzed using DAVID were determined using Fisher's Exact Test. Only data sets with a False Discovery Rate (FDR) of less than 5 % were included. P-values < 0.05 were considered significant for all experiments.
Study Approvals
The Partners Human Research Committee approved use of tissues for this study. All human tissue studies used exclusively de-identified and discarded material collected in the course of routine clinical care, for which the Partners Human Research Committee determined that signed informed consent was not required. All animals were housed and treated in accordance with protocols approved by the Harvard Medical Area (HMA) Standing Committee on Animals.
Supplementary Material
Acknowledgments
We wish to thank Rebeca Cardoso, Sandra King and Pritesh Karia for technical support; James Rheinwald, Chuck Dimitroff, Tobias Schatton, and Norman Sharpless for comments on the manuscript; and James Rheinwald for advice, reagents and support during the establishment of the Ramsey Laboratory. This work was supported by NCI K99/R00CA157730 (MRR) and the BWH Department of Dermatology Fund for New Investigators.
Abbreviations
- CCAR2
Cell Cycle activator and Apoptosis Regulator 2)
- SCC
Squamous Cell carcinoma
- RFX1
Regulatory Factor X, 1
- CREB
CAMP Responsive Element Binding Protein 1
- SIRT1
Sirtuin 1
Footnotes
CONFLICT OF INTEREST
The authors have declared that no conflict of interest exists.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha EJ, Noh SJ, Kwon KS, Kim CY, Park BH, Park HS, et al. Expression of DBC1 and SIRT1 is associated with poor prognosis of gastric carcinoma. Clin Cancer Res. 2009;15:4453–4459. doi: 10.1158/1078-0432.CCR-08-3329. [DOI] [PubMed] [Google Scholar]
- Chen GQ, Tian H, Yue WM, Li L, Li SH, Qi L, et al. SIRT1 expression is associated with lymphangiogenesis, lymphovascular invasion and prognosis in pN0 esophageal squamous cell carcinoma. Cell Biosci. 2014;4:48. doi: 10.1186/2045-3701-4-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini CC, Escande C, Nin V, Chini EN. DBC1 (Deleted in Breast Cancer 1) modulates the stability and function of the nuclear receptor Rev-erbalpha. Biochem J. 2013;451:453–461. doi: 10.1042/BJ20121085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini EN, Chini CC, Nin V, Escande C. Deleted in breast cancer-1 (DBC-1) in the interface between metabolism, aging and cancer. Biosci Rep. 2013:33. doi: 10.1042/BSR20130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho D, Park H, Park SH, Kim K, Chung M, Moon W, et al. The expression of DBC1/CCAR2 is associated with poor prognosis of ovarian carcinoma. J Ovarian Res. 2015;8:2. doi: 10.1186/s13048-015-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Du P, Kibbe WA, Lin SM. lumi: a pipeline for processing Illumina microarray. Bioinformatics. 2008;24:1547–1548. doi: 10.1093/bioinformatics/btn224. [DOI] [PubMed] [Google Scholar]
- Escande C, Chini CC, Nin V, Dykhouse KM, Novak CM, Levine J, et al. Deleted in breast cancer-1 regulates SIRT1 activity and contributes to high-fat diet-induced liver steatosis in mice. J Clin Invest. 2010;120:545–558. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglia-Mari G, Sarasin A. TP53 mutations in human skin cancers. Hum Mutat. 2003;21:217–228. doi: 10.1002/humu.10179. [DOI] [PubMed] [Google Scholar]
- Huan Y, Wu D, Zhou D, Sun B, Li G. DBC1 promotes anoikis resistance of gastric cancer cells by regulating NF-kappaB activity. Oncol Rep. 2015;34:843–849. doi: 10.3892/or.2015.4007. [DOI] [PubMed] [Google Scholar]
- Karia PS, Han J, Schmults CD. Cutaneous squamous cell carcinoma: estimated incidence of disease, nodal metastasis, and deaths from disease in the United States, 2012. J Am Acad Dermatol. 2013;68:957–966. doi: 10.1016/j.jaad.2012.11.037. [DOI] [PubMed] [Google Scholar]
- Kim HJ, Kim SH, Yu EJ, Seo WY, Kim JH. A positive role of DBC1 in PEA3-mediated progression of estrogen receptor-negative breast cancer. Oncogene. 2014 doi: 10.1038/onc.2014.381. [DOI] [PubMed] [Google Scholar]
- Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- Kim JR, Moon YJ, Kwon KS, Bae JS, Wagle S, Yu TK, et al. Expression of SIRT1 and DBC1 is associated with poor prognosis of soft tissue sarcomas. PLoS One. 2013;8:e74738. doi: 10.1371/journal.pone.0074738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim JH, Yu EJ, Lee KW, Park CK. The overexpression of DBC1 in esophageal squamous cell carcinoma correlates with poor prognosis. Histol Histopathol. 2012;27:49–58. doi: 10.14670/HH-27.49. [DOI] [PubMed] [Google Scholar]
- Koyama S, Wada-Hiraike O, Nakagawa S, Tanikawa M, Hiraike H, Miyamoto Y, et al. Repression of estrogen receptor beta function by putative tumor suppressor DBC1. Biochem Biophys Res Commun. 2010;392:357–362. doi: 10.1016/j.bbrc.2010.01.025. [DOI] [PubMed] [Google Scholar]
- Lee BH, Mathews MB. Transcriptional coactivator cAMP response element binding protein mediates induction of the human proliferating cell nuclear antigen promoter by the adenovirus E1A oncoprotein. Proc Natl Acad Sci U S A. 1997;94:4481–4486. doi: 10.1073/pnas.94.9.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Wang R, Lozada E, Fan W, Orren DK, Luo J. Acetylation of WRN protein regulates its stability by inhibiting ubiquitination. PLoS One. 2010;5:e10341. doi: 10.1371/journal.pone.0010341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Lee BH, Mathews MB. Involvement of RFX1 protein in the regulation of the human proliferating cell nuclear antigen promoter. J Biol Chem. 1999;274:15433–15439. doi: 10.1074/jbc.274.22.15433. [DOI] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Nin V, Chini CC, Escande C, Capellini V, Chini EN. Deleted in breast cancer 1 (DBC1) protein regulates hepatic gluconeogenesis. J Biol Chem. 2014;289:5518–5527. doi: 10.1074/jbc.M113.512913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Riley Pt, Frisch SM. Regulation of anoikis by deleted in breast cancer-1 (DBC1) through NF-kappaB. Apoptosis. 2013;18:949–962. doi: 10.1007/s10495-013-0847-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz JC, Park S, Phillips N, Matsumura S, Tsai WW, Kasper L, et al. Combinatorial regulation of a signal-dependent activator by phosphorylation and acetylation. Proc Natl Acad Sci U S A. 2014;111:17116–17121. doi: 10.1073/pnas.1420389111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin B, Minter-Dykhouse K, Yu J, Zhang J, Liu T, Zhang H, et al. DBC1 functions as a tumor suppressor by regulating p53 stability. Cell Rep. 2015;10:1324–1334. doi: 10.1016/j.celrep.2015.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey MR, He L, Forster N, Ory B, Ellisen LW. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res. 2011;71:4373–4379. doi: 10.1158/0008-5472.CAN-11-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenberg J, Rishi V, Orosz A, Moitra J, Glick A, Vinson C. Inhibition of CREB function in mouse epidermis reduces papilloma formation. Mol Cancer Res. 2009;7:654–664. doi: 10.1158/1541-7786.MCR-08-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurabashi A, Wada-Hiraike O, Hirano M, Fu H, Isono W, Fukuda T, et al. CCAR2 negatively regulates nuclear receptor LXRalpha by competing with SIRT1 deacetylase. J Steroid Biochem Mol Biol. 2015;149:80–88. doi: 10.1016/j.jsbmb.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Shim UJ, Lee IS, Kang HW, Kim J, Kim WT, Kim IY, et al. Decreased DBC1 Expression Is Associated With Poor Prognosis in Patients With Non-Muscle-Invasive Bladder Cancer. Korean J Urol. 2013;54:631–637. doi: 10.4111/kju.2013.54.9.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- Song NY, Surh YJ. Janus-faced role of SIRT1 in tumorigenesis. Ann N Y Acad Sci. 2012;1271:10–19. doi: 10.1111/j.1749-6632.2012.06762.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanikawa M, Wada-Hiraike O, Yoshizawa-Sugata N, Shirane A, Hirano M, Hiraike H, et al. Role of multifunctional transcription factor TFII-I and putative tumour suppressor DBC1 in cell cycle and DNA double strand damage repair. Br J Cancer. 2013;109:3042–3048. doi: 10.1038/bjc.2013.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Won KY, Cho H, Kim GY, Lim SJ, Bae GE, Lim JU, et al. High DBC1 (CCAR2) expression in gallbladder carcinoma is associated with favorable clinicopathological factors. Int J Clin Exp Pathol. 2015;8:11440–11445. [PMC free article] [PubMed] [Google Scholar]
- Yu EJ, Kim SH, Kim HJ, Heo K, Ou CY, Stallcup MR, et al. Positive regulation of beta-catenin-PROX1 signaling axis by DBC1 in colon cancer progression. Oncogene. 2015 doi: 10.1038/onc.2015.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu XM, Liu Y, Jin T, Liu J, Wang J, Ma C, et al. The Expression of SIRT1 and DBC1 in Laryngeal and Hypopharyngeal Carcinomas. PLoS One. 2013;8:e66975. doi: 10.1371/journal.pone.0066975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Luo K, Liu T, Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 2012;26:791–796. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zannini L, Buscemi G, Kim JE, Fontanella E, Delia D. DBC1 phosphorylation by ATM/ATR inhibits SIRT1 deacetylase in response to DNA damage. J Mol Cell Biol. 2012;4:294–303. doi: 10.1093/jmcb/mjs035. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gu Y, Sha S, Kong X, Zhu H, Xu B, et al. DBC1 is over-expressed and associated with poor prognosis in colorectal cancer. Int J Clin Oncol. 2014;19:106–112. doi: 10.1007/s10147-012-0506-5. [DOI] [PubMed] [Google Scholar]
- Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451:587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Yang L, Peng L, Izumi V, Koomen J, Seto E, et al. hMOF acetylation of DBC1/CCAR2 prevents binding and inhibition of SirT1. Mol Cell Biol. 2013;33:4960–4970. doi: 10.1128/MCB.00874-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.