Abstract
Objectives
Aging is an important osteoarthritis (OA) risk factor and compromised stress defense responses may mediate this risk. The Sestrins (Sesn) promote cell survival under stress conditions and regulate AMPK and mTOR signaling. This study examined Sesn expression in normal and OA cartilage and functions of Sesn in chondrocytes.
Methods
Sesn expression in human and mouse normal and OA cartilage was analyzed by quantitative PCR and immunohistochemistry. Sesn function was investigated by using siRNA mediated Sesn knockdown and overexpression with analysis of cell survival, gene expression, autophagy, and AMPK and mTOR activation.
Results
Sesn mRNA levels were significantly reduced in human OA cartilage and immunohistochemistry of human and mouse OA cartilage also showed a corresponding reduction in protein levels. In cultured human chondrocytes Sesn1, 2 and 3 were expressed and increased by tunicamycin, an endoplasmic reticulum stress response inducer and 2-deoxyglucose (2DG), a metabolic stress inducer. Sesn1 and 2 were increased by tBHP, an oxidative stress inducer.
Sesn knockdown by siRNA reduced chondrocyte viability under basal culture conditions and in the presence of 2DG. Sesn overexpression enhanced LC3-II formation and autophagic flux, and this was related to changes in mTOR but not AMPK activation.
Conclusion
These findings are the first to show that Sesn expression is suppressed in OA affected cartilage. Sesn support chondrocyte survival under stress conditions and promote autophagy activation through modulating mTOR activity. Suppression of Sesn in OA cartilage contributes to deficiency in an important cellular homeostasis mechanism.
Keywords: Sestrins, osteoarthritis, mTOR, autophagy
Introduction
Osteoarthritis is the most prevalent joint disease 1 with aging, obesity and joint injury as its major risk factors 1. In established disease, all joint tissues are affected 2 while disease initiation appears to occur in cartilage and cartilage damage also appears to be a main driver of OA progression 3.
Pathogenesis mechanisms involved in cartilage degradation include chondrocyte death, the increased expression of extracellular matrix (ECM) degrading enzymes, inflammatory mediators, and reduced synthesis of new ECM components. There is also evidence for abnormal chondrocyte differentiation 4.
A recent concept that has emerged is a deficiency in important cellular defense mechanisms in aging and OA-affected cartilage contributes to the initiation and progression of tissue damage 4, 5. There is reduced expression of antioxidant enzymes, which in the setting of increased reactive oxygen and nitrogen species aggravates oxidative damage 6. There is also a dysfunction in the removal of damaged organelles such as mitochondria and macromolecules due to defects in autophagy 5. Intracellular signaling mechanisms that mediate these defective functions are reduced expression and activation of Forkhead-box class O (FoxO) transcription factors 7, AMP-activated protein kinase (AMPK) 8 and an apparent hyperactivation of mammalian target of rapamycin (mTOR) 9. This is a central axis of cell signaling in OA that is linked to the Phosphatidylinositol-4,5-Bisphosphate 3-Kinase (PI3K) pathway and is involved in the cellular response to diverse types of stress 10.
Sestrins (Sesn) were originally identified as downstream effectors of p53 11, and are involved in regulation of cell viability under different stress conditions. Sesn expression is induced by DNA damage in a pathway dependent on p53, and by oxidative stress, independent of p53. Sesn regulate cell function mainly through negative control of mTOR through AMPKα1 phosphorylation 12, 13. Sesn also can have direct control over autophagy by binding to p62 and regulating degradation of its substrate 14. Sesn2 and p62 physically associate with Unc-51-like protein kinase 1 (ULK1), an autophagy-initiating protein kinase 15. Sesn thus regulate autophagy through mTOR but also through different mTOR independent mechanisms.
This study is the first to analyze which Sesn are expressed in normal cartilage and to determine changes in aging and OA. We also examine the role of Sesn in the regulation of chondrocyte viability, mTOR activation and autophagy.
Methods
Cartilage donors
Normal human knee cartilage tissues obtained from tissue banks from 3 young normal female (age 35–46, mean 39.3±5.8), 6 aging female (age 57–89, mean 71.8±13.1) and 13 young normal male (age 18–49, mean 32.3±10.4),10 aging male (age 50–86, mean 66.1±13.2) donors (approved by Scripps Institutional Review Board) and processed within 24–72 hours post mortem. The cartilage samples from these young and aged donors were macroscopically normal. Full thickness cartilage was harvested for RNA isolation from identical locations on the medial femoral condyles. OA-affected cartilage was harvested from the tissue removed during knee replacement surgery from 6 female (age 42–75, mean 59.3±11.3) and 5 male (age 60–71, mean 66.6±4.34) donors.
Tissue processing and RNA isolation
Cartilage was stored at −20ºC in Allprotect Tissue Reagent (Qiagen, Valencia, CA) immediately after resection from the subchondral bone. For RNA isolation, cartilage was pulverized in a 6770 Freezer/Mill Cryogenic Grinder (SPEX SamplePrep, Metuchen, NJ), and homogenized in Qiazol Lysis Reagent (Qiagen) using 25mg tissue per 700ul Qiazol. RNA was isolated using the miRNeasy Mini kit (Qiagen) with on-column DNAse digestion, followed by removal of proteoglycans using RNAmate (BioChain Institute, Newark, CA).
Quantitative polymerase chain reaction (qPCR)
RNA from cultured chondrocytes using Direct-zol RNA MiniPrep kits and TRI-Reagent (Zymo Research). Quantitative PCR was performed using a LightCycler 480 instrument (Roche Diagnostics). The following pre-designed TaqMan gene expression assays (Life Technologies) were used: Sesn1 (Hs00902787), Sesn2 (Hs00230241), Sesn3 (Hs00914870), Gapdh (Hs02758991_g1).
Immunohistochemistry
Immunohistochemistry was performed to assess protein expression patterns in human and mouse cartilage using the following rabbit antibodies: Sesn1 (Proteintech), Sesn2 (human: Abcam, mouse: Proteintech), Sesn3 (Proteintech). Rabbit IgG (1 μg/ml) was used as a negative control in all experiments. For human cartilage, expression patterns were compared between normal and OA samples. The methods for tissue processing and immunohistochemistry were described earlier 7.
Mouse knee joints
All animal experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee (IACUC) at The Scripps Research Institute. In mice, we analyzed young normal and aged knees as a model of aging-related OA as described 16. We also analyzed knees from animals with surgically induced OA 7. The surgical OA model was induced in 4 months old normal wild-type C57BL/6J mice by transection of the medial meniscotibial ligament and the medial collateral ligament (MMTL+MCL) and animals were euthanized 10 weeks later.
In the spontaneous aging-related OA model, male and female C57BL/6J mice were kept under normal conditions and knee joints were collected at 6 and 18 months of age from at least 12 mice per time point. The surgical OA model was induced in 4 months old C57BL/6J mice by transection of the medial meniscotibial ligament and the medial collateral ligament (MMTL+MCL) as described 17 and animals were euthanized 10 weeks later. Knee joints from both murine models were resected from both hind legs, fixed in 10% zinc-buffered formalin for 2 days, decalcified in TBD-2 for 24 hours. Serial sections (4-μm-thick) were cut, and expression of Sesn proteins was analyzed by immunohistochemistry.
Chondrocyte culture
Chondrocytes were isolated from human knee cartilage as described previously 18. In brief, cartilage slices were removed from the femoral condyles and washed in Dulbecco’s minimal essential medium (DMEM). Tissues were then minced with a scalpel, transferred into a digestion buffer containing DMEM, 5% fetal bovine serum, L-glutamine, antibiotics, and 2 mg/ml clostridial collagenase (Sigma Chemical Co., St. Louis, MO) and incubated on a gyratory shaker at 37°C until the fragments were digested.
The isolated chondrocytes were plated at high density in DMEM with 10% CS and antibiotics and allowed to attach to the culture flasks. The cells were incubated at 37°C in a humidified gas mixture containing 5% of CO2 balanced with air. The chondrocytes were used in the experiments at confluence (2–3 weeks in primary culture).
The immortalized human chondrocyte cell line T-C/28 19 was obtained from Dr. Mary Goldring. Cells were cultured in DMEM containing 10% CS and only cells that had been maintained for fewer than 20 passages were used in all experiments.
Cells were exposed to different stress conditions. We used 2-deoxy-glucose (2DG) as an inducer of metabolic stress, tunicamycin for endoplasmic reticulum (ER) stress and tert-Butyl hydroperoxide (tBHP) to model oxidative stress 18.
Plasmid and small interfering RNA (siRNA)
The siRNAs targeting Sesn1, Sesn2 or Sesn3 (s26032, s44570 and s38097, respectively; Life Technologies) or control siRNAs were transfected into chondrocytes using Lipofectamine RNAiMAX reagent (Invitrogen). All transfections were performed following the manufacturer’s protocol. Experiments were carried out 24–48 hours post-transfection, and cells were harvested 18–24 hours thereafter for PCR and immunoblot analysis.
For overexpression, the Sesn2 (SC320354, OriGene) or mRFP-GFP-LC3 (ptfLC3, #21074, Addgene) or control plasmid were transfected into immortalized human T-C/28 chondrocytes 19 using Lipofectamine 3000 reagent (Invitrogen). All transfections were performed following the manufacturer’s protocol. Experiments were carried out 24–72 hours post-transfection, and cells were harvested for PCR and western blot analysis.
Immunofluorescence staining
Cells were fixed with 4% formaldehyde in PBS for 15 min. After washing three times with PBS, cells were counterstained with DAPI and then mounted in antifade agent on glass slides and visualized with an inverted fluorescence microscope (Olympus).
Cell viability assays
The MTT assay is a colorimetric assay for measuring cell metabolic activity. NAD(P)H-dependent cellular oxidoreductase enzymes typically reflect the number of viable cells present. Following transfection with siRNA and incubation for 72 hours, cell viability was determined using MTT assay as described 18.
The CellTiter-Glo Luminescent Cell Viability Assay (Promega) is used to determine the number of viable cells based on quantitation of the ATP present. Following transfection with siRNA and incubation for 72 hours, cell viability was determined using the CellTiter-Glo Luminescent assay, according to the manufacturer’s protocol. The results shown represent the mean ± 95% confidence interval of triplicate wells.
Analysis of autophagy
Autophagy activation was analyzed by the standard Western blotting for LC3 conversion 20 as described 21.
To detect the formation of autophagosomes and autolysosomes 22, cells were transfected with the mRFP-GFP-LC3 plasmid 23. Immunofluorescence images of mRFP-GFP-LC3 were analyzed by standard protocols 23. Because GFP is more sensitive than RFP to the acidic environment of lysosomes, the tandem RFP-GFP-LC3 protein will label autophagosomes as yellow (GFP/RFP) puncta, and autolysosomes as red (RFP) puncta 23. Thus, quantification of yellow and red puncta allows the determination of autophagosomes and autolysosomes, respectively. The number of LC3-positive red or yellow puncta was counted in at least 20 cells per experimental condition.
Western blotting
At indicated time points, cultured human chondrocytes were lysed in RIPA buffer supplemented with Halt protease inhibitor cocktail and phosphatase inhibitor cocktail (Thermo Scientific) and samples were analyzed by western blotting as previously described 7. The following antibodies were used: Sesn1, Sesn2, Sesn3 (Proteintech), pAMPK, AMPK, pS6, S6, LC3 (Cell Signaling), and Gapdh (Ambion).
Statistical analysis
For all data sets on human chondrocytes, at least three independent experiments with cells from different donors were performed. Each condition in each of the experiments was tested in duplicate (for PCR and western blotting) or triplicate (cell viability). When using the T-C/28 cell line at least three separate experiments, each in duplicate or triplicate were performed.
Summary statistics for quantitative data are reported as means ± 95% confidence interval (CI), and for qualitative data as frequencies and percentages. Parametric t-tests or analyses of variance were used to assess differences between groups relating to quantitative variables, and chi-square tests were utilized with qualitative variables. P-values <0.05 were considered statistically significant. All analyses were done with SPSS statistical software, version 23.0 for Windows.
Results
Sesn expression in normal, aging and OA cartilage
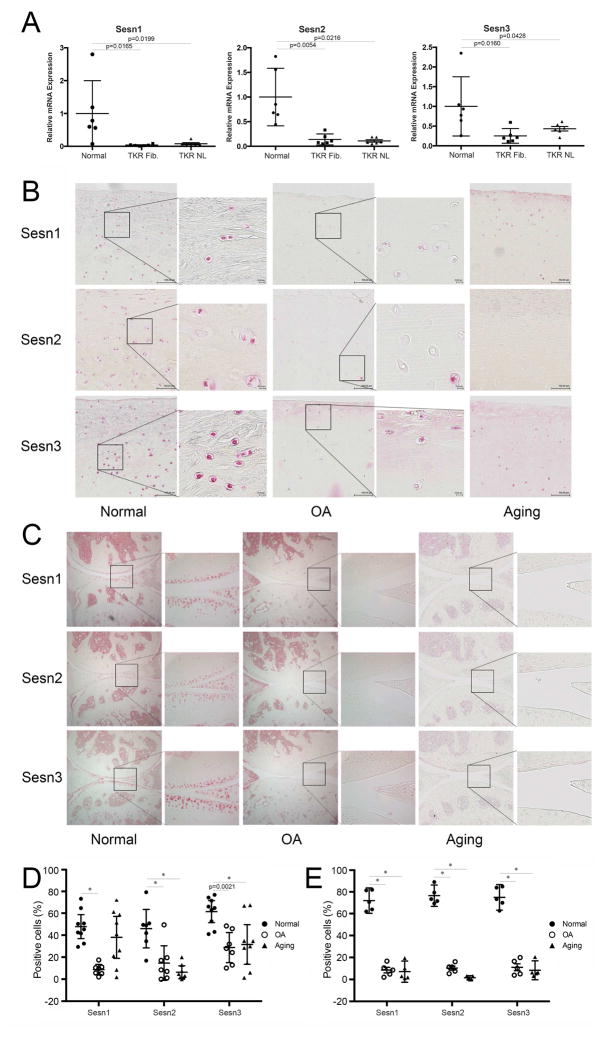
The expression of Sesn mRNAs was analyzed by qPCR and this showed a significant reduction in Sesn1, Sesn2 and Sesn3 in human OA compared with normal cartilage (Figure 1A). Next, changes in Sesn protein expression in human cartilage were assessed by immunohistochemistry (Figure 1B,D). In young normal cartilage, approximately 50–70% of cells were positive for Sesn and the Sesn expressing cells were evenly distributed throughout all zones of cartilage. In old normal donors with no diagnosis or macroscopic evidence of OA, the percentage of Sesn2 and Sesn3 positive cells were significantly decreased in the superficial and medial zone compared with young normal cartilage. Sesn1, Sesn2 and Sesn3 positive cells in OA cartilage were also significantly decreased compared with normal cartilage.
Figure 1. Sesn mRNA and protein expression in articular cartilage.
A) Sesn1, Sesn 2, and Sesn 3 mRNA levels were analyzed by quantitative PCR in cartilage samples from normal (N=6) and OA (N=6) donors. TKR Fib: total knee replacement, fibrillated area. TKR NL: total knee replacement, normal appearing area. Data are expressed as mean ± 95% confidence interval (CI). Data were analyzed by paired-samples t-test. * = P < 0.05 versus normal group.
B) Immunohistochemical analysis of Sesn expression in human cartilage collected from normal (N=9), aging (N=9) and OA (N=7) knee joints. Representative images are shown. Images are 10× and insets on the right are 40× magnification.
C) Immunohistochemical analysis of Sesn protein expression in mouse joints from normal (6 months old, N=5), aging (18 months old, N=5) and DMM induced OA (N=5). Representative images are shown. Images are 10×and insets are 40× magnification.
D) Quantification of Sesn1, Sesn2 and Sesn3 immunopositive cells in human cartilage superficial and mid zones. Data are expressed as % positive cells. Dara were analyzed by chi-square test. * = P < 0.05 versus normal group.
E) Quantification of Sesn1, Sesn2 and Sesn3 immunopositive cells in mouse articular cartilage. Data are expressed as % positive cells. Data were analyzed by chi-square test. * = P < 0.05 versus normal group.
In normal mouse knee joints, the percentages of positive cells were similar for Sesn1, Sesn2 and Sesn3 (Figure 1C,E). In C57Bl/6 mice with surgically induced OA, there was a significant reduction in Sesn1, Sesn2 and Sesn3 positive cells compared to normal knees (Figure 1C,E). In normal C57Bl/6 wild-type mice there was also a significant reduction in the expression of all three Sesn with aging (Figure 1C,E).
Regulation of Sesn expression in cultured chondrocytes
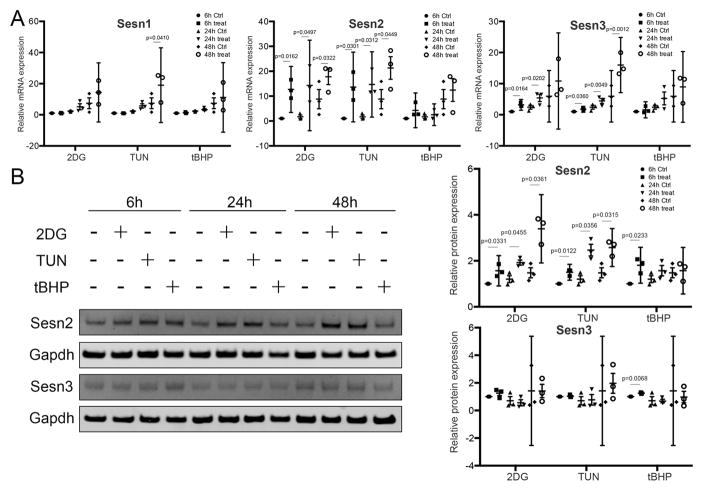
Normal human articular chondrocytes were exposed to different stress conditions to determine regulation of Sesn expression. Under basal conditions, low levels of expression were detected for all Sesn. Tunicamycin and 2-DG significantly increased mRNA levels of Sesn2 and Sesn3 at 6, 24 and 48 hours (Figure 2). Moreover, tunicamycin significantly increased RNA levels of Sesn1 at 48 hours. No changes in Sesn1, Sesn2 and Sesn3 RNA levels were detected in tBHP-treated samples at any time point. Concerning Sesn protein expression, tunicamycin and 2-DG significantly increased protein level of Sesn2 at 6, 24 and 48 hours. Moreover, tBHP increased protein level of Sesn2 and 3 at 6 hours. Western blot analysis showed similar to the mRNA levels, that the protein levels of Sesn changed in response to the stress stimuli (Figure 2).
Figure 2. Changes in Sesn expression in response to stress inducers in cultured human chondrocytes.
A) mRNA levels for Sesn1, Sesn2 and Sesn3 assessed by quantitative PCR in human articular chondrocytes treated with 2-deoxy-D-glucose (2-DG; 20mM), tunicamycin (1ug/ml), or tBHP (250uM) for 6, 24 and 48 hours. Data are expressed as mean ± 95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05. Graphs represent quantitative PCR data from three different experiments using cells from different donors, each performed in duplicate.
B) Protein levels for Sesn2 and Sesn3 assessed by Western blotting of human articular chondrocytes treated with the indicated stress inducers for 6, 24 and 48 hours. Representative western blot images are shown. Graphs represent quantification of three different experiments each performed in duplicate. Data are expressed as mean ± 95% CI. Data were analyzed by paired-samples t-test * = P < 0.05.
Sesn and chondrocyte viability
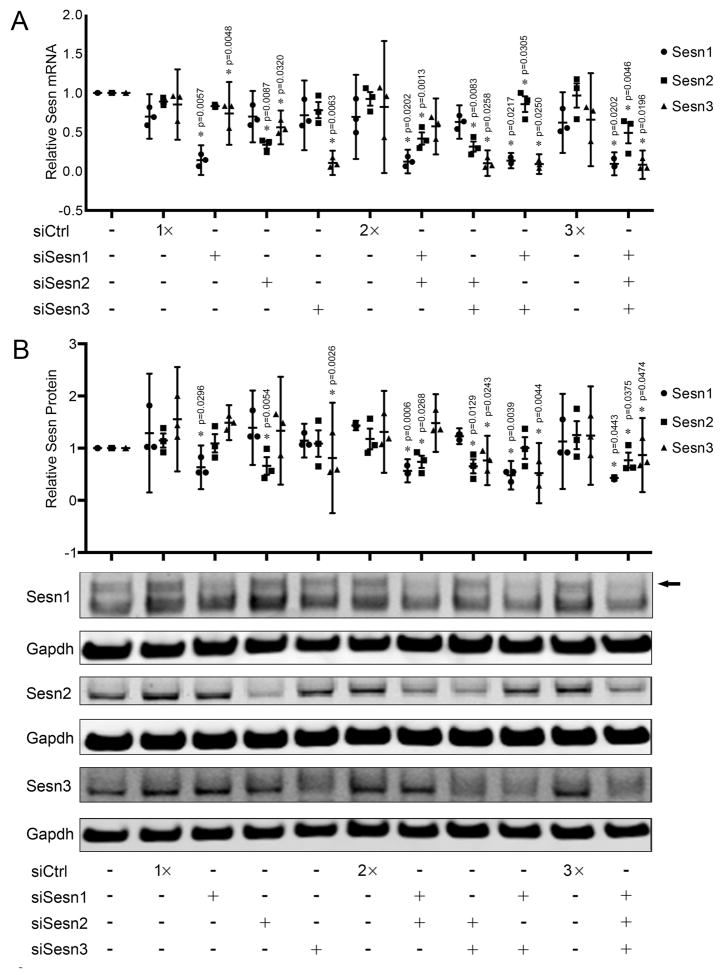
Individual and combinations of Sesn siRNAs were transfected into chondrocytes to determine the role of Sesn in regulating cell viability. Transfection with siRNAs reduced Sesn1 mRNA levels by 80%, Sesn2 mRNA levels by 60% and Sesn3 by 88% (Figure 3A). Protein levels were reduced by 50% for Sesn1, 60% for Sesn2 and by 65% for Sesn3 (Figure 3B). Sesn3 mRNA levels were consistently decreased by transfection with siRNAs targeting Sesn1 by 10% and Sesn2 by 30%. This effect was not detected at the protein level. Sesn1 or Sesn2 mRNA or protein levels were not affected by transfection of siRNAs targeting the other Sesn.
Figure 3. Specificity of Sesn siRNAs.
A) Sesn1, Sesn2 and Sesn3 mRNA levels analyzed by quantitative PCR in human articular chondrocytes transfected with small interfering RNA (siRNA) targeting Sesn (siSesn). Data are expressed as mean ±95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05 versus siCtrl. Graphs represent quantitative PCR data from three different experiments using cells from different donors, each performed in duplicate.
B) Sesn1, Sesn2 and Sesn3 protein expression in human chondrocytes transfected with siSesn. Representative western blot images are shown. Graphs represent quantification of three different experiments. Data are expressed as mean ±95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05 versus siSesnCtrl. Graphs represent quantification of western blots from three different experiments using cells from different donors, each performed in duplicate. Arrow marks Sesn1 specific band.
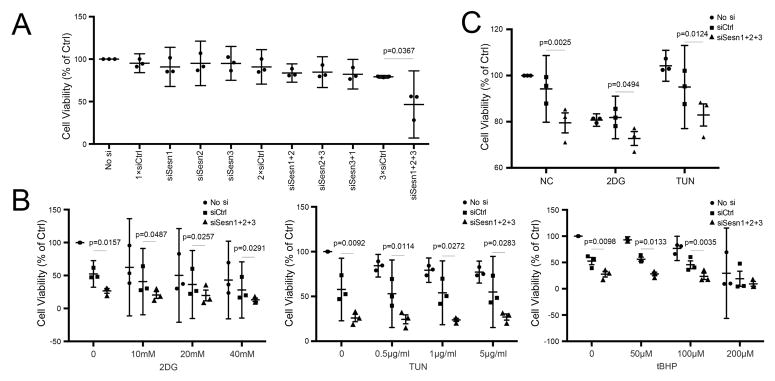
Under basal culture conditions, only the combination of siRNAs for the three Sesns but not the individual Sesn siRNAs significantly reduced cell viability (Figure 4A). In response to treatment with the stress inducers tunicamycin, 2DG and tBHP, there was a dose-dependent and significant reduction in cell viability and this was further reduced by siRNA for Sesn1, Sesn2 and Sesn3 (Figure 4B,C).
Figure 4. Sesn and chondrocyte viability.
A) Cell viability in cultured human chondrocytes transfected with individual and combinations siSesn assessed by MTT assay. Data are expressed as mean ±95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05 versus siCtrl. Graphs represent quantification of MTT assay from three different experiments using cells from different donors, each performed in triplicate.
B) Cell viability in cultured human chondrocytes transfected with siRNA under treatment with different concentrations of 2-DG, TUN and tBHP. Viability was assessed using MTT assay. Data are expressed as mean ±95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05 versus siCtrl. Graphs represent quantification by MTT assay from three different experiments using cells from different donors, each performed in triplicate.
C) Cell viability in cultured human chondrocytes transfected with combinations of siSesn under treatment with 2-DG (20mM), TUN (1ug/ml) and tBHP (250 uM) assessed by Celltiter-Glo assay. Data are expressed as mean ±95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05 versus siCtrl. Graphs represent quantification of MTT assay from three different experiments using cells from different donors, each performed in triplicate.
Sesns control of mTOR activity and autophagy
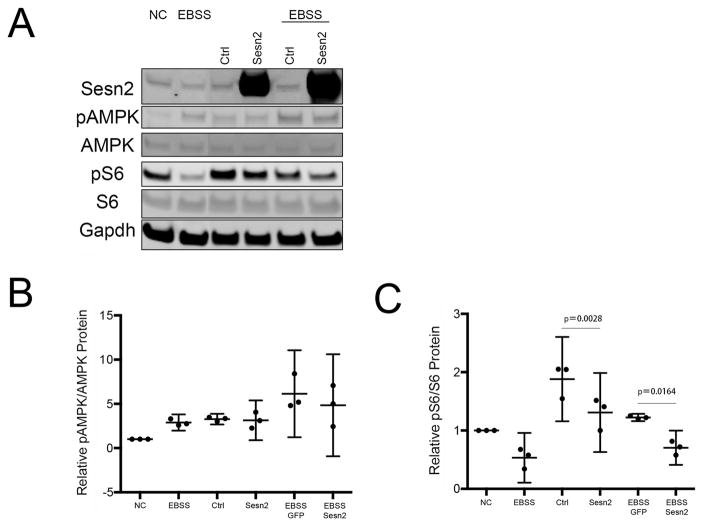
We analyzed mTOR signaling upon knocking down all Sesn or overexpressing Sesn2 in the human T-C/28 chondrocyte cell line 19 under basal conditions and upon treatment with Earle’s balanced salt solution (EBSS) to induce starvation. We focused in Sesn2 as this Sesn showed the largest differences in expression following exposure to stress conditions (Figure 2) and Sesn 2 inhibits mTOR and might play a more important role in stress responses than other Sesn family members 13, 15, 24, 25. As shown in Figure 5A,C, S6 phosphorylation levels were significantly increased in cells treated with siSesn2. Conversely, Sesn2 overexpression with or without EBSS treatment also decreased phosphorylation of S6 (Figure 5A and 5C). However, knockdown of all Sesn or overexpression of Sesn2 did not affect AMPK phosphorylation.
Figure 5. Sesn effects on mTOR and AMPK signaling pathway in human chondrocytes.
A) T-C/28 chondrocytes were transfected with indicated constructs for indicated time with or without EBSS and cell lysates were analyzed by western blotting. Representative western blot images are shown.
B) Graphs represent quantification of relative pAMPK/AMPK protein levels by western blots from three different experiments. Data are expressed as mean ±95% CI. Data were analyzed by paired-samples t-test.* = P < 0.05.
C) Graphs represent quantification of relative pS6/S6 protein levels by western blots from three different experiments. Data are expressed as mean ±95% CI. Data were analyzed paired-samples t-test. * = P < 0.05.
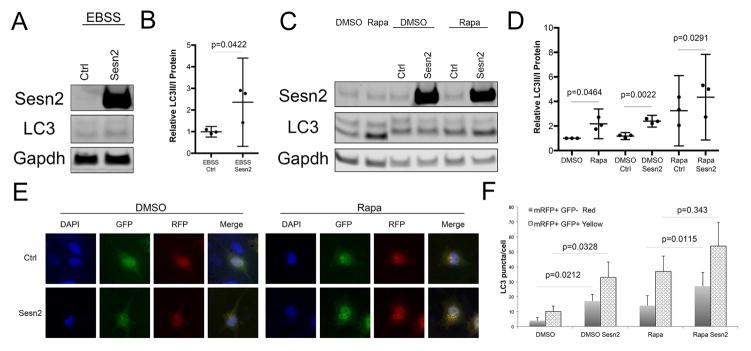
As mTOR is a negative regulator of autophagy 26, we assessed whether Sesn affect autophagy through controlling mTOR activity. Firstly, we monitored changes in the levels of the autophagy marker LC3 in T-C/28 cells after knockdown or overexpression of Sesn, followed by treatment with the autophagy inducers EBSS or rapamycin (Figure 6A–D). Treatment of cells with control siRNAs significantly increased LC3 as compared to the DMSO only control. This is likely due to effects of the transfection reagent lipofectamine 27. Notably, overexpression of Sesn2 increased the relative amount of LC3-II under normal conditions or after EBSS or rapamycin treatment. However, knocking down Sesn2 or all Sesn did not affect LC3-II formation (Figure 6A–D).
Figure 6. Sesn effects on autophagy in human chondrocytes.
A,B) T-C/28 cells transfected with the indicated constructs were treated with EBSS for 1 hour. Western blotting was performed with indicated antibodies. Representative western blot images are shown. Graphs represent quantification of western blots from three different experiments. Data are expressed as mean ± 95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05.
C,D) T-C/28 cells transfected with the indicated constructs were treated with rapamycin (10uM, 6 hours). Western blotting was performed with indicated antibodies. Representative western blot images are shown. Graphs represent quantification of western blots from three different experiments. Data are expressed as mean ± 95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05.
E,F) Autophagosome and autolysosome formation in T-C/28 cells following Sesn2 overexpression. The mRFP-GFP-LC3 plasmid was transfected into T-C/28 cells and 24 hours later cells were transfected with, Sesn2 plasmid. After 24 hours, the cells were treated with rapamycin (10uM, 6 hours). Representative images are shown. Graphs represent quantification of puncta in at least 20 cells from two different experiments. Data are expressed as mean ±95% CI. Data were analyzed by paired-samples t-test. * = P < 0.05.
To analyze autophagic flux, we also monitored the formation of autophagosomes and autolysosomes following transfection of mRFP-GFP-LC3 23. As shown in Figure 6E,F, overexpression of Sesn2 under basal conditions or under rapamycin treatment led to a 2-fold increase of LC3 puncta (both yellow and red), indicative of increased autophagic flux.
Discussion
This study is the first to report that normal articular cartilage expresses Sesn and that OA and aging are associated with reduced expression of Sesn. In cultured chondrocytes, Sesn are induced by various types of extracellular stress and regulate cell survival, mTOR signaling and autophagy activation.
Sesn were originally identified as p53 induced genes 11. In certain cancer cells, Sesn2 can also be induced in a p53-independent and JNK-dependent manner 11, 28. The major physiological functions of Sesn are in the regulation of metabolic homeostasis via control of mTOR activation 29 and their potential function of antioxidants 11. Sesn1 and 2 reduce oxidative stress by rescuing the peroxidase activity of overoxidized peroxiredoxins 30 and by activating the Nrf2 (nuclear factor erythroid 2-related factor 2)-Keap1 (Kelch-like ECH-associated protein 1) pathway 14, which induces antioxidant genes. Our data show that Sesn expression in chondrocytes is induced by various stressors, which also can induce cell death. Knockdown of Sesn under treatment with these stressors significantly reduces cell viability in cultured chondrocytes, indicating that they convey essential cellular protective functions.
Sesn2 inhibits activation of mTOR 25, a prometabolic serine/threonine kinase that controls protein synthesis, cell growth, autophagy and cell death and conveys stress signals for the reprogramming of cellular metabolism and the restoration of organismal homeostasis 29. Genetic deficiency of Sesn2 in mice is not associated with any gross developmental abnormalities 31 but exacerbates obesity-induced mTOR activation, glucose intolerance, insulin resistance and results in diverse age- and obesity-associated metabolic pathologies such as accumulation of lipid droplets and protein aggregates, mitochondrial dysfunction, and muscle degeneration 24, 29. Sesn2-deficient mice fail to inactivate mammalian target of rapamycin complex 1 (mTORC1) in the liver during fasting 14, and spontaneously elevated mTORC1 signaling is observed in mice devoid of both Sesn2 and Sesn3 29. Mice deficient in all Sesn exhibit reduced postnatal survival associated with defective mTORC1 inactivation in multiple organs during fasting 32. Persistent mTOR activation is associated with diverse diseases and metabolic disorders 33. mTOR hyperactivation has been observed in OA cartilage 9 and mTOR inhibition with rapamycin 34 or cartilage specific deletion 35 reduced the severity of experimental OA in mice. Our present data show that Sesn overexpression or knockdown in chondrocytes modulates mTOR activation as indicated by changes in the phosphorylation of the mTOR target S6.
A potential mechanism for mTOR hyperactivation is misregulation of AMP-activated protein kinase (AMPK). AMPK is a central nutrient and energy-sensor that is activated under stress conditions such as hypoxia, glucose starvation 36. AMPK regulates the activity of proteins associated with mammalian target of rapamycin complex 1 (mTORC1), including tuberous sclerosis protein complex 2 (TSC2) and raptor, thereby regulating the activity of mTORC1. Sesn2 associates with AMPK and promotes its activation by an upstream kinase LKB1 13, 25. This activation of AMPK can result in mTORC1 inhibition and subsequent autophagy activation 37. Sesn2-deficient cells are unable to promote autophagy under certain stress conditions 37. AMPK protein and phosphorylation levels are reduced in OA 8. The abnormal Sesn expression and AMPK activation are two mechanisms that converge to lead to mTOR hyperactivation in OA. However, in the present study the modulation in mTOR activation following Sesn knockdown or overexpression was not accompanied by changes in AMPK phosphorylation, suggesting that, under the experimental conditions used, the effect of Sesn on mTOR is not dependent on AMPK in chondrocytes. Our findings are in concordance with recent reports showing that Sesn also inhibit mTORC1 activity in the absence of AMPK 32. This AMPK-independent mechanism involves Sesn binding to the heterodimeric RagA/B-RagC/D GTPases resulting in suppression of mTOR lysosomal localization 38.
The present results also show that Sesn knockdown are not only associated with reduced mTOR activation as measured by phosphorylation of S6 but also of with changes in autophagy. We observed differences in the levels of LC3II as well as differences in the levels of autophagosomes and autolysosomes following modulation of Sesn levels by overexpression of knockdown in chondrocytes. Defects in autophagy have previously been reported in OA-affected and aging cartilage 16, 39.
Although a role for Sesn in cellular aging has been postulated based on findings in model organisms with Sesn gene deletion 40, the present findings are to our knowledge the first to show an aging related reduction in the expression of Sesn. This was observed in articular cartilage during aging of human and mouse knee joints. Sesn expression was also reduced in the surgical knee destabilization model in mice. Since Sesn1 and Sesn3 are target genes of FoxO transcription factors 24, 41, the reduced Sesn expression in aging and OA cartilage could be in part due to a reduction in FoxO expression 7.
The present study has the following limitations. The results on the role of Sesn in regulating cell survival under stress conditions are obtained with primary chondrocytes where Sesn were knocked down with siRNA. In our hands, the transfection efficiency in human chondrocytes is too low to achieve sufficient overexpression of proteins using plasmids and regulation of cell survival in chondrocyte cell lines is very different from primary chondrocytes in which we performed these key experiments.
In conclusion, Sesn mRNA and protein levels are reduced in OA-affected and aging human and mouse cartilage. Sesn support chondrocyte survival under stress conditions, promote autophagy activation and modulate mTOR activity. Suppression of Sesn in OA cartilage contributes to deficiency in an important cellular homeostasis mechanism.
Acknowledgments
Role of Funding Source
This study was supported by National Institutes of Health grants AG049617, AG007996 and the Sam and Rose Stein Endowment Fund.
Footnotes
Author Contributions
All authors were involved in drafting the article or revising it critically for important intellectual content, and all authors approved the final version to be published. Dr. Lotz had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study conception and design: Shen, Alvarez-Garcia, Lotz
Acquisition of data: Shen, Alvarez-Garcia, Olmer
Analysis and interpretation of data: Shen, Alvarez-Garcia, Olmer, Li, Lotz
Conflict of Interest
The authors have no conflicts of interest.
Ethics approval
This study was conducted with the approval of the Human Subjects Committee and the Institutional Animal Care and Use Committee at The Scripps Research Institute.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lawrence RC, Felson DT, Helmick CG, Arnold LM, Choi H, Deyo RA, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part II. Arthritis Rheum. 2008;58:26–35. doi: 10.1002/art.23176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loeser RF, Goldring SR, Scanzello CR, Goldring MB. Osteoarthritis: a disease of the joint as an organ. Arthritis Rheum. 2012;64:1697–707. doi: 10.1002/art.34453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldring MB, Goldring SR. Articular cartilage and subchondral bone in the pathogenesis of osteoarthritis. Ann N Y Acad Sci. 2010;1192:230–7. doi: 10.1111/j.1749-6632.2009.05240.x. [DOI] [PubMed] [Google Scholar]
- 4.Lotz M, Loeser RF. Effects of aging on articular cartilage homeostasis. Bone. 2012;51:241–8. doi: 10.1016/j.bone.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lotz MK, Carames B. Autophagy and cartilage homeostasis mechanisms in joint health, aging and OA. Nature reviews Rheumatology. 2011;7:579–87. doi: 10.1038/nrrheum.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Henrotin Y, Kurz B, Aigner T. Oxygen and reactive oxygen species in cartilage degradation: friends or foes? Osteoarthritis Cartilage. 2005;13:643–54. doi: 10.1016/j.joca.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 7.Akasaki Y, Hasegawa A, Saito M, Asahara H, Iwamoto Y, Lotz MK. Dysregulated FOXO transcription factors in articular cartilage in aging and osteoarthritis. Osteoarthritis Cartilage. 2014;22:162–70. doi: 10.1016/j.joca.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Terkeltaub R, Yang B, Lotz M, Liu-Bryan R. Chondrocyte AMP-activated protein kinase activity suppresses matrix degradation responses to proinflammatory cytokines interleukin-1beta and tumor necrosis factor alpha. Arthritis Rheum. 2011;63:1928–37. doi: 10.1002/art.30333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pal B, Endisha H, Zhang Y, Kapoor M. mTOR: A Potential Therapeutic Target in Osteoarthritis? Drugs in R&D. 2015 doi: 10.1007/s40268-015-0082-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beier F, Loeser RF. Biology and pathology of Rho GTPase, PI-3 kinase-Akt, and MAP kinase signaling pathways in chondrocytes. J Cell Biochem. 2010;110:573–80. doi: 10.1002/jcb.22604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budanov AV, Shoshani T, Faerman A, Zelin E, Kamer I, Kalinski H, et al. Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene. 2002;21:6017–31. doi: 10.1038/sj.onc.1205877. [DOI] [PubMed] [Google Scholar]
- 12.Alexander A, Walker CL. The role of LKB1 and AMPK in cellular responses to stress and damage. FEBS Lett. 2011;585:952–7. doi: 10.1016/j.febslet.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 13.Sanli T, Linher-Melville K, Tsakiridis T, Singh G. Sestrin2 modulates AMPK subunit expression and its response to ionizing radiation in breast cancer cells. PLoS One. 2012;7:e32035. doi: 10.1371/journal.pone.0032035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bae SH, Sung SH, Oh SY, Lim JM, Lee SK, Park YN, et al. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013;17:73–84. doi: 10.1016/j.cmet.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 15.Ro SH, Semple IA, Park H, Park H, Park HW, Kim M, et al. Sestrin2 promotes Unc-51-like kinase 1 mediated phosphorylation of p62/sequestosome-1. FEBS J. 2014;281:3816–27. doi: 10.1111/febs.12905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carames B, Olmer M, Kiosses WB, Lotz M. The relationship of autophagy defects and cartilage damage during joint aging in a mouse model. Arthritis Rheumatol. 2015 doi: 10.1002/art.39073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otsuki S, Hanson SR, Miyaki S, Grogan SP, Kinoshita M, Asahara H, et al. Extracellular sulfatases support cartilage homeostasis by regulating BMP and FGF signaling pathways. Proc Natl Acad Sci U S A. 2010;107:10202–7. doi: 10.1073/pnas.0913897107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Akasaki Y, Alvarez-Garcia O, Saito M, Carames B, Iwamoto Y, Lotz MK. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014;66:3349–58. doi: 10.1002/art.38868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldring MB, Birkhead JR, Suen LF, Yamin R, Mizuno S, Glowacki J, et al. Interleukin-1 beta-modulated gene expression in immortalized human chondrocytes. J Clin Invest. 1994;94:2307–16. doi: 10.1172/JCI117595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carames B, Taniguchi N, Seino D, Blanco FJ, D’Lima D, Lotz M. Mechanical injury suppresses autophagy regulators and pharmacologic activation of autophagy results in chondroprotection. Arthritis Rheum. 2012;64:1182–92. doi: 10.1002/art.33444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–26. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–60. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 24.Lee JH, Budanov AV, Park EJ, Birse R, Kim TE, Perkins GA, et al. Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science. 2010;327:1223–8. doi: 10.1126/science.1182228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Budanov AV, Karin M. p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell. 2008;134:451–60. doi: 10.1016/j.cell.2008.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim YC, Guan KL. mTOR: a pharmacologic target for autophagy regulation. J Clin Invest. 2015;125:25–32. doi: 10.1172/JCI73939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mo RH, Zaro JL, Ou JH, Shen WC. Effects of Lipofectamine 2000/siRNA complexes on autophagy in hepatoma cells. Mol Biotechnol. 2012;51:1–8. doi: 10.1007/s12033-011-9422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XY, Wu XQ, Deng R, Sun T, Feng GK, Zhu XF. Upregulation of sestrin 2 expression via JNK pathway activation contributes to autophagy induction in cancer cells. Cell Signal. 2013;25:150–8. doi: 10.1016/j.cellsig.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Lee JH, Budanov AV, Talukdar S, Park EJ, Park HL, Park HW, et al. Maintenance of metabolic homeostasis by Sestrin2 and Sestrin3. Cell Metab. 2012;16:311–21. doi: 10.1016/j.cmet.2012.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Budanov AV, Sablina AA, Feinstein E, Koonin EV, Chumakov PM. Regeneration of peroxiredoxins by p53-regulated sestrins, homologs of bacterial AhpD. Science. 2004;304:596–600. doi: 10.1126/science.1095569. [DOI] [PubMed] [Google Scholar]
- 31.Wempe F, De-Zolt S, Koli K, Bangsow T, Parajuli N, Dumitrascu R, et al. Inactivation of sestrin 2 induces TGF-beta signaling and partially rescues pulmonary emphysema in a mouse model of COPD. Dis Model Mech. 2010;3:246–53. doi: 10.1242/dmm.004234. [DOI] [PubMed] [Google Scholar]
- 32.Peng M, Yin N, Li MO. Sestrins function as guanine nucleotide dissociation inhibitors for Rag GTPases to control mTORC1 signaling. Cell. 2014;159:122–33. doi: 10.1016/j.cell.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–84. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 34.Carames B, Hasegawa A, Taniguchi N, Miyaki S, Blanco FJ, Lotz M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann Rheum Dis. 2012;71:575–81. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, Vasheghani F, Li YH, Blati M, Simeone K, Fahmi H, et al. Cartilage-specific deletion of mTOR upregulates autophagy and protects mice from osteoarthritis. Ann Rheum Dis. 2015;74:1432–40. doi: 10.1136/annrheumdis-2013-204599. [DOI] [PubMed] [Google Scholar]
- 36.Gwinn DM, Shackelford DB, Egan DF, Mihaylova MM, Mery A, Vasquez DS, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30:214–26. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maiuri MC, Kroemer G. Autophagy in stress and disease. Cell Death Differ. 2015;22:365–6. doi: 10.1038/cdd.2014.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parmigiani A, Nourbakhsh A, Ding B, Wang W, Kim YC, Akopiants K, et al. Sestrins inhibit mTORC1 kinase activation through the GATOR complex. Cell Rep. 2014;9:1281–91. doi: 10.1016/j.celrep.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carames B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JH, Budanov AV, Karin M. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab. 2013;18:792–801. doi: 10.1016/j.cmet.2013.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen CC, Jeon SM, Bhaskar PT, Nogueira V, Sundararajan D, Tonic I, et al. FoxOs inhibit mTORC1 and activate Akt by inducing the expression of Sestrin3 and Rictor. Dev Cell. 2010;18:592–604. doi: 10.1016/j.devcel.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]