Abstract
Francisella tularensis is an intracellular bacterium and as such is expected to encounter a continuous attack by reactive oxygen species (ROS) in its intracellular habitat and efficiently coping with oxidative stress is therefore essential for its survival. The oxidative stress response system of F. tularensis is complex and includes multiple antioxidant enzymes and pathways, including the transcriptional regulator OxyR and the H2O2-decomposing enzyme catalase, encoded by katG. The latter is regulated by OxyR. A deletion of either of these genes, however, does not severely compromise the virulence of F. tularensis and we hypothesized that if the bacterium would be deficient of both catalase and OxyR, then the oxidative defense and virulence of F. tularensis would become severely hampered. To test this hypothesis, we generated a double deletion mutant, ΔoxyR/ΔkatG, of F. tularensis LVS and compared its phenotype to the parental LVS strain and the corresponding single deletion mutants. In accordance with the hypothesis, ΔoxyR/ΔkatG was distinctly more susceptible than ΔoxyR and ΔkatG to H2O2, ONOO−, and , moreover, it hardly grew in mouse-derived BMDM or in mice, whereas ΔkatG and ΔoxyR grew as well as F. tularensis LVS in BMDM and exhibited only slight attenuation in mice. Altogether, the results demonstrate the importance of catalase and OxyR for a robust oxidative stress defense system and that they act cooperatively. The lack of both functions render F. tularensis severely crippled to handle oxidative stress and also much attenuated for intracellular growth and virulence.
Keywords: Francisella tularensis, OxyR, KatG, oxidative stress, virulence
Introduction
Francisella tularensis, a Tier 1 select agent and the causative agent of tularemia, is a zoonotic, facultative intracellular bacterium with two clinically relevent subspecies, tularensis and holarctica, the former of which causes an aggressive disease with high mortality if left untreated (Oyston et al., 2004). Although there is no licensed vaccine against this potential bioterrorism agent, the subspecies holarctica live vaccine strain, LVS, is used to vaccinate laboratory workers, and is widely used in Francisella research as it is attenuated in humans, but retains its virulence in mice (Sjöstedt, 2006; Conlan, 2011).
Francisella tularensis is capable of infecting numerous cell types, including professional phagocytes, like macrophages. Upon phagocytosis, it transiently resides within the phagosome before escaping into the cytosol to replicate (Bröms et al., 2010; Chong and Celli, 2010). Phagocytes constitute a hostile environment utilizing a wide array of anti-bacterial mechanisms, such as phagosome acidification, disruption of pathogen membrane integrity, removal or sequestration of nutrients, and the production of reactive oxygen species (ROS) (Flannagan et al., 2009) and since F. tularensis is an intracellular bacterium, it will encounter a continuous exposure to ROS. Vital macromolecules, such as proteins and DNA, will react with ROS, thereby disrupting their functions (Fridovich, 1998; Schaible and Kaufmann, 2004; Flannagan et al., 2009). There are several ROS with potent antibacterial effects, such as superoxide and H2O2. The former is produced at high levels by the phagocyte oxidase (phox) and it rapidly combines with nitric oxide (NO), which is produced at high levels by inducible nitric oxide synthase (iNOS), to form peroxynitrite, a highly reactive compound. H2O2 is toxic per se, but the damage it exerts can be exacerbated in combination with intracellular ferrous iron, resulting in the formation of hydroxyl radicals (HO•) and hydroxide anions (OH−) through the Fenton reaction.
Reactive oxygen species (ROS) are not only formed during host attack, but low levels are also formed as by-products of normal aerobic metabolism. Thus, pathogens, in particular intracellular pathogens, have a pressing need for defense mechanisms to combat the ever present levels of ROS, but even more so to combat the assault of ROS experienced within a host (Betteridge, 2000). The critical roles of ROS and NO for the host defense against tularemia are illustrated by the extreme susceptibility of phox-deficient and iNOS-deficient mice to an F. tularensis infection (Lindgren et al., 2004). Moreover, ex vivo, it has been demonstrated that the requirements for host protection vary depending on the cell type investigated, since killing of F. tularensis by mouse peritoneal cells is NO-dependent, but NO-independent by mouse pulmonary cells (Anthony et al., 1992; Polsinelli et al., 1994; Lindgren et al., 2005).
The oxidative stress defense system of Escherichia coli has been extensively studied and includes numerous detoxifying enzymes, such as catalase, superoxide dismutases (SODs), alkyl hydroperoxide reductase (Ahp), and the H2O2-activated transcriptional regulator OxyR. The latter combats the effect of H2O2 by dual mechanisms, since it regulates the expression of both catalase and the ferric uptake regulator (Fur) (Farr and Kogoma, 1991; Zheng et al., 1998, 1999; Pomposiello and Demple, 2001). Catalase renders H2O2 harmless by degrading it to oxygen and water, whilst Fur down-regulates the expression of genes involved in iron uptake, thus limiting the amount of iron with which H2O2 can combine in the Fenton reaction (Andrews et al., 2003; Troxell and Hassan, 2013). Catalase, SODs, AhpC and other detoxifying enzymes are employed as oxidative stress defense mechanisms also by F. tularensis (Bakshi et al., 2006; Lindgren et al., 2007; Melillo et al., 2009; Binesse et al., 2015). The F. tularensis catalase, encoded by katG, mediates H2O2 tolerance and is known to be important for the virulence of F. tularensis LVS (Lindgren et al., 2007). SodB, FeSOD, and SodC, CuZnSOD, are both known to be important for the dismutation of in F. tularensis, and SodB further acts in the defense against oxidative stress by harnessing iron (Bakshi et al., 2006; Melillo et al., 2009). The F. tularensis AhpC enzyme is important for the detoxification of and peroxynitrite (ONOO−), but not of H2O2, in the highly virulent SCHU S4 strain (Binesse et al., 2015), but the importance in the LVS strain is yet unknown. F. tularensis also encodes an oxyR homolog, the role of which has been studied recently (Ma et al., 2016). It was found that the absence of OxyR rendered LVS defective for oxidative stress defense, growth in macrophages and epithelial cells, and virulence in mice. Moreover, it was demonstrated that OxyR regulates the expression of the ahpC, katG, and sodB genes, with the most pronounced regulatory effect exerted on ahpC.
A more thorough understanding of the F. tularensis antioxidant system will undoubtedly reveal virulence mechanisms of this bacterium, since ROS constitute such an essential threat to the pathogen. As aforementioned, antioxidant enzymes, such as catalase, AhpC, SodC, and SodB, all contribute to the virulence of F. tularensis in mice, although each appears to render the bacterium only moderately attenuated and this indicates that the antioxidant system of F. tularensis is complex and may in part possess overlapping functions (Lindgren et al., 2007; Ma et al., 2016). Indeed, a double deletion mutant of katG and ahpC has not been possible to generate in F. tularensis (Binesse et al., 2015) and this demonstrates that the cooperative functions of these enzymes are crucial, although either one is not essential. The aim of the present study was to better understand this interconnecting web of antioxidants in F. tularensis. To this end, a double deletion mutant, ΔoxyR/ΔkatG, was generated since this mutant, besides lack of catalase activity, should have a repressed expression of OxyR-regulated antioxidant genes, one of which is AhpC (Ma et al., 2016). We hypothesized that the lack of both KatG and OxyR would lead to a severely impaired phenotype of F. tularensis LVS. We therefore characterized the phenotypes of single deletion mutants, ΔoxyR and ΔkatG, and a double deletion mutant, ΔoxyR/ΔkatG, in comparison to the parental LVS strain.
Materials and methods
Bacterial strains
The F. tularensis LVS strain was obtained from the Francisella strain collection (FSC) at FOI, Swedish Defense Research Agency. The katG deletion mutant (ΔkatG) has been described previously (Lindgren et al., 2007).
The ΔoxyR and ΔoxyR/ΔkatG mutants of the LVS strain were generated by allelic replacement as described previously (Golovliov et al., 2003). Briefly, sequences up- and down-stream of oxyR were amplified by PCR. The fragments contained complementary sequences, which were joined together by a second PCR. The resulting fragment was cloned into the pDM4 suicide-vector, which was transformed into Escherichia coli S17-λpir and thereafter transferred to LVS by conjugation. Clones with a successful recombination event were selected on plates supplemented with Cm and polymyxin B. Correct integration was confirmed by PCR. Positive clones were subjected to sucrose selection to select for a second recombination event and clones were screened by PCR to identify successful deletion mutants. The double deletion mutant ΔoxyR/ΔkatG was generated using the same procedure, apart from using the pDMK3 plasmid carrying kanamycin resistance. The deletions were verified by sequencing 1500 bp on each side of the deleted region.
Aerobic and microaerobic growth
Bacteria were cultivated overnight on plates based on modified GC-agar (MC plates) and then inoculated to an OD600 of 0.1 in Chamberlain's chemically defined medium (CDM). All cultures were split into triplicates and were incubated at 37°C and 200 rpm in an aerobic (normal air) or a microaerobic (10% O2 and 10% CO2) milieu up to 48 h with monitoring of the OD600.
H2O2 susceptibility assay
Bacteria were cultivated overnight on MC plates, inoculated to an OD600 of 0.1 in CDM and H2O2 was added to the final concentration of 0.02, 0.1, or 0.5 mM, respectively. Controls were grown without the addition of H2O2. All cultures were split into triplicates and were incubated at 37°C and 200 rpm up to 24 h with monitoring of the OD600.
Catalase activity assay
Catalase degrades H2O2 to O2 and H2O. H2O2 absorbs light at 240 nm and degradation of H2O2 can therefore be measured as a reduction of A240 nm over time.
Strains were cultivated overnight after being diluted to an OD600 of 0.1 in CDM. For each strain, one set of tubes were left untreated and another set of tubes were supplemented with H2O2 to a final concentration of 0.02, 0.1, or 0.2 mM. All cultures were split into triplicates and incubated at 37°C, 200 rpm for 2, 4, and 24 h before sampling for evaluation of catalase activity. Depending on the density and growth phase of the culture, a volume of 10–50 μl were withdrawn and diluted in PBS to reach a final volume of 120 μl in UV-clear 96-well plates (Greiner Bio-one, Frickenhausen, Germany). Then, 80 μl 100 mM H2O2 in PBS was added to each sample immediately before placing the plate in a Tecan Infinite 200 pro plate reader and measuring the reduction in absorption at 240 nm for 10 min. A molar extinction coefficient of H2O2 at 240 nm of 43.6 M−1cm−1 was used to calculated the concentration of H2O2 using the Beer-Lambert law, A = εcl. One unit of catalase is defined as the amount that decomposes 1 μmol of H2O2 per minute per OD600 at 25°C. The catalase units were normalized against the OD of the culture.
Paraquat susceptibility assay
Susceptibility of F. tularensis strains to was determined by use of the generating compound paraquat dichloride hydrate (Sigma-Aldrich, St. Louis, USA) in a disc diffusion assay. Paraquat generates through reacting with parts of the respiratory chain in bacteria, causing the reduction of O2 to (Hassan and Fridovich, 1979). Bacterial strains were cultivated on MC plates overnight, re-suspended in phosphate-buffered saline (PBS) and approximately 3 × 105 CFU were plated onto MC plates. Sterile filter discs (Oxoid Blank Antimicrobial Susceptibility Discs, Thermo Scientific, MA, USA) were placed in the center the plates once they had dried, and 10 μl of MQ-water, 1.25 mM, 5 mM or 20 mM paraquat solution was added to each disc. The plates were incubated for 4 days at 37°C, 5% CO2 before the size of the growth inhibition zone surrounding each disc was determined.
Peroxynitrite susceptibility assay
3-morpholinosydnonimine hydrochloride (SIN-1) (Molecular Probes, Oregon, USA) spontaneously releases (NO) and under physiological conditions, thereby generating peroxynitrite (ONOO−). Under physiological conditions 1 mM SIN-1 generates 10 μM ONOO−/min (Lindgren et al., 2007).
Strains were cultivated in CDM to logarithmic growth phase and diluted to a density of approximately 2 × 106 bacteria/ml in PBS. The bacterial suspensions were incubated with or without the addition of 0.48 mM SIN-1 with equal amounts of SIN-1 added at the start of the experiment and again after 1.5 h to ensure stable levels of ONOO− (Lindgren et al., 2005). After 3 h samples were collected, diluted and plated on MC plates for determination of viable bacteria.
Analysis of gene expression by real time PCR
Bacteria were cultivated overnight on MC plates, inoculated to an OD600 of 0.1 in CDM and incubated at 37°C, 5% CO2 for 10 h before sampling. RNA extraction, cDNA synthesis and Real Time PCR (RT-PCR) were all performed as described previously (Honn et al., 2012).
Briefly, RNA was extracted using Trizol reagent (Invitrogen, CA, USA) from pelleted bacteria, 3 × 109 CFU/sample. Contaminating DNA was removed using the DNA-free kit (Ambion, Inc, Austin, TX, USA) and RNA was quantified by Nanodrop (Thermo Fisher Scientific, Wilmington, DE, USA). cDNA was synthesized from 1 μg RNA/sample using iScript (BioRad, Hemel, Hampstead, UK), RT-PCR was performed using the Power SYBR green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) and the ABI Prism 7900Ht Sequence Detection System (Applied Biosystems) as described (Honn et al., 2012). Trizol, DNA-free, iScript and Power SYBR green were all used in accordance with the instructions provided by the manufacturers. Forward and reverse primers were obtained from Invitrogen and have been published previously for fslA (FTL_1832), fslB (FTL_1833), fslC (FTL_1834), fslD (FTL_1835), fslE (FTL_1836), fupA (FTL_0439), furA (FTL_1831), (Lindgren et al., 2009), tul4, iglC (FTL_0113), (Bröms et al., 2009), mglA (FTL_0260), feoB (FTL_0133), and katG (FTL_1504) (Honn et al., 2012), sequences for, grxA (FTL_0985), grxB (FTL_1792), gpx (FTL_1383), sspA (FTL_1606), ahpC1 (FTL_0542), ahpC2 (FTL_1191), sodB (FTL_0380), sodC (FTL_1791), clpB (FTL_0094), groES (FTL_1715), groEL (FTL_1714), and dnaK (FTL_1191) are available upon request.
The Ct values of the selected genes were normalized to the Ct value of the house keeping gene FTT0901 (lpnA) and relative copy numbers (RCN) were calculated according to the following equation: RCN = 2−ΔCt × 100, where ΔCt is Ct(target)−Ct(FTT0901) (Gavrilin et al., 2006). Thus, the copy number of a given gene is related to the copy number of FTT0901. Normalized Ct values were used for statistical evaluation of the data by One way ANOVA followed by Tukey's honest significant difference (HSD).
Preparation and infection of BMDM
The capacity of LVS and the mutants to proliferate intracellularly were assessed in bone marrow-derived macrophages (BMDMs). BMDMs were generated from C57BL/6 mice essentially as described previously (Bröms et al., 2011).
The day before infection, BMDM cells were seeded at a density of 4 × 105 cells/ml in 24-well tissue-culture plates and incubated at 37°C, 5% CO2 with or without murine recombinant 1000 U/ml of IFN-γ (Peprotech, Rocky Hill, NJ, USA) The next day, the cells were washed and reconstituted with fresh, pre-warmed culture media. Bacteria were grown overnight on MC plates and re-suspended in PBS to a density of approximately 3 × 109 bacteria/ml. Bacteria were diluted in DMEM and added to each well at multiplicity of infection of 30 and bacterial uptake was allowed to occur for 90 min at 37°C, 5% CO2. Remaining extracellular bacteria were removed by rinsing the monolayers three times with DMEM and incubating with gentamicin for 45 min followed by rinsing the monolayers three times. This time-point was defined as 0 h. After 0, 4 and 24 h incubation the macrophages were lysed in 0.1% deoxycholate in PBS. The lysate were serially diluted in PBS and plated on MC plates for determination of viable bacteria.
Mouse experiments
Virulence of the mutant strains was determined by subcutaneous infection of female C57BL/6 mice with 4 × 103 CFU/mouse of LVS, ΔoxyR, ΔkatG, and ΔoxyR/ΔkatG. Mice were monitored for signs of illness and were euthanized by inhalation of isoflurane followed by CO2 asphyxiation after 3 or 6 days, whereupon the number of viable bacteria in spleens and livers were determined by homogenizing the organs in PBS and plating dilutions on MC plates. All animal experiments were approved by the Local Ethical Committee on Laboratory Animals, Umeå, Sweden (no. A 1-09, A 99-11, and A 67-14).
Statistical analysis
One way ANOVA followed by Tukey's HSD test was used to determine statistical significant difference between groups.
Results
Growth under aerobic vs. microaerobic conditions
CDM effectively supports growth of LVS. We therefore compared growth of the bacterial strains, LVS, ΔoxyR, ΔkatG, and ΔoxyR/ΔkatG. The former three strains all replicated to the same extent, whereas ΔoxyR/ΔkatG showed intact growth to late log phase, but impaired growth thereafter. Therefore, it did not reach as high densities as LVS and the other strains at 24 h (P < 0.001; Figure 1A). To explore if a reduced oxygen tension could rescue the growth of ΔoxyR/ΔkatG, the strains were cultivated under microaerobic conditions, i.e., 10% O2 and 10% CO2. Indeed, ΔoxyR/ΔkatG grew as well as the other strains and reached an optical density of > 2.0 within 48 h (Figure 1A). As noted before (Honn et al., 2012), the growth rate of LVS under microaerobic conditions was reduced compared to aerobic conditions (Figure 1A).
Figure 1.
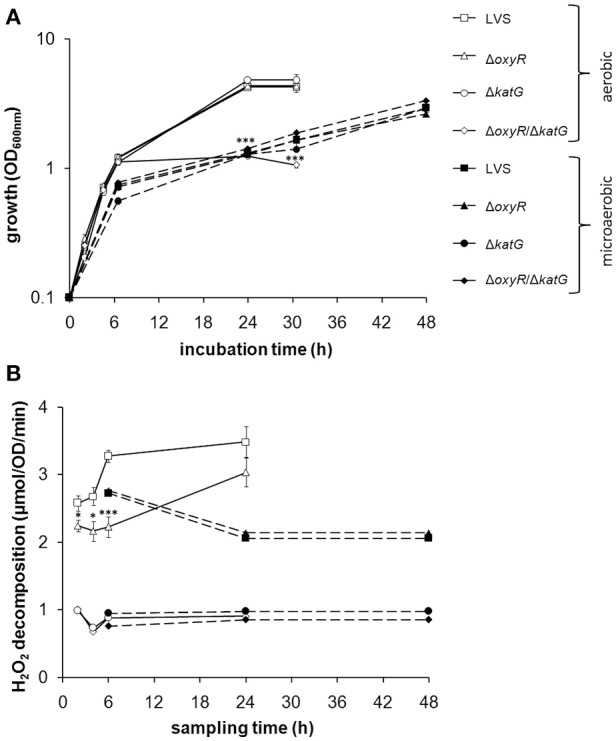
(A) Growth of F. tularensis strains in CDM under aerobic or microaerobic conditions and (B) catalase activity of the strains at indicated time-points during growth. (A) Shows a representative experiment of at least three performed and (B) the average from six to nine separate observations for each time point and growth condition. Error bars represent the SEM. *P < 0.05, ***P < 0.001 vs. LVS.
Catalase activity under aerobic vs. microaerobic conditions
The results so far suggested that LVS experienced oxidative stress during growth in an aerobic environment and to handle this stress, required either the function of catalase, or the expression of OxyR-regulated detoxifying mechanisms. OxyR is known to respond to oxidative stress by inducing antioxidant enzymes, such as catalase. As an indicator of oxidative stress and to investigate if catalase is under the regulation of oxyR in LVS, we measured the activity of the enzyme during growth of the bacteria in CDM. The catalase activity in LVS gradually increased during the two to 24 h period, whereas the catalase activity in ΔoxyR was sustained at a constant, but lower level compared to LVS from two to six h (P < 0.05 at 2 and 4 h and P < 0.001 at 6 h; Figure 1B). However, the catalase activity of the two strains was similar at 24 h (Figure 1B). In the microaerobic environment, the catalase activity of LVS and ΔoxyR was similar, but for both lower than in the aerobic environment (Figure 1B). The H2O2 decomposition in samples containing ΔkatG or ΔoxyR/ΔkatG was below 1 μmol, regardless of growth condition and time point, indicating the absence of catalase activity (Figure 1B).
In summary, ΔoxyR demonstrated a basal catalase activity, but did not induce this activity further during the aerobic logarithmic growth phase as LVS did. ΔoxyR/ΔkatG, which lacks this basal catalase activity, failed to grow to high densities under the aerobic condition, but grew as well as LVS in the microaerobic milieu.
H2O2 tolerance
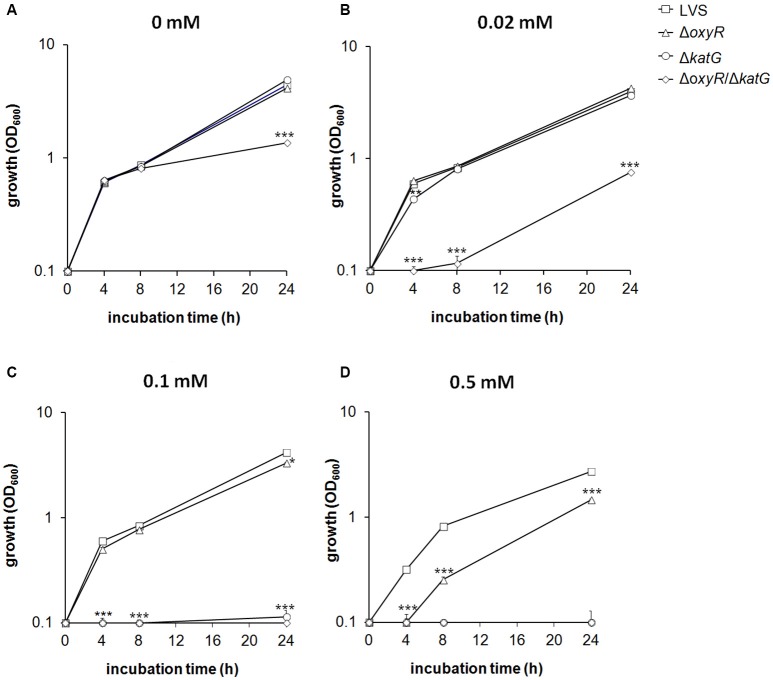
ΔoxyR and ΔkatG grew as well as LVS in CDM despite the reduced, or lack of catalase activity (Figure 2A). To investigate their adaptation to stress, H2O2, the substrate of catalase, was added to the cultures. Growth of LVS or ΔoxyR was not affected by 0.02 mM H2O2, whereas, initially, the growth rate of ΔkatG was reduced (P < 0.01) and growth of ΔoxyR/ΔkatG almost completely inhibited (P < 0.001; Figure 2B). At 0.1 mM of H2O2, LVS and ΔoxyR still grew rapidly, in contrast to ΔkatG and ΔoxyR/ΔkatG that did not grow at all (P < 0.001; Figure 2C). Growth of ΔoxyR was significantly reduced in the presence of 0.5 mM of H2O2 compared to LVS (P < 0.001; Figure 2D). Exposure of the strains to H2O2 did not significantly change their catalase activity (data not shown).
Figure 2.
Growth of F. tularensis strains in CDM during exposure to various concentrations of H2O2 (A) 0 mM, (B) 0.02 mM, (C) 0.1 mM, and (D) 0.5 mM. The results shown illustrate one representative experiment of at least three performed. Each value represents the average for triplicate samples and error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. LVS.
In summary, the mutant strains displayed increased susceptibility to H2O2 as compared to LVS, with the effect being most pronounced for ΔoxyR/ΔkatG, followed by ΔkatG, and the least affected strain being ΔoxyR.
Susceptibility to paraquat-mediated killing
is continuously generated as a by-product of the respiratory chain during growth of bacteria. To investigate the capacity of the bacteria to defend against such ROS, LVS, ΔoxyR, ΔkatG, and ΔoxyR/ΔkatG were exposed to paraquat in a disc diffusion assay (Figure 3). Paraquat dichloride hydrate generates through a reaction with parts of the respiratory chain in bacteria, causing the reduction of O2 to (Hassan and Fridovich, 1979). ΔoxyR displayed a significantly larger zone of inhibition than did LVS in the presence of 1.25 and 5 mM paraquat (P < 0.001 and 0.01, respectively), but the zones were similar when exposed to 20 mM (Figure 3). The zone of inhibition for ΔkatG was larger compared to LVS at 1.25 mM (P < 0.05), but similar at the two higher concentrations (Figure 3). A significantly larger zone of inhibition was observed for ΔoxyR/ΔkatG vs. LVS and ΔkatG at all three concentrations of paraquat (P < 0.001 for 1.25 and 5 mM and P < 0.01 for 20 mM) and also larger compared to ΔoxyR at 1.25 and 5 mM (P < 0.01; Figure 3).
Figure 3.
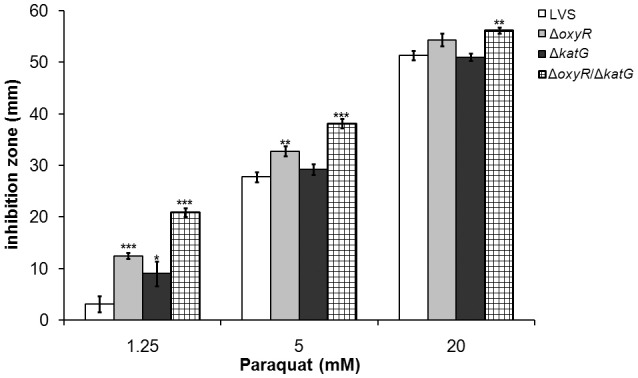
F. tularensis strains were exposed to the -generating compound paraquat in a disc diffusion assay. Each bar represents the average from three separate experiments with triplicate samples in each and error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. LVS for each concentration.
In summary, the results demonstrated that ΔoxyR and ΔoxyR/ΔkatG were more susceptible to paraquat-mediated killing compared to LVS, with ΔoxyR/ΔkatG being the most susceptible, whereas ΔkatG was only slightly more susceptible than LVS.
Susceptibility to SIN-1-mediated killing
Peroxynitrite (ONOO−) is a highly reactive and bactericidal ROS formed through the reaction between (NO) and and it is active against F. tularensis in activated macrophages (Lindgren et al., 2005). Experimentally, SIN-1 can be used to mimic a continuous exposure to ONOO−. SIN-1 slowly decomposes, thereby releasing both NO and that combine to form ONOO−, which quickly is internalized since it passes through lipid bilayers (Hogg et al., 1992; Murphy et al., 1998).
The exposure to 0.48 mM SIN-1 for 3 h reduced the viability of all strains in comparison to un-treated cultures (P < 0.001 for all strains), but affected the mutant strains to a greater extent compared to LVS (P < 0.001 vs. LVS for all; Figure 4). The viability of LVS decreased approximately 0.8 log10, of ΔoxyR 2.8 log10, of ΔkatG 3.0 log10, and of ΔoxyR/ΔkatG 4.6 log10 CFU. The latter was significantly more susceptible than any of the other strains (P < 0.001; Figure 4).
Figure 4.
F. tularensis strains were exposed to the peroxynitrite generating compound SIN-1 for 3 h. After 1.5 h of incubation, additional SIN-1 was added to the tubes to ensure a constant generation of peroxynitrite during the whole incubation period. Each bar represents the average from three separate experiments with triplicate samples in each and error bars represent the SEM. ***P < 0.001 vs. LVS.
In summary, all mutant strains displayed increased susceptibility to ONOO− as compared to LVS, with the effect being similar for ΔoxyR and ΔkatG and most pronounced for ΔoxyR/ΔkatG.
Gene expression
ΔoxyR/ΔkatG did not grow after the late logarithmic growth phase (Figure 1A), and we therefore found it of interest to explore the gene expression of the strains at 10 h, i.e., during the late logarithmic growth phase. The analysis was focused on genes expressing proteins influencing the oxidative stress response of the bacterium, such as antioxidant enzymes, chaperones and iron-related proteins. Genes found to be differentially expressed vs. LVS are shown in Figure 5. Of all genes examined, ahpC was the sole gene significantly repressed in ΔoxyR (P < 0.001; Figure 5A). A similar degree of repression, about 3-fold, was observed in ΔoxyR/ΔkatG, which in addition, had a 1.5 to 2-fold increased expression of sodB, sodC and FTT0086 (P < 0.001 for all genes; Figure 5A). ahpC was not repressed in ΔkatG and as expected, katG transcripts were not detected in either ΔkatG or in ΔoxyR/ΔkatG (Figure 5A). All chaperone genes examined were upregulated 1.6 to 2.5-fold in ΔoxyR and ΔkatG (P < 0.001 for all genes; Figure 5B). In contrast, these genes, except for clpB, were suppressed 2.4 to 3.1-fold in ΔoxyR/ΔkatG (P < 0.001 for all genes).
Figure 5.
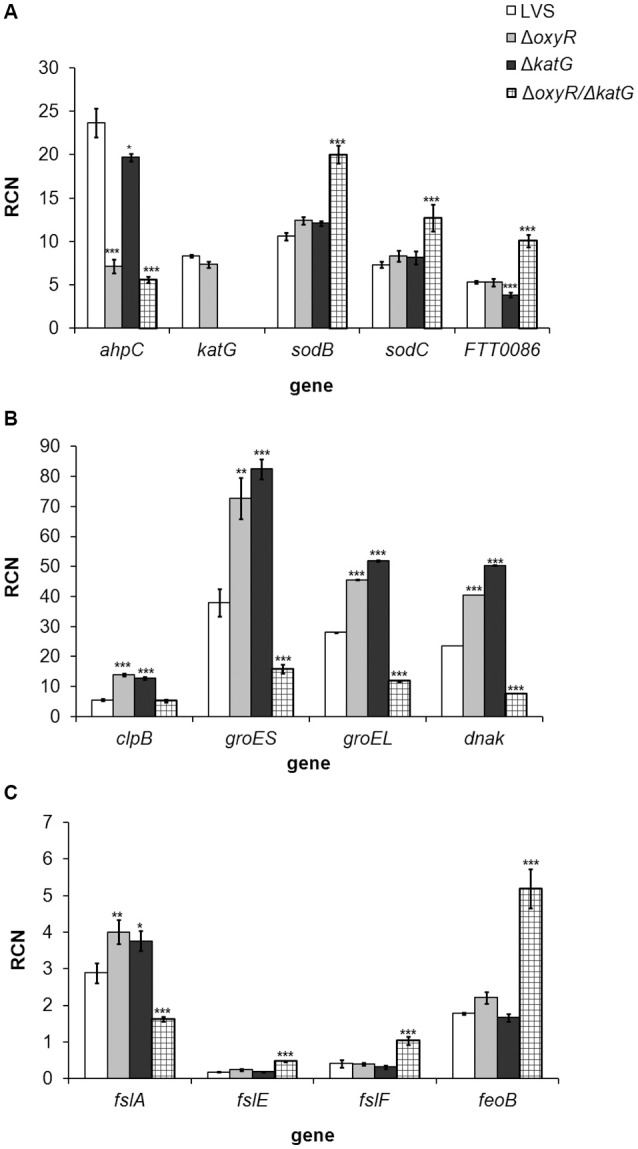
Real-time PCR analysis of genes expressing (A) antioxidant enzymes, (B) chaperones and (C) iron-related genes in F. tularensis strains cultivated in CDM for 10 h. Copy numbers of the respective gene in relation to the housekeeping gene FTT0901 is shown (RCN). The bars represent the average from three separate experiments with triplicate samples in each and error bars represent the SEM. *P < 0.05, **P < 0.01, ***P < 0.001 vs. LVS.
fslA, the first gene of the siderophore operon, was slightly up-regulated in ΔoxyR and ΔkatG, although only about 1.2-fold, whereas the other iron-related genes were expressed at similar levels as in LVS. In contrast, fslA was suppressed 1.8-fold in ΔoxyR/ΔkatG and fslE, fslF and feoB were upregulated 2.5 to 2.9-fold (P < 0.001; Figure 5C).
In summary, the absence of OxyR resulted in a suppressed expression of ahpC and an up-regulated expression of genes encoding chaperone proteins. Except for ahpC, the expression profile of ΔkatG was similar to ΔoxyR. In contrast, loss of both oxyR and katG changed the expression profile and low expression of chaperone-encoding genes was observed in ΔoxyR/ΔkatG, together with high expression of antioxidant genes, except for ahpC and katG, and an altered expression of genes related to iron-uptake.
Intracellular replication in BMDM
Based on the increased susceptibility to various ROS displayed by ΔoxyR, ΔkatG, and ΔoxyR/ΔkatG, it was of interest to test whether the strains were defective for replication in professional phagocytes. Non-stimulated or IFN-γ-stimulated BMDMs were infected with LVS, ΔoxyR, ΔkatG, or ΔoxyR/ΔkatG at an MOI of 30, and the viability of internalized bacteria was determined after 0 h, 4 h, and 24 h. In non-stimulated BMDM, LVS grew from approximately 2.5 log10 CFU to more than 5.0 log10 CFU within 24 h and also ΔoxyR and ΔkatG grew to similar extent (Figure 6A). ΔoxyR/ΔkatG grew in non-stimulated cells, but reached approximately 10-fold lower numbers compared to the other strains after 24 h (P < 0.001; Figure 6A).
Figure 6.
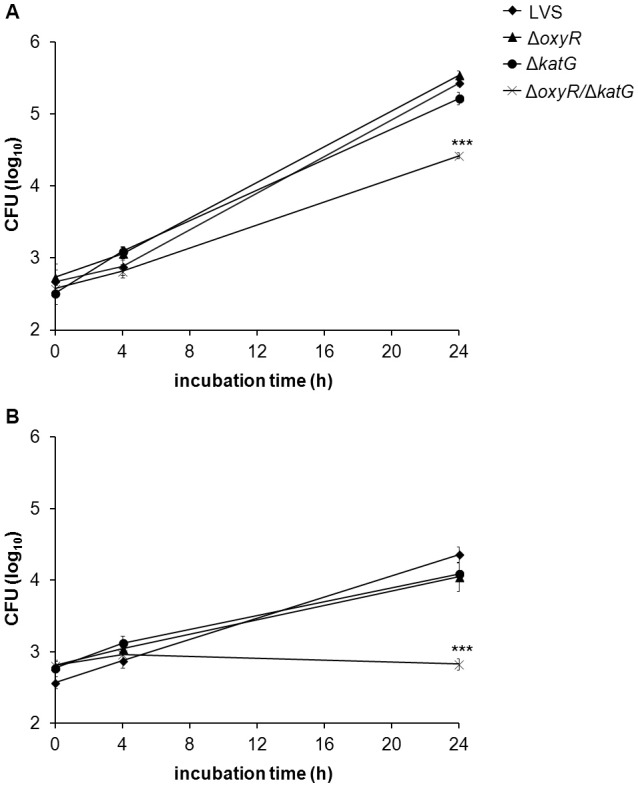
Growth of LVS, ΔoxyR, ΔkatG, and ΔoxyR/ΔkatG in (A) non-stimulated or (B) IFN-γ–stimulated BMDM. Monolayers of BMDM were infected with the various strains at an MOI of 30 and at indicated time points, intracellular bacteria were enumerated by lysis of the cultures and plating 10-fold serial dilutions on MC plates. The diagrams show one representative experiment, with the average of triplicate samples at each time point and treatment. Error bars represent the SEM. Similar results were observed in two additional experiments. ***P < 0.001 vs. LVS.
IFN-γ-stimulation of BMDM prior to infection reduced the numbers of LVS, ΔkatG, and ΔoxyR about 10-fold at 24 h vs. the numbers in non-stimulated cultures (P < 0.001; Figures 6A,B). There was no growth of ΔoxyR/ΔkatG in IFN-γ-stimulated cultures and, thus, significantly lower bacterial numbers compared to non-stimulated cultures at 24 h (P < 0.001; Figures 6A,B) and vs. all the other strains exposed to IFN-γ (P < 0.001; Figure 6B).
Thus, the ΔoxyR and ΔkatG mutants showed intact capacity of intracellular replication, whereas the ΔoxyR/ΔkatG mutant showed impaired replication in BMDM, both in the presence and absence of IFN-γ.
Virulence in mice
The virulence of LVS, ΔoxyR, ΔkatG, and ΔoxyR/ΔkatG was determined by subcutaneous infection of C57BL/6 mice with 4 × 103 CFU/mouse, a non-lethal dose, and enumeration of viable bacteria in spleen and liver on day 3 and 6 of infection. Compared to LVS, there were lower numbers of both ΔoxyR and ΔkatG on day 3 in the liver of the mice (P < 0.05; Figure 7A), whereas there were no differences between these strains in either the liver or spleen at the other time points (Figures 7A,B). Numbers of ΔoxyR/ΔkatG in both organs were at least 100-fold lower vs. all other strains at both time points (P < 0.001). Thus, both ΔoxyR and ΔkatG showed slight attenuation in mice, whereas ΔoxyR/ΔkatG was highly attenuated.
Figure 7.
Bacterial burden of mice at (A) 3 and (B) 6 days after subcutaneous inoculation of 4 × 103 CFU of either LVS, ΔoxyR, ΔkatG or ΔoxyR/ΔkatG. The average of four mice and SEM are shown. *P < 0.05, ***P < 0.001 vs. LVS.
Discussion
Francisella tularensis is a versatile bacterium capable of surviving in many different hosts, vectors and in various cell types, including the normally bactericidal macrophages. Upon phagocytosis, F. tularensis is encased in a phagosome, a membrane-bound compartment designed for the annihilation of phagocytosed microbes, which is rich in antimicrobial molecules, such as reactive oxygen and nitrogen species. Although F. tularensis only transiently resides in this compartment, it must still muster defenses against highly reactive species in order to survive and escape to the cytosol, where it proceeds to replicate. By entering the cytosol, F. tularensis gains access to a nutrient-rich, protected niche in which it multiplies. As survival and replication in the intracellular niche is essential for the life cycle of F. tularensis, a thorough understanding of how the bacterium survives intracellularly is essential to fully grasp its defense mechanisms against oxidative stress. To this end, the study focused on understanding the interplay between catalase and OxyR, the latter being important for the expression of several antioxidant enzymes, in the defense against ROS and their impact on the survival of the bacterium in professional phagocytes.
To investigate if OxyR is involved in the oxidative stress response of LVS, we constructed an in-frame deletion of oxyR. A similar investigation has been performed recently by Ma et al. which studied the role of OxyR in LVS (Ma et al., 2016). It was found that OxyR controlled transcription of katG and the findings agree with the reduced catalase activity of ΔoxyR observed in the present study. Nevertheless, our study revealed that even in the absence of OxyR, there was still prominent catalase activity. Overall, it appears that OxyR, as expected, regulates katG in the LVS strain, however, the regulation does not completely abolish its expression as is the case observed for various other bacterial species, e.g., E. coli (Michán et al., 1999), Salmonella enterica (Morgan et al., 1986), Haemophilus influenza (Whitby et al., 2012), or Moraxella catarrhalis (Hoopman et al., 2011). In both the present study, and in the previous study, it was observed that the lack of OxyR led to marked suppression of ahpC2 (Ma et al., 2016). In addition Ma et al. demonstrated suppressed expression of both katG and sodB in ΔoxyR by real-time PCR and demonstrated that OxyR binds to the upstream promoter regions of each gene. In contrast, there was no down-regulation of katG or sodB observed in the present study. Likely, this is a consequence of the rapid on/off switch of the promoter binding capacity of OxyR in response to the oxidative levels in the bacteria leading to a limited window when elevated mRNA levels can be detected (Wei et al., 2012).
Besides antioxidant genes, our study revealed an aberrant expression of genes encoding chaperone proteins of the mutants. Such proteins are induced in response to various stresses, including oxidative stress (Hartl et al., 2011). Thus, the induced expression of these genes in ΔoxyR and in ΔkatG, also observed by Ma et al. (2016), likely is a reflection of oxidative stress encountered by the mutants. The chaperone network likely helps the bacterium to handle this stress through unfolding and/or degradation of mis-folded/damaged proteins. The reason behind the suppressed expression of multiple chaperone genes in ΔoxyR/ΔkatG is obscure, but should lead to an accumulation of damaged or mis-folded proteins and may explain why it was so impaired for growth in broth. The intact growth of ΔoxyR/ΔkatG under microaerobic conditions likely reflects that reduced levels of ROS are formed and therefore that antioxidant defenses are less important. The aberrant expression of genes related to iron-uptake did not result in a skewed iron content of ΔoxyR/ΔkatG (data not shown) and it is therefore not obvious that this would influence the susceptibility of the strain to various ROS.
The F. tularensis ahpC2 gene is divergently transcribed from the oxyR promoter, a feature commonly seen for genes transcriptionally regulated by OxyR (Hahn et al., 2002; Maddocks and Oyston, 2008). AhpC belongs to the peroxiredoxin family, which is ubiquitously found in nature (Rhee et al., 2005) and is known to be involved in defenses against peroxides in E. coli (Storz et al., 1989), and both peroxides and peroxynitrite in, e.g., Salmonella typhimurium (Bryk et al., 2000), and in the defense against superoxide and peroxynitrite in the virulent SCHU S4 strain of F. tularensis subsp. tularensis (Binesse et al., 2015). In agreement with this, and in view of the reduced expression of AhpC in ΔoxyR, this mutant was also highly susceptible to ONOO−. ΔkatG was as susceptible as ΔoxyR to ONOO− and in view of the substantial catalase activity remaining in ΔoxyR, this result implies that the function of catalase overlaps with other OxyR-regulated detoxifying mechanisms, presumably AhpC, to protect against ONOO−. Further corroborating the importance of AhpC and catalase was the failure to generate a katG and ahpC double deletion mutant and even an ahpC mutant in LVS. Hence, AhpC seems indispensable to LVS, which is in stark contrast to SCHU S4, where deletion of ahpC resulted in only slight attenuation (Binesse et al., 2015). This indicates that there is a disparity regarding the importance of the enzyme between the SCHU S4 and LVS strains, possibly a factor that to some extent explains the difference in virulence between the strains, since it implies that the detoxifying mechanisms of SCHU S4 are much more elaborate. Nevertheless, as for LVS, it has not been possible to generate a katG and ahpC double deletion mutant of SCHU S4 (Kadzhaev et al., 2009; Binesse et al., 2015). Collectively, this indicates that the mechanisms of protection conferred by these enzymes may be overlapping and the lack of both is detrimental to the survival of both LVS and highly virulent F. tularensis strains.
Based on the failure to generate a katG and ahpC double deletion mutant and the marked suppression of ahpC in the ΔoxyR mutant, we hypothesized that the absence of OxyR together with the absence of catalase would severely disarm the capability of the bacterium to handle ROS. Indeed, we observed that the ΔoxyR/ΔkatG mutant was hyper-susceptible to H2O2, ONOO−, and ; much more so than either ΔoxyR or ΔkatG. Collectively, the results demonstrate that the roles of OxyR-regulated antioxidant enzymes and catalase overlap to protect LVS against various ROS. We find it likely that the reduced activity of catalase and expression of ahpC observed in oxyR contributed to the increased susceptibility of the mutant to H2O2, , and ONOO− through the increase of both Fenton-mediated toxicity and direct - and ONOO−-mediated damage. We further suggest that the reduced levels of AhpC together with the lack of catalase in the ΔoxyR/ΔkatG strain, despite an increased expression of sodB, sodC and FTT0086, resulted in enhanced Fenton-mediated toxicity and ONOO−-mediated damage, which likely account for the extreme susceptibility of the double mutant to , H2O2, and ONOO−. Our findings concur with those of Ma et al. (2016), and, in addition, demonstrate that the combined activity of catalase and OxyR-regulated detoxifying mechanisms are critical for ROS detoxification by F. tularensis.
Despite the enhanced susceptibility of both ΔoxyR and ΔkatG to various ROS, the strains replicated as efficiently as LVS in mouse BMDM, but importantly, the capacity to replicate in professional phagocytes required either OxyR or catalase, since ΔoxyR/ΔkatG failed to replicate. IFN-γ-activation of BMDM restricted growth of LVS, ΔkatG, and ΔoxyR to a similar degree and completely blocked the growth of ΔoxyR/ΔkatG. The majority of F. tularensis LVS escapes the phagosome of IFN-γ-activated macrophages (Lindgren et al., 2004), but the mechanism of growth inhibition appears to vary depending on the cell model used (Edwards et al., 2010). IFN-γ-mediated inhibition of intracellular growth of F. novicida is dependent on the expression of IRGB10 and various guanylate-binding proteins (Meunier et al., 2015; Man et al., 2016), however, the role of this pathway is unknown for other F. tularensis species.
Our results reveal elaborate interconnecting roles between OxyR-regulated ROS-detoxifying mechanisms and catalase and demonstrate that either needs to be intact for the bacterium to thrive in professional phagocytes. The roles of the anti-oxidative mechanisms could be to protect the bacterium from direct damage by various ROS, such as ONOO−, which has been demonstrated to be crucial for killing of F. tularensis in peritoneal cells (Lindgren et al., 2005). Alternatively, or additionally, the antioxidants may restrict macrophage activation through their ability to preserve phosphatase activity required for kinase signaling and proinflammatory cytokine production (Melillo et al., 2010).
Our finding that ΔoxyR replicated as efficiently as LVS in BMDM is in contrast to findings in a previous study, which reported that an oxyR mutant of LVS was markedly impaired with regard to escape from the phagosome, replication in professional phagocytes, and virulence in the mouse model (Ma et al., 2016). Notably, the LVS strain used by Ma et al. replicated less than 10-fold during 24 h in C57BL/6 BMDM, whereas the LVS strain used in the present study replicated about 500-fold. Isolates of LVS with different virulence are used in the research community (Griffin et al., 2015) and the distinct differences in the intracellular growth of these two LVS strains are additional examples of such distinct phenotypes. The phenotypic differences between the two LVS strains likely explain the discrepant findings of the two studies. The observation in the present study of the intact growth of the single mutants in BMDM was corroborated by findings in vivo, since the ΔoxyR and ΔkatG mutants showed essentially intact growth in organs of mice, whereas ΔoxyR/ΔkatG hardly grew at all. Despite their effective growth in the organs, a previous study demonstrated a more distinct growth defect of the ΔkatG mutant, most likely because a 100-fold higher dose was given (Lindgren et al., 2007). Moreover, by the intranasal route, ΔoxyR was demonstrated to be moderately attenuated (Ma et al.). Based on these collective findings, it can be concluded that both OxyR and KatG contribute to the virulence of F. tularensis LVS and that the concomitant loss is detrimental to the virulence of the bacterium.
Altogether, the results presented in this study clearly demonstrate the mutual importance of catalase and OxyR for a robust oxidative stress defense system and that either of these systems is vital for the intracellular replication of F. tularensis and for its virulence.
Author contributions
Conceived and designed the experiments: AS, MH, and HL. Performed the experiments: MH, HL, and GB. Analyzed the data: AS, MH, HL, and GB. Wrote the paper: HL, MH, and AS.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Grant support was obtained from the Swedish Medical Research Council (K2010-9485, K2012-3469, and K2013-8621), and the Medical Faculty, Umeå University, Umeå, Sweden.
References
- Andrews S. C., Robinson A. K., Rodríguez-Quiñones F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. 10.1016/S0168-6445(03)00055-X [DOI] [PubMed] [Google Scholar]
- Anthony L. S., Morrissey P. J., Nano F. E. (1992). Growth inhibition of Francisella tularensis live vaccine strain by IFN-gamma-activated macrophages is mediated by reactive nitrogen intermediates derived from L-arginine metabolism. J. Immunol. 148, 1829–1834. [PubMed] [Google Scholar]
- Bakshi C. S., Malik M., Regan K., Melendez J. A., Metzger D. W., Pavlov V. M., et al. (2006). Superoxide dismutase B gene (sodB)-deficient mutants of Francisella tularensis demonstrate hypersensitivity to oxidative stress and attenuated virulence. J. Bacteriol. 188, 6443–6448. 10.1128/JB.00266-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betteridge D. J. (2000). What is oxidative stress? Metab. Clin. Exp. 49, 3–8. 10.1016/S0026-0495(00)80077-3 [DOI] [PubMed] [Google Scholar]
- Binesse J., Lindgren H., Lindgren L., Conlan W., Sjöstedt A. (2015). Roles of reactive oxygen species-degrading enzymes of Francisella tularensis SCHU S4. Infect. Immun. 83, 2255–2263. 10.1128/IAI.02488-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Lavander M., Meyer L., Sjöstedt A. (2011). IglG and IglI of the Francisella pathogenicity island are important virulence determinants of Francisella tularensis LVS. Infect. Immun. 79, 3683–3696. 10.1128/IAI.01344-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Lavander M., Sjöstedt A. (2009). A conserved alpha-helix essential for a type VI secretion-like system of Francisella tularensis. J. Bacteriol. 191, 2431–2446. 10.1128/JB.01759-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bröms J. E., Sjöstedt A., Lavander M. (2010). The role of the Francisella tularensis pathogenicity island in type VI secretion, intracellular survival, and modulation of host cell signaling. Front. Microbiol. 1:136. 10.3389/fmicb.2010.00136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk R., Griffin P., Nathan C. (2000). Peroxynitrite reductase activity of bacterial peroxiredoxins. Nature 407, 211–215. 10.1038/35025109 [DOI] [PubMed] [Google Scholar]
- Chong A., Celli J. (2010). The Francisella intracellular life cycle: toward molecular mechanisms of intracellular survival and proliferation. Front. Microbiol. 1:138. 10.3389/fmicb.2010.00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlan J. W. (2011). Tularemia vaccines: recent developments and remaining hurdles. Future Microbiol. 6, 391–405. 10.2217/fmb.11.22 [DOI] [PubMed] [Google Scholar]
- Edwards J. A., Rockx-Brouwer D., Nair V., Celli J. (2010). Restricted cytosolic growth of Francisella tularensis subsp. tularensis by IFN-gamma activation of macrophages. Microbiology 156, 327–339. 10.1099/mic.0.031716-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr S. B., Kogoma T. (1991). Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55, 561–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flannagan R. S., Cosio G., Grinstein S. (2009). Antimicrobial mechanisms of phagocytes and bacterial evasion strategies. Nat. Rev. Microbiol. 7, 355–366. 10.1038/nrmicro2128 [DOI] [PubMed] [Google Scholar]
- Fridovich I. (1998). Oxygen toxicity: a radical explanation. J. Exp. Biol. 201, 1203–1209. [DOI] [PubMed] [Google Scholar]
- Gavrilin M. A., Bouakl I. J., Knatz N. L., Duncan M. D., Hall M. W., Gunn J. S., et al. (2006). Internalization and phagosome escape required for Francisella to induce human monocyte IL-1β processing and release. Proc. Natl. Acad. Sci. U.S.A. 103, 141–146. 10.1073/pnas.0504271103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovliov I., Sjöstedt A., Mokrievich A., Pavlov V. (2003). A method for allelic replacement in Francisella tularensis. FEMS Microbiol. Lett. 222, 273–280. 10.1016/S0378-1097(03)00313-6 [DOI] [PubMed] [Google Scholar]
- Griffin A. J., Crane D. D., Wehrly T. D., Bosio C. M. (2015). Successful protection against tularemia in C57BL/6 mice is correlated with expansion of Francisella tularensis-specific effector T cells. Clin. Vaccine Immunol. 22, 119–128. 10.1128/CVI.00648-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J. S., Oh S. Y., Roe J. H. (2002). Role of OxyR as a peroxide-sensing positive regulator in Streptomyces coelicolor A3(2). J. Bacteriol. 184, 5214–5222. 10.1128/JB.184.19.5214-5222.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F. U., Bracher A., Hayer-Hartl M. (2011). Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hassan H. M., Fridovich I. (1979). Paraquat and Escherichia coli. Mechanism of production of extracellular superoxide radical. J. Biol. Chem. 254, 10846–10852. [PubMed] [Google Scholar]
- Hogg N., Darley-Usmar V. M., Wilson M. T., Moncada S. (1992). Production of hydroxyl radicals from the simultaneous generation of superoxide and nitric oxide. Biochem. J. 281, 419–424. 10.1042/bj2810419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honn M., Lindgren H., Sjöstedt A. (2012). The role of MglA for adaptation to oxidative stress of Francisella tularensis LVS. BMC Microbiol. 12:14. 10.1186/1471-2180-12-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoopman T. C., Liu W., Joslin S. N., Pybus C., Brautigam C. A., Hansen E. J. (2011). Identification of gene products involved in the oxidative stress response of Moraxella catarrhalis. Infect. Immun. 79, 745–755. 10.1128/IAI.01060-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadzhaev K., Zingmark C., Golovliov I., Bolanowski M., Shen H., Conlan W., et al. (2009). Identification of genes contributing to the virulence of Francisella tularensis SCHU S4 in a mouse intradermal infection model. PLoS ONE 4:e5463. 10.1371/journal.pone.0005463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Honn M., Golovlev I., Kadzhaev K., Conlan W., Sjöstedt A. (2009). The 58-kilodalton major virulence factor of Francisella tularensis is required for efficient utilization of iron. Infect. Immun. 77, 4429–4436. 10.1128/IAI.00702-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Shen H., Zingmark C., Golovliov I., Conlan W., Sjöstedt A. (2007). Resistance of Francisella tularensis strains against reactive nitrogen and oxygen species with special reference to the role of KatG. Infect. Immun. 75, 1303–1309. 10.1128/IAI.01717-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren H., Stenman L., Tärnvik A., Sjöstedt A. (2005). The contribution of reactive nitrogen and oxygen species to the killing of Francisella tularensis LVS by murine macrophages. Microbes Infect. 7, 467–475. 10.1016/j.micinf.2004.11.020 [DOI] [PubMed] [Google Scholar]
- Lindgren H., Stenmark S., Chen W., Tärnvik A., Sjöstedt A. (2004). Distinct roles of reactive nitrogen and oxygen species to control infection with the facultative intracellular bacterium Francisella tularensis. Infect. Immun. 72, 7172–7182. 10.1128/IAI.72.12.7172-7182.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Russo V. C., Rabadi S. M., Jen Y., Catlett S. V., Bakshi C. S., et al. (2016). Elucidation of a mechanism of oxidative stress regulation in Francisella tularensis live vaccine strain. Mol. Microbiol. 101, 856–878. 10.1111/mmi.13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks S. E., Oyston P. C. (2008). Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology 154, 3609–3623. 10.1099/mic.0.2008/022772-0 [DOI] [PubMed] [Google Scholar]
- Man S. M., Karki R., Sasai M., Place D. E., Kesavardhana S., Temirov J., et al. (2016). IRGB10 liberates bacterial ligands for sensing by the AIM2 and Caspase-11-NLRP3 inflammasomes. Cell 167, 382–396.e17. 10.1016/j.cell.2016.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo A. A., Bakshi C. S., Melendez J. A. (2010). Francisella tularensis antioxidants harness reactive oxygen species to restrict macrophage signaling and cytokine production. J. Biol. Chem. 285, 27553–27560. 10.1074/jbc.M110.144394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melillo A. A., Mahawar M., Sellati T. J., Malik M., Metzger D. W., Melendez J. A., et al. (2009). Identification of Francisella tularensis live vaccine strain CuZn superoxide dismutase as critical for resistance to extracellularly generated reactive oxygen species. J. Bacteriol. 191, 6447–6456. 10.1128/JB.00534-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier E., Wallet P., Dreier R. F., Costanzo S., Anton L., Rühl S., et al. (2015). Guanylate-binding proteins promote activation of the AIM2 inflammasome during infection with Francisella novicida. Nat. Immunol. 16, 476–484. 10.1038/ni.3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michán C., Manchado M., Dorado G., Pueyo C. (1999). In vivo transcription of the Escherichia coli oxyR regulon as a function of growth phase and in response to oxidative stress. J. Bacteriol. 181, 2759–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan R. W., Christman M. F., Jacobson F. S., Storz G., Ames B. N. (1986). Hydrogen peroxide-inducible proteins in Salmonella typhimurium overlap with heat shock and other stress proteins. Proc. Natl. Acad. Sci. U.S.A. 83, 8059–8063. 10.1073/pnas.83.21.8059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. P., Packer M. A., Scarlett J. L., Martin S. W. (1998). Peroxynitrite: a biologically significant oxidant. Gen. Pharmacol. 31, 179–186. 10.1016/S0306-3623(97)00418-7 [DOI] [PubMed] [Google Scholar]
- Oyston P. C., Sjöstedt A., Titball R. W. (2004). Tularaemia: bioterrorism defence renews interest in Francisella tularensis. Nat. Rev. Microbiol. 2, 967–978. 10.1038/nrmicro1045 [DOI] [PubMed] [Google Scholar]
- Polsinelli T., Meltzer M. S., Fortier A. H. (1994). Nitric oxide-independent killing of Francisella tularensis by IFN-gamma-stimulated murine alveolar macrophages. J. Immunol. 153, 1238–1245. [PubMed] [Google Scholar]
- Pomposiello P. J., Demple B. (2001). Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19, 109–114. 10.1016/S0167-7799(00)01542-0 [DOI] [PubMed] [Google Scholar]
- Rhee S. G., Chae H. Z., Kim K. (2005). Peroxiredoxins: a historical overview and speculative preview of novel mechanisms and emerging concepts in cell signaling. Free Radic. Biol. Med. 38, 1543–1552. 10.1016/j.freeradbiomed.2005.02.026 [DOI] [PubMed] [Google Scholar]
- Schaible U. E., Kaufmann S. H. (2004). Iron and microbial infection. Nat. Rev. Microbiol. 2, 946–953. 10.1038/nrmicro1046 [DOI] [PubMed] [Google Scholar]
- Sjöstedt A. (2006). Intracellular survival mechanisms of Francisella tularensis, a stealth pathogen. Microbes Infect. 8, 561–567. 10.1016/j.micinf.2005.08.001 [DOI] [PubMed] [Google Scholar]
- Storz G., Jacobson F. S., Tartaglia L. A., Morgan R. W., Silveira L. A., Ames B. N. (1989). An alkyl hydroperoxide reductase induced by oxidative stress in Salmonella typhimurium and Escherichia coli: genetic characterization and cloning of ahp. J. Bacteriol. 171, 2049–2055. 10.1128/jb.171.4.2049-2055.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B., Hassan H. M. (2013). Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell. Infect. Microbiol. 3:59. 10.3389/fcimb.2013.00059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Q., Minh P. N., Dötsch A., Hildebrand F., Panmanee W., Elfarash A., et al. (2012). Global regulation of gene expression by OxyR in an important human opportunistic pathogen. Nucleic Acids Res. 40, 4320–4333. 10.1093/nar/gks017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby P. W., Morton D. J., Vanwagoner T. M., Seale T. W., Cole B. K., Mussa H. J., et al. (2012). Haemophilus influenzae OxyR: characterization of its regulation, regulon and role in fitness. PLoS ONE 7:e50588. 10.1371/journal.pone.0050588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Aslund F., Storz G. (1998). Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279, 1718–1721. 10.1126/science.279.5357.1718 [DOI] [PubMed] [Google Scholar]
- Zheng M., Doan B., Schneider T. D., Storz G. (1999). OxyR and SoxRS regulation of fur. J. Bacteriol. 181, 4639–4643. [DOI] [PMC free article] [PubMed] [Google Scholar]