Abstract
The long-range movement of organelles, vesicles, and macromolecular complexes by microtubule-based transport is crucial for cell growth and survival. The canonical view of intracellular transport is that each cargo directly recruits molecular motors via cargo-specific adaptor molecules. Recently, a new paradigm called ‘hitchhiking’ has emerged: some cargos can achieve motility by interacting with other cargos that have already recruited molecular motors. In this way, cargos are co-transported together and their movements are directly coupled. Cargo hitchhiking was discovered in fungi. However, the observation that organelle dynamics are coupled in mammalian cells suggests that this paradigm may be evolutionarily conserved. We review here the data for hitchhiking and discuss the biological significance of this non-canonical mode of microtubule-based transport.
Hitchhiking Is a Novel Mechanism Used To Achieve Cargo Motility
Precise spatiotemporal organization of intracellular cargos is important for cellular development, maturation, and survival. In most eukaryotic cells, long-range movement of cargos is driven by the molecular motors kinesin and dynein, which display processive motility (see Glossary) along polarized microtubule tracks. Dynein moves towards the microtubule minus-end, and most kinesins move towards the microtubule plus-end. In humans, a single dynein (cytoplasmic dynein-1; ‘dynein’ here), and at least 15 kinesins are responsible for the long-range movements of cargo during interphase (Box 1) [1,2]. The number of cargos far exceeds the number of molecular motors, and ranges from organelles to large macromolecular complexes to viruses. Thus, elucidating the mechanisms that are used to recruit molecular motors to each cargo type is of great interest in the transport field. In addition, defects in microtubule-based transport and mutations in the motors themselves lead to neurodevelopmental and neurodegenerative disorders in humans [3–6].
The canonical view of microtubule-based transport is that specific cargo adaptors recruit molecular motors to cargos. Adaptors for endosomes and other cargos, including mitochondria, autophagosomes, lysosomes, and mRNA, have been identified (Box 1; reviewed in [2,7–11]). However, for many cargos the mechanism of motor recruitment remains unknown. Recently, a novel mechanism of cargo motility, termed ‘hitchhiking’, has been described. Evidence suggests that ribosomes, mRNA, peroxisomes, endoplasmic reticulum (ER), and lipid droplets can achieve motility not by directly recruiting the molecular motor machinery themselves but by ‘hitchhiking’ on organelles that are already moving (Table 1, Key Table). Thus, these organelles act as ‘vehicles’ to support the movement of other cargos.
Table 1.
Key Table Examples of Hitchhikinga
| Organism | Biological process | Hitchhiker | Vehicle | Tether/linker | Refs |
|---|---|---|---|---|---|
| Saccharomyces cerevisiae | RNA localization in daughter cell | She2-mRNPs | Cortical ER | She2 | [32] |
| RNA retention in daughter cell | Pab1-mRNPs | COP1 vesicles | Pab1–Arf1 | [36] | |
| Ustilago maydis | RNA/polysome distribution in hyphae | Rrm4-mRNPs | EEs | Rrm4–Upa1 | [22–24,27] |
| Organelle distribution in hyphae | Peroxisomes, ER, and lipid droplets | EEs | ? | [41,55] | |
| Aspergillus nidulans | Peroxisome distribution in hyphae | Peroxisomes | EEs | PxdA | [40] |
| Xenopus laevis | Asymmetric localization of RNA during oogenesis | Vg1 mRNA | ER | Vg1RBP/Vera | (reviewed in [38]) |
| Drosophila melanogaster | Asymmetric localization of RNA during oogenesis | oskar mRNA | Rab11- recycling endosomes? | ? | (reviewed in [38]) |
| Candida albicans | Asymmetric localization of RNA at hyphal tip | SEC2 mRNA | Secretory vesicles | Sec2 protein | [39] |
Bold text indicates that these examples have experimental evidence for bona fide hitchhiking; characterized as (i) observable, co-transporting cargo; (ii) no membrane fusion; (iii) perturbations that affect the movement of vehicle cargo perturb the hitchhiker, but mutating the assembly and/or function of the hitchhiker does not disrupt vehicle movement.
We propose three main criteria for hitchhiking. First, hitchhiking cargo and their vehicle cargo comigrate during long-range movement. Second, the interactions do not involve membrane fusion, maintaining the biochemical identity of each cargo. Third, the movement of the hitchhiking cargo relies on the vehicle cargo, but not vice versa. In this review we summarize the evidence for cargo hitchhiking as we have defined it. Given these criteria, vesicular trafficking pathways involving membrane fusion, including the trafficking of cell surface receptors to and from the plasma membrane, viral hijacking via endocytosis, and the trafficking of components to cilia, are not considered to be hitchhiking. We also discuss observations that have not yet been shown to be hitchhiking based on our criteria, but may indeed represent bona fide hitchhiking after further experimentation. Finally, we will discuss the biological significance of this novel form of motility.
Endosomes are Hitchhiking Vehicles
Many examples of hitchhiking use highly-motile endosomes as the vehicle cargo. Endosomes are dynamic membrane-bound compartments that are crucial for the sorting, recycling, and degradation of internalized components from the plasma membrane. In addition to these fundamental processes, endosomes and endocytosis have a variety of other functions. (i) Endocytosis is used as a means to terminate signaling at the cell surface. (ii) Signaling endosomes containing internalized cell-surface receptors can be used as long-range signaling foci in other subcellular regions. (iii) Endosomes can also serve as ‘multipurpose platforms’ for the recruitment and assembly of signaling complexes important for cell division, polarization, and migration (reviewed in [12]). (iv) Finally, as highlighted here, endosomes can act as vehicles on which cargos can hitchhike over long distances in the cell. To execute these versatile functions, endosomes are highly motile and associate with a variety of cargo adaptors and molecular motors (Box 1).
Hitchhiking of mRNA-Containing Complexes
The subcellular localization of mRNA enables local protein translation and is crucial for polarized growth in a wide range of biological processes including budding during cell division in S. cerevisiae, axis specification in Drosophila and Xenopus oocytes, and growth cone development in neurons (reviewed in [13]). These mRNAs move within large messenger ribonucleoprotein (mRNP) complexes that include RNA-binding proteins (RBPs) and can also include ribosomes and proteins that are important for RNA stability and translational regulation [14,15]. For mRNA to move and distribute precisely, some RBPs function as cargo adaptors for molecular motors [16]. For instance, apical transport of mRNA in the Drosophila oocyte is achieved by an RBP, Egalitarian (Egl), which can bind directly to the dynein transport complex [8,16–18]. However, the mode of transport for most mRNAs is unknown.
mRNA and Polysome Hitchhiking on Early Endosomes in Filamentous Fungi
One of the first examples of hitchhiking involved mRNPs attaching to Rab5-positive early endosomes (EEs) in the filamentous fungus Ustilago maydis. EEs in filamentous fungi exhibit long-distance bidirectional movement dependent on kinesin-3 and dynein [19–21], and this is important for polarized growth of hyphae (Box 2). Recent studies in U. maydis demonstrated that motile EEs are required for the movement of an RBP (Rrm4) [22–24] which binds to mRNAs encoding proteins needed for polarized growth (Figure 1A) [25,26]. Similarly to EEs, Rrm4-containing mRNPs also move bidirectionally on microtubules using kinesin-3 and dynein [23]. Initially it was assumed that this motility was due to the ability of Rrm4 to directly recruit these motors, similarly to Egl in Drosophila. Surprisingly, several lines of evidence demonstrated that Rrm4-mRNPs move by hitching a ride on moving EEs. First, nearly all Rrm4 puncta colocalized with EEs, and its associated mRNAs comigrated with moving EEs [22–24]. Second, EEs were required for the movement of Rrm4 [22,24]; this phenomenon fits our definition of hitchhiking because the converse is not true: EE movement, identity, and function are not affected in cells lacking Rrm4 [22–24,27].
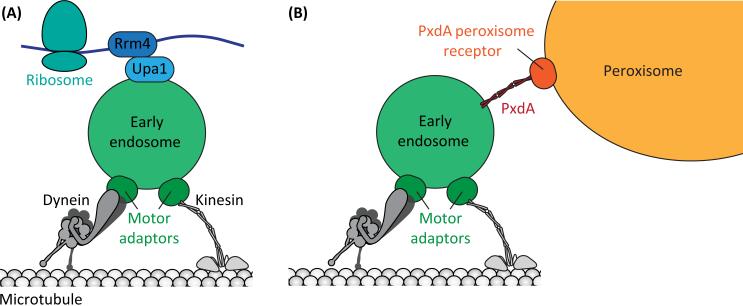
Figure 1. Mechanisms of Cargo Hitchhiking.
(A) In U. maydis ribosome-associated mRNAs hitchhike on EEs. The RNA-binding protein (RBP) Rrm4 links the mRNA to the early endosome-binding protein, Upa1. Kinesin-3 and dynein/dynactin motors are recruited via the motor adaptor Hok1 (related to mammalian Hook3). (B) In both U. maydis and A. nidulans peroxisomes hitchhike on EEs. The endosome-interacting protein, PxdA, is required for peroxisome hitchhiking and may act as a tether between the two organelles. The putative peroxisome receptor for PxdA has not yet been identified. Kinesin-3 and dynein/dynactin motors are recruited via the motor adaptor Hok1/HookA. Although not depicted here, Hook associates with endosomes via a larger protein complex that contains the homologs of the fused toes (FTS) and fused toes and hook interacting protein (FHIP) proteins.
How do Rrm4 and its associated RNAs hitchhike on EEs? To accomplish endosomal hitchhiking, Rrm4 interacts with a FYVE domain protein, Upa1, which directly binds to phosphatidylinositol 3-phosphate (PI3P) found on early endosomal membranes [27]. The association of Rrm4 with EEs, via its interaction with Upa1, is not affected by the absence of dynein or kinesin-3 [23,27]. Thus, Upa1 provides the tether between Rrm4 and EEs (Figure 1A).
Why do Rrm4-bound mRNAs hitchhike on EEs? One possibility is that EEs deliver mRNAs to specific subcellular regions, enabling local protein translation. However, this does not appear to be the case for Rrm4-bound mRNAs (including the septin cdc3 and the small GTPase rho3). Instead, hitchhiking is required for the even distribution of these mRNAs along hyphae because deleting the rrm4 gene or its RNA-recognition motif (RRM) domain results in their non-uniform distribution [22,25]. Fluorescence recovery after photobleaching (FRAP) experiments for the Cdc3 septin protein also revealed that the recovery kinetics are too fast to account for local translation at the hyphal tip (where septins are abundant), suggesting that translation may be occurring coincidently with transport [22]. In support of this, Rrm4-associated EEs also contain multiple ribosomes, and additional evidence suggests that Rrm4-bound mRNAs are translationally active on the surface of EEs during movement [22,24]. A recent study also demonstrated that newly translated septins are assembled on EEs and offloaded near the hyphal tip [28]. Taken together, the current model in the field proposes that the rapid bidirectional movement of EEs provides a convenient mixing platform for the distribution of polysomes along hyphae (see below) [24], as well as providing a platform for the assembly and movement of newly synthesized protein complexes [22,28].
mRNA Hitchhiking in Budding Yeast
A subset of mRNAs also hitchhikes on organelles in the yeast Saccharomyces cerevisiae, where mother cells divide by budding to form a daughter cell. Unlike filamentous fungi and animal cells, which primarily use microtubules to transport intracellular cargos over long distances [29], budding yeasts use actin filaments and myosin motors (reviewed in [30]). Similarly to dynein- and kinesin-based motility (Box 1), myosin motors also use adaptor proteins to engage with their cargos [30]. For example, cortical endoplasmic reticulum is transported to the budding daughter cell using the cargo adaptor She3, which recruits the type V myosin, Myo4 [31]. In an early example of hitchhiking, some mRNAs encoding polarity and exocytosis factors were found to comigrate with cortical ER [13,32–34]. These mRNAs, which are destined for the bud, are tethered to the cortical ER by an RBP called She2 [32,33,35]. In the absence of She2, cortical ER localizes to the bud normally, but mRNAs do not properly localize. Furthermore, specifically disrupting cortical ER movement also perturbs asymmetric mRNA localization in the bud [33]. She2-mRNP hitchhiking on cortical ER may allow for precise timing of asymmetric protein translation at the budding daughter cell. Consistent with this, She2-mRNPs contain the translational repressors, Khd1 and Puf6, and are not translationally active (in contrast to Rrm4-mRNPs) until reaching the bud [13].
COPI (coat protein) vesicles are also required for proper bud localization of mRNA in S. cerevisiae [36]. In this case, the small GTPase Arf1 forms a complex with the RBP Pab1 on the surface of COPI vesicles, an interaction that requires mRNA. The movement of COPI vesicles is independent of the Myo4/She2/She3 machinery, but instead relies on a second type V myosin in yeast, Myo2 [37]. The association of Pab1-mRNP with COPI vesicles seems to be important for the asymmetric retention of mRNAs in the budding daughter cell subsequent to hitchhiking on cortical ER [36].
Hitchhiking of mRNA-Containing Complexes in other Organisms
Although the specific mechanisms are less clear than the aforementioned examples, two early examples suggested that hitchhiking of mRNAs also occurs during Xenopus and Drosophila oogenesis (reviewed in [38]). In both organisms, the asymmetric localization and translation of mRNAs is crucial for axis specification and oocyte development. In Xenopus, vg1 mRNA [which encodes a transforming growth factor (TGF) β-like protein] co-transports with ER to the vegetal cortex by binding to the ER-associated RBP Vg1RBP/Vera. In Drosophila, mutations in Rab11, which marks recycling endosomes, disrupts the posterior localization of an mRNA called oskar. Because Rab11 mutants also disrupt microtubule organization, it is unclear whether oskar tethers to recycling endosomes or if its mislocalization is an indirect effect [13,38].
Similar mechanisms have recently been identified in the dimorphic fungus Candida albicans. During filamentous growth, apical transport of secretory vesicles (driven by Myo2) supports the asymmetric localization of polarity proteins including the small GTPase Sec4 and its guanine nucleotide exchange factor Sec2 [13,39]. Sec2 protein is capable of binding its own mRNA on the vesicle membrane, and this binding is regulated by phosphorylation [39]. The regulated binding of Sec2 to its own mRNA likely ensures precise spatiotemporal regulation of translation near sites of apical growth [39], but the specific mechanisms of how this is achieved are unclear.
Organelle Hitchhiking
The adaptor proteins that mediate motor recruitment to many organelles are unknown. The fact that non-membranous cargos such as mRNP complexes hitchhike on endosomes and other organelles set up an intriguing question: can membrane-bound organelles also hitchhike? Two recent studies in the filamentous fungi Aspergillus nidulans and U. maydis (Box 2) demonstrated that this is indeed the case for peroxisomes, lipid droplets, and ER [40,41].
Peroxisomes Move by Hitchhiking on Early Endosomes
Peroxisomes are single-membrane enclosed organelles that have diverse functions in different species, but are conserved in their ability to breakdown long-chain fatty acids and metabolize reactive oxygen species such as hydrogen peroxide [42]. Long-range peroxisome movement is dependent on microtubule-based transport in many eukaryotes, with the exception of plants and budding yeast which use actin-based mechanisms [43]. In both filamentous fungi and animal cells, the majority of peroxisomes undergo oscillatory and diffusive movements with little to no overall displacement, while the remaining ~5–20% exhibit long-range bidirectional movements [40,43,44]. To date, no microtubule-based cargo adaptor for peroxisomes has been identified [43], but their movement and distribution require microtubules and microtubule-based motors [20,45–49].
Surprisingly, two recent studies in filamentous fungi demonstrated that long-range movement of peroxisomes required the motility of EEs, providing the first examples of organelle hitchhiking [40,41]. In both U. maydis and A. nidulans the subset of moving peroxisomes comigrate with EEs; this is in contrast to Rrm4 where nearly all puncta colocalize with moving EEs. In addition, EEs lead peroxisomes during co-movement, suggesting that EEs dictate the directional movement of peroxisomes. Perturbations that specifically disrupt EE function and movement also affect the movement and distribution of peroxisomes [41,50].
Identification of the Early Endosome–Peroxisome Tether
As described above, for some forms of mRNA hitchhiking, the tethering proteins have been identified (Figure 1A). What tether(s) mediates organelle hitchhiking? Recently a large coiled coil-containing protein named PxdA (peroxisome distribution mutant A) was shown to link peroxisomes to EEs in A. nidulans [40]. The pxdA gene is required for peroxisome motility, but not for movement of other microtubule motor-based cargos such as nuclei and endosomes. PxdA protein colocalizes with moving EEs and comigrates with the subset of moving peroxisomes. A C-terminal region of the long coiled coil of PxdA mediates EE association, and removal of this sequence causes defects in the movement and distribution of peroxisomes. Together these data strongly support a role for PxdA in mediating the association between EEs and peroxisomes.
The details of how PxdA mediates hitchhiking remain to be determined. One possibility is that the coiled coil of PxdA acts as a physical tether that directly couples EEs to peroxisomes. The region of PxdA that mediates EE interaction contains a BAR domain, which is a module capable of binding to curved membranes including endosomes [51]. The identity of other proteins that mediate the interaction of PxdA with either EEs or peroxisomes is not yet known. It is also formally possible that PxdA is not a physical tether but instead marks a subpopulation of EEs (PxdA is not found on all EEs [40]) that interact with peroxisomes. In this case an additional factor (s) would serve as the tether between the two organelles.
Other Hitchhiking Organelles in Filamentous Fungi
Recent work in U. maydis demonstrated that both lipid droplets and ER, but not mitochondria, also comigrate with moving EEs [41]. As with peroxisomes, EEs lead lipid droplet and ER movements, suggesting that these organelles also hitchhike on EEs [41]. Peroxisomes, lipid droplets, and ER appear to independently hitchhike on EEs because the three hitchhiking cargos do not comigrate with each other [41]. It is not yet known if PxdA is also required for hitchhiking of lipid droplets and ER, or if other tethering proteins are involved.
Why do Organelles Hitchhike on Early Endosomes?
Why do peroxisomes, lipid droplets, and ER use this non-canonical mode of microtubule-based transport to achieve motility? All three organelles have roles in lipid homeostasis, and have been shown to interact for metabolic exchange [52,53]. One possibility is that motile EEs facilitate these interactions by serving as an assembly platform for cargo, similar to the assembly of Rrm4-bound mRNAs with ribosomes. However, as described above, ER, lipid droplets, and peroxisomes move independently of each other during hitchhiking on EEs [41]. Another possibility is that bidirectionally-moving EEs serve as a convenient platform to achieve a uniform distribution of organelles throughout the cell. In the case of peroxisomes, homogenous mixing would aid in guarding against toxic reactive oxygen species and catabolizing fatty acids in a spatiotemporal manner [43,54], as well as combating hyphal lysis in A. nidulans (Box 2). In support, mathematical modeling revealed that EE motility is essential to achieve uniform mixing of peroxisomes in hyphae [55].
Organelle Dynamics Are Coupled in Mammalian Cells
Endosomes and Mitochondria Associate with Dynamic ER Movements
Similarly to the ER in fungal cells, the mammalian ER is dynamic. Two types of movements, ER tubule sliding and ER ring rearrangements occur along microtubules and are dependent on molecular motors [56]. Two recent studies in mammalian cells demonstrate that mitochondria and endosomes associate with the ER during these dynamic movements [57,58]. These endosomes comigrate with the ER and their movements are coupled; this interaction likely facilitates endosomal fission and maturation [59]. However, further work will be necessary to determine if endosomes dictate the long-distance movement of ER in mammalian cells and represent hitchhiking as we have defined it (see above).
Endosomes Share Their Motors with Autophagosomes and Internalized Exosomes
As we have defined hitchhiking, cargos transiently connect to vehicle cargos, and, in the case of membranous cargos, do not fuse with each other. Two recent examples do not fit these criteria because the membranes of the interacting organelles fuse, but we highlight them here because one organelle shares its transport machinery with another to achieve motility. In the first example, neuronal autophagosomes fuse with late endosomes [60], leading to dynein-based motility towards the lysosome [61]. Disrupting this fusion decreases autophagosome movement [61]. In the second example, exosomes are endocytosed and encapsulated in endosomes as intact vesicles; this is important for their movement and subsequent fusion with lysosomes [62].
Concluding Remarks and Future Directions
Hitchhiking is a non-canonical mode of intracellular transport that has been described primarily in fungi (Table 1). Although bona fide hitchhiking has yet to be observed in mammalian cells, it likely exists given that dynamic interactions between organelles at membrane contact sites have been identified in a variety of different cell types [63]. Future research will need to address this and other important questions (see Outstanding Questions). While the biological purpose of hitchhiking remains unclear, it has been suggested that endosomes act as multipurpose platforms [12,64]. Why are endosomes a vehicle used for hitchhiking? One possibility is that bidirectionally-moving endosomes can act as a convenient mixing method to evenly distribute cargo throughout the cell [24,55]. Another is that endosomes receive extracellular signals via endocytosis, and this could drive transport to particular regions of the cell. Finally, co-movement of cargo may facilitate the interactions at membrane contact sites (i.e., assembly platforms); these sites are important for organelle maturation and metabolic exchange of lipids and ions [63]. In addition to endosomes, other vesicles and cortical ER serve as vehicles during hitchhiking. Identifying other hitchhiking organelles, elucidating the molecular mechanisms involved in hitchhiking, and exploring the biological significance of hitchhiking is important future work.
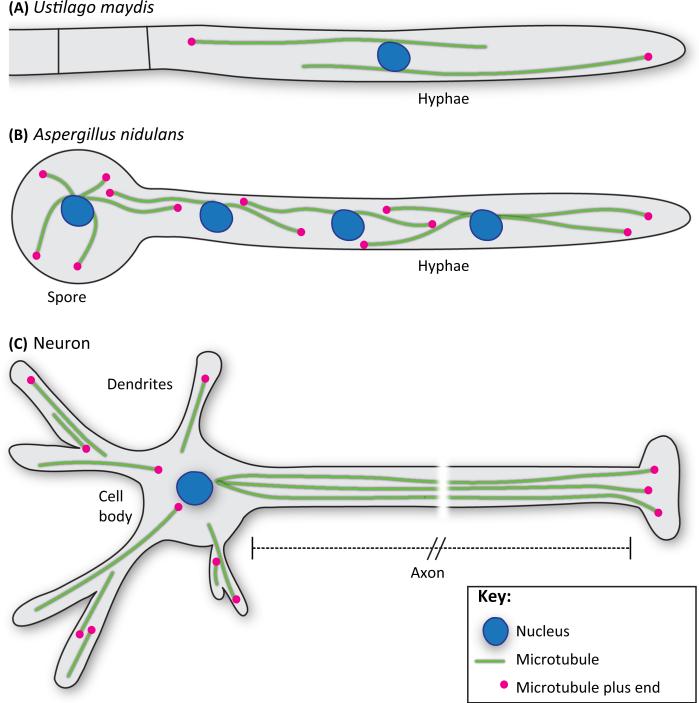
Figure I. Filamentous Fungi are Model Polarized Cells.
(A) Ustilago maydis is a pathogenic fungus that causes corn smut. Before infection it switches from yeast-like growth to filamentous growth. Researchers in the transport field primarily study haploid U. maydis that have been induced (via activation of a transcription factor) to grow in the filamentous, unipolar form. (B) A. nidulans has been a model organism in the transport field since classical genetic screens identified several genes required for mitosis and nuclear positioning [79,84,85]. Most researchers in the field perform experiments on haploid hyphae. Both U. maydis and A. nidulans have uniformly polarized microtubules close to the hyphal tip; therefore the directionality of cargos and motors can easily be determined in this region. (C) Neurons also have highly polarized microtubule cytoskeletons, particularly in axons, where plus-ends are located at the axon terminus.
Trends.
Microtubule-based transport can be achieved by ‘hitchhiking’, where some cargos move by connecting to motile ‘vehicle’ cargos.
Hitchhiking cargos include mRNA, proteins, peroxisomes, endoplasmic reticulum, and lipid droplets. Vehicle cargo includes endosomes and the endoplasmic reticulum.
PxdA and Upa1 are linker proteins that mediate endosomal hitchhiking of peroxisomes and mRNAs, respectively.
Hitchhiking has been described primarily in fungi, but emerging evidence suggests that organelle dynamics may also be coupled in mammalian cells.
Box 1. Microtubule-Based Motors and Cargo Adaptors.
Dynein and kinesins are responsible for the movement of endosomes and other cargos [7,10]. These motors use ATP hydrolysis to move along microtubules. Dynein is a ~1.4 MDa complex composed of two heavy chains, containing the ATP- and microtubule-binding domains, as well as two copies of intermediate chains, light intermediate chains, and dimers of three different light chains [2]. Dynein also interacts with the dynactin complex (the mammalian complex contains 23 polypeptides), which is required for most dynein functions. Mammalian dynein, unlike the well-characterized S. cerevisiae dynein [65], requires dynactin and a coiled-coil cargo adaptor (or ‘activator’) to achieve processive motility [66,67]. Five activators have been confirmed so far: Bicaudal-D2 (BicD2), Hook3, Hook1, Spindly, and Rab11–FIP3 [66–68]. While some cargo adaptors serve as activators, others are involved only in recruiting dynein to its cargo [7,8].
The mammalian kinesin superfamily contains 45 genes separated into 15 subfamilies [1,10]. Kinesin-1, -2, and -3 (encoded by 15 genes) are responsible for the majority of microtubule-based transport towards the microtubule plus-end [1]. Kinesin-1 is a heterotetramer consisting of two kinesin heavy chains, which contain the microtubule- and ATP-binding domains, and two kinesin light chains. The kinesin-2 family assembles as a dimer and interacts with kinesin-associated protein/KAP. Kinesin-3 also likely functions as a dimer in vivo [1]. Members of each of these three kinesin families have been shown to exist in an autoinhibited state, which is relieved upon cargo adaptor binding [1].
In canonical trafficking, both kinesin and dynein use adaptor proteins to associate with their cargos. There is a large literature on this topic (reviewed in [2,7–11]); we focus here on endosomes, a key vehicle organelle for hitchhiking in filamentous fungi. Distinct populations of endosomes are marked by small GTPases of the Rab family (>60 proteins in humans), which recruit effectors important for vesicle function, formation, and maturation [69]. Distinct populations of endosomes are marked by different Rabs, including early (Rab4/5/22), late (Rab7/9), and recycling endosomes (Rab11). In some examples, Rab effectors recruit the motor machinery to endosomes and other vesicles. For example, late endosomes associate with the cargo-adaptors RILP (Rab7-interacting lysosomal protein) and snapin (SNARE-associated protein), which subsequently interact with p150 and dynein IC, respectively [70–72]. Early and recycling endosomes associate with Hook-family proteins, which can recruit both kinesin-3 and dynein/dynactin [50,66,73–75]. Hook proteins associate with endosomes via a larger protein complex that contains fused toes (FTS) and fused toes and hook interacting protein (FHIP) proteins [74,76,77]. Rab6-positive vesicles (predominantly involved in Golgi trafficking) associate with various members of the BicD family, which interact with dynein/dynactin and kinesin-1 and -3 [8]. The array of cargo-adaptors has likely evolved to distinguish between different molecular motors and vesicle subtypes, as well as to execute diverse spatiotemporal requirements among vesicle populations.
Box 2. Filamentous Fungi: Polarized Model Systems for Studying Microtubule-based Transport.
The filamentous fungi Aspergillus nidulans and Ustilago maydis (which also has a yeast-like form in its life cycle) are well-studied polarized model organisms that are genetically and biochemically tractable, as well as amenable to live-cell imaging [78]. Unlike budding yeast, which predominantly uses actin and myosin for long-distance transport, both A. nidulans and U. maydis are more similar to metazoans and use microtubules, dynein, and kinesin for long-distance cargo motility [78]. Furthermore, their genomes include conserved dynein and dynactin complexes, the dynein activator Hook, and cargo-carrying kinesin-3s and kinesin-1 [11,50,74,78].
Filamentous fungi have long hyphal compartments, which make them ideal model polarized cells, reminiscent of some metazoan cells including neurons. Similarly to neuronal axons, hyphae contain uniformly polarized microtubules at the apical poles (hyphal tip; Figure IA,B). Neuronal dendrites and regions of hyphae away from the poles contain microtubules of mixed polarity (Figure I). During filamentous growth the long hyphae of A. nidulans and U. maydis elongate by apical extension at the polar tip [29,79]. A combination of endocytosis, membrane insertion, and secretion is crucial for polarized growth [29,79]. These processes require EEs because perturbations in their movement, function, or maturation cause defects in hyphal length and colony formation [11,29,64,78,79]. In U. maydis, EE motility is required for early plant infection (leading to corn smut) by facilitating the transcription (via retrograde signaling) and secretion (via anterograde signaling) of effectors crucial for suppressing the host defense reaction [80]. A single kinesin-3 and dynein motor, as well as the cargo adaptor Hook, are responsible for the movement of EEs and vesicles along hyphae [19–21,50,74]. As described in Box 1, Hook proteins (HookA in A. nidulans and Hok1 in U. maydis) associate with endosomes via the homologs of FTS and FHIP [74,76].
Peroxisomes are also important for the polarized growth and maintenance of hyphae. Peroxisomes are crucial for filamentous growth when the sole carbon source comprises fatty acids such as butyrate [81]. In some filamentous fungi, including A. nidulans, a peroxisome-derived organelle called the Woronin body is crucial for plugging the pores of septa in response to hyphal wounding or lysis [82]. Another subclass of peroxisomes has also been shown to support the localization of ApsB, a protein crucial for the formation of microtubule-organizing centers (MTOCs) at septa in A. nidulans [83].
Outstanding Questions.
Why do some cargos use hitchhiking rather than traditional modes of microtubule-based transport?
What is the biological purpose of hitchhiking?
How does PxdA link early endosomes to peroxisomes?
What tethers and tethering complexes mediate hitchhiking of lipid droplets and ER?
What are the signals that initiate and stop hitchhiking?
Do other cargos hitchhike?
Is hitchhiking conserved in mammalian cells?
Acknowledgments
We would like to thank Morgan DeSantis, William Bret Redwine, and Vivek S. Gowda for helpful feedback on this review.
Glossary
- Autophagosome
double-membrane enclosed organelle that delivers damaged or toxic proteins, organelles, and other cytosolic materials to the lysosome for degradation during autophagy.
- COPI (coat protein) vesicles
vesicles derived from the Golgi apparatus containing coat protein complexes that mainly carry proteins from the Golgi to the ER (retrogradely) and between Golgi compartments.
- Cortical endoplasmic reticulum
specialized endoplasmic reticulum (ER) in yeast, plants and some metazoans whose tubules and cisternae physically associate with the cell cortex. In S. cerevisiae, cortical ER moves into the bud in a She3–Myo4-dependent manner and is retained there (cortical ER inheritance) during cell division.
- ER ring rearrangements
dynamic ring-like structures that propagate along ER tubules [57].
- ER tubule sliding
ER tubules slide along microtubules in a kinesin/dynein-dependent manner [58]
- Exosome
extracellular vesicles containing proteins and RNA important for cell–cell communication in a variety of organisms. They are released by a host cell and can induce signaling in a recipient cell [62].
- FYVE domain
zinc-finger domain that inserts into membranes including endosomes; named after localization sequences from four proteins: Fab1, YOTB, Vac1, and EEA1.
- Hypha (plural, hyphae)
in filamentous fungi, hyphae are long, branched filaments enclosed within a chitin-containing cell wall and plasma membrane. Hyphae exhibit filamentous growth at the cellular apex, including the addition of cell wall and membrane. A typical hypha is divided into multiple cells along its length by septa.
- Lipid droplet
organelle that stores neutral lipids and sequesters toxic lipids. It is crucial for lipid metabolism and energy homeostasis. They have also been shown to participate in fatty acid trafficking and act as an assembly platform for proteins in the immune system. Lipid droplet perturbations have been linked to neurodegenerative diseases [86].
- Lysosome
large organelle that contains degradative enzymes.
- Membrane contact site
membranes from two cargos tethered in close apposition (<30 nm). These sites exist in organisms ranging from budding yeast to mammalian cells [63].
- Polysome
a group of translationally-active ribosomes attached to an mRNA.
- Processive motility
the ability of a single motor molecule to take consecutive steps along its cytoskeletal track.
- Septa
internal cross-walls or pores along hyphae that form at regular intervals and partition the hypha into distinct cells. These septa do not completely close, leaving a small pore between hyphal compartments, and are capable of cytosolic exchange of most organelles and other intracellular components.
- Septins
GTP-binding proteins that form cytoskeletal filaments or rings and help to compartmentalize subcellular regions. In U. maydis, septins Cdc3, Cdc10, Cdc11, and Cdc12 are crucial for unipolar filamentous growth. They assemble as hetero-octamers that are building blocks for higher-order filaments that form a gradient emanating from the hyphal tip [28].
- Small GTPase
small cytosolic G-proteins (~20–25 kDa) homologous to Ras that hydrolyze GTP on membranes. Its GDP- (‘off’) and GTP- (‘on’) bound states act as a bidirectional switch capable of activating downstream effectors during signaling.
Footnotes
Uncited Reference
[86]
References
- 1.Verhey KJ, et al. Kinesin assembly and movement in cells. Annu. Rev. Biophys. 2011;40:267–288. doi: 10.1146/annurev-biophys-042910-155310. [DOI] [PubMed] [Google Scholar]
- 2.Cianfrocco MA, et al. Mechanism and regulation of cytoplasmic dynein. Annu. Rev. Cell Dev. Biol. 2015;31:83–108. doi: 10.1146/annurev-cellbio-100814-125438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Franker MA, Hoogenraad CC. Microtubule-based transport – basic mechanisms, traffic rules and role in neurological pathogenesis. J. Cell Sci. 2013;126:2319–2329. doi: 10.1242/jcs.115030. [DOI] [PubMed] [Google Scholar]
- 4.Millecamps S, Julien JP. Axonal transport deficits and neurodegenerative diseases. Nat. Rev. Neurosci. 2013;14:161–176. doi: 10.1038/nrn3380. [DOI] [PubMed] [Google Scholar]
- 5.Schiavo G, et al. Cytoplasmic dynein heavy chain: the servant of many masters. Trends Neurosci. 2013;36:641–651. doi: 10.1016/j.tins.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maday S, et al. Axonal transport: cargo-specific mechanisms of motility and regulation. Neuron. 2014;84:292–309. doi: 10.1016/j.neuron.2014.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fu MM, Holzbaur EL. Integrated regulation of motor-driven organelle transport by scaffolding proteins. Trends Cell Biol. 2014;24:564–574. doi: 10.1016/j.tcb.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoogenraad CC, Akhmanova A. Bicaudal D ramily of motor adaptors: linking dynein motility to cargo binding. Trends Cell Biol. 2016;26:327–340. doi: 10.1016/j.tcb.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 9.Akhmanova A, Hammer JA., 3rd Linking molecular motors to membrane cargo. Curr. Opin. Cell Biol. 2010;22:479–487. doi: 10.1016/j.ceb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirokawa N, et al. Kinesin superfamily motor proteins and intracellular transport. Nat. Rev. Mol. Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- 11.Xiang X, et al. Cytoplasmic dynein and early endosome transport. Cell Mol. Life Sci. 2015;72:3267–3280. doi: 10.1007/s00018-015-1926-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gould GW, Lippincott-Schwartz J. New roles for endosomes: from vesicular carriers to multi-purpose platforms. Nat. Rev. Mol. Cell Biol. 2009;10:287–292. doi: 10.1038/nrm2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haag C, et al. Membrane-coupled mRNA trafficking in fungi. Annu. Rev. Microbiol. 2015;69:265–281. doi: 10.1146/annurev-micro-091014-104242. [DOI] [PubMed] [Google Scholar]
- 14.Jansen RP, et al. mRNA transport meets membrane traffic. Trends Genet. 2014;30:408–417. doi: 10.1016/j.tig.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Dreyfuss G, et al. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- 16.Bullock SL. Messengers, motors and mysteries: sorting of eukaryotic mRNAs by cytoskeletal transport. Biochem. Soc. Trans. 2011;39:1161–1165. doi: 10.1042/BST0391161. [DOI] [PubMed] [Google Scholar]
- 17.Bullock SL, et al. Guidance of bidirectional motor complexes by mRNA cargoes through control of dynein number and activity. Curr. Biol. 2006;16:1447–1452. doi: 10.1016/j.cub.2006.05.055. [DOI] [PubMed] [Google Scholar]
- 18.Navarro C, et al. Egalitarian binds dynein light chain to establish oocyte polarity and maintain oocyte fate. Nat. Cell Biol. 2004;6:427–435. doi: 10.1038/ncb1122. [DOI] [PubMed] [Google Scholar]
- 19.Abenza JF, et al. Long-distance movement of Aspergillus nidulans early endosomes on microtubule tracks. Traffic. 2009;10:57–75. doi: 10.1111/j.1600-0854.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- 20.Egan MJ, et al. Lis1 is an initiation factor for dynein-driven organelle transport. J. Cell Biol. 2012;197:971–982. doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wedlich-Soldner R, et al. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 2002;21:2946–2957. doi: 10.1093/emboj/cdf296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baumann S, et al. Endosomal transport of septin mRNA and protein indicates local translation on endosomes and is required for correct septin filamentation. EMBO Rep. 2014;15:94–102. doi: 10.1002/embr.201338037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baumann S, et al. Kinesin-3 and dynein mediate microtubule-dependent co-transport of mRNPs and endosomes. J. Cell Sci. 2012;125(Pt 11):2740–2752. doi: 10.1242/jcs.101212. [DOI] [PubMed] [Google Scholar]
- 24.Higuchi Y, et al. Early endosome motility spatially organizes polysome distribution. J. Cell Biol. 2014;204:343–357. doi: 10.1083/jcb.201307164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Konig J, et al. The fungal RNA-binding protein Rrm4 mediates long-distance transport of ubi1 and rho3 mRNAs. EMBO J. 2009;28:1855–1866. doi: 10.1038/emboj.2009.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Becht P, et al. Role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis. Eukaryot Cell. 2005;4:121–133. doi: 10.1128/EC.4.1.121-133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohlmann T, et al. A FYVE zinc finger domain protein specifically links mRNA transport to endosome trafficking. Elife. 2015;4:e06041. doi: 10.7554/eLife.06041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zander S, et al. Endosomal assembly and transport of heteromeric septin complexes promote septin cytoskeleton formation. J. Cell Sci. 2016;129:2778–2792. doi: 10.1242/jcs.182824. [DOI] [PubMed] [Google Scholar]
- 29.Steinberg G, Perez-Martin J. Ustilago maydis, a new fungal model system for cell biology. Trends Cell Biol. 2008;18:61–67. doi: 10.1016/j.tcb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Hammer JA, 3rd, Sellers JR. Walking to work: roles for class V myosins as cargo transporters. Nat. Rev. Mol. Cell Biol. 2012;13:13–26. doi: 10.1038/nrm3248. [DOI] [PubMed] [Google Scholar]
- 31.Estrada P, et al. Myo4p and She3p are required for cortical ER inheritance in Saccharomyces cerevisiae. J. Cell Biol. 2003;163:1255–1266. doi: 10.1083/jcb.200304030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmid M, et al. Coordination of endoplasmic reticulum and mRNA localization to the yeast bud. Curr. Biol. 2006;16:1538–1543. doi: 10.1016/j.cub.2006.06.025. [DOI] [PubMed] [Google Scholar]
- 33.Aronov S, et al. mRNAs encoding polarity and exocytosis factors are cotransported with the cortical endoplasmic reticulum to the incipient bud in Saccharomyces cerevisiae. Mol. Cell Biol. 2007;27:3441–3455. doi: 10.1128/MCB.01643-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fundakowski J, et al. Localization of a subset of yeast mRNAs depends on inheritance of endoplasmic reticulum. Traffic. 2012;13:1642–1652. doi: 10.1111/tra.12011. [DOI] [PubMed] [Google Scholar]
- 35.Genz C, et al. Association of the yeast RNA-binding protein She2p with the tubular endoplasmic reticulum depends on membrane curvature. J. Biol. Chem. 2013;288:32384–32393. doi: 10.1074/jbc.M113.486431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trautwein M, et al. Arf1p provides an unexpected link between COPI vesicles and mRNA in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:5021–5037. doi: 10.1091/mbc.E04-05-0411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pon LA. Golgi inheritance: rab rides the coat-tails. Curr. Biol. 2008;18:R743–R745. doi: 10.1016/j.cub.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 38.Cohen RS. The role of membranes and membrane trafficking in RNA localization. Biol. Cell. 2005;97:5–18. doi: 10.1042/BC20040056. [DOI] [PubMed] [Google Scholar]
- 39.Caballero-Lima D, et al. In Candida albicans hyphae, Sec2p is physically associated with SEC2 mRNA on secretory vesicles. Mol. Microbiol. 2014;94:828–842. doi: 10.1111/mmi.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salogiannis J, et al. Peroxisomes move by hitchhiking on early endosomes using the novel linker protein PxdA. J. Cell Biol. 2016;212:289–296. doi: 10.1083/jcb.201512020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guimaraes SC, et al. Peroxisomes, lipid droplets, and endoplasmic reticulum ‘hitchhike’ on motile early endosomes. J. Cell Biol. 2015;211:945–954. doi: 10.1083/jcb.201505086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Islinger M, et al. Be different – the diversity of peroxisomes in the animal kingdom. Biochim. Biophys. Acta. 2010;1803:881–897. doi: 10.1016/j.bbamcr.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 43.Neuhaus A, et al. Why do peroxisomes associate with the cytoskeleton? Biochim. Biophys. Acta. 2016;1863:1019–1026. doi: 10.1016/j.bbamcr.2015.11.022. [DOI] [PubMed] [Google Scholar]
- 44.Rapp S, et al. Microtubule-based peroxisome movement. J. Cell Sci. 1996;109(Pt 4):837–849. doi: 10.1242/jcs.109.4.837. [DOI] [PubMed] [Google Scholar]
- 45.Kural C, et al. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 2005;308:1469–1472. doi: 10.1126/science.1108408. [DOI] [PubMed] [Google Scholar]
- 46.Dietrich D, et al. Identification of the kinesin KifC3 as a new player for positioning of peroxisomes and other organelles in mammalian cells. Biochim. Biophys. Acta. 2013;1833:3013–3024. doi: 10.1016/j.bbamcr.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Schrader M, et al. Real time imaging reveals a peroxisomal reticulum in living cells. J. Cell Sci. 2000;113(Pt 20):3663–3671. doi: 10.1242/jcs.113.20.3663. [DOI] [PubMed] [Google Scholar]
- 48.Ally S, et al. Opposite-polarity motors activate one another to trigger cargo transport in live cells. J. Cell Biol. 2009;187:1071–1082. doi: 10.1083/jcb.200908075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kulic IM, et al. The role of microtubule movement in bidirectional organelle transport. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10011–10016. doi: 10.1073/pnas.0800031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang J, et al. HookA is a novel dynein–early endosome linker critical for cargo movement in vivo. J. Cell Biol. 2014;204:1009–1026. doi: 10.1083/jcb.201308009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Weering JR, Cullen PJ. Membrane-associated cargo recycling by tubule-based endosomal sorting. Semin. Cell Dev. Biol. 2014;31:40–47. doi: 10.1016/j.semcdb.2014.03.015. [DOI] [PubMed] [Google Scholar]
- 52.Schrader M, et al. Peroxisome interactions and cross-talk with other subcellular compartments in animal cells. Subcell Biochem. 2013;69:1–22. doi: 10.1007/978-94-007-6889-5_1. [DOI] [PubMed] [Google Scholar]
- 53.Barbosa AD, et al. Lipid drople–organelle interactions: emerging roles in lipid metabolism. Curr. Opin. Cell Biol. 2015;35:91–97. doi: 10.1016/j.ceb.2015.04.017. [DOI] [PubMed] [Google Scholar]
- 54.Schrader M, et al. Peroxisomal motility and interaction with microtubules. Microsc. Res. Tech. 2003;61:171–178. doi: 10.1002/jemt.10326. [DOI] [PubMed] [Google Scholar]
- 55.Lin C, et al. Active diffusion and microtubule-based transport oppose myosin forces to position organelles in cells. Nat. Commun. 2016;7:11814. doi: 10.1038/ncomms11814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wozniak MJ, et al. Role of kinesin-1 and cytoplasmic dynein in endoplasmic reticulum movement in VERO cells. J. Cell Sci. 2009;122(Pt 12):1979–1989. doi: 10.1242/jcs.041962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Friedman JR, et al. Endoplasmic reticulum–endosome contact increases as endosomes traffic and mature. Mol. Biol. Cell. 2013;24:1030–1040. doi: 10.1091/mbc.E12-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Friedman JR, et al. ER sliding dynamics and ER–mitochondrial contacts occur on acetylated microtubules. J. Cell Biol. 2010;190:363–375. doi: 10.1083/jcb.200911024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rowland AA, et al. ER contact sites define the position and timing of endosome fission. Cell. 2014;159:1027–1041. doi: 10.1016/j.cell.2014.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Maday S, Holzbaur EL. Autophagosome assembly and cargo capture in the distal axon. Autophagy. 2012;8:858–860. doi: 10.4161/auto.20055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng XT, et al. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 2015;209:377–386. doi: 10.1083/jcb.201412046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heusermann W, et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J. Cell Biol. 2016;213:173–184. doi: 10.1083/jcb.201506084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prinz WA. Bridging the gap: membrane contact sites in signaling, metabolism, and organelle dynamics. J. Cell Biol. 2014;205:759–769. doi: 10.1083/jcb.201401126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gohre V, et al. Microtubule-dependent membrane dynamics in Ustilago maydis: trafficking and function of Rab5a-positive endosomes. Commun. Integr. Biol. 2012;5:485–490. doi: 10.4161/cib.21219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reck-Peterson SL, et al. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenney RJ, et al. Activation of cytoplasmic dynein motility by dynactin–cargo adapter complexes. Science. 2014;345:337–341. doi: 10.1126/science.1254198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schlager MA, et al. In vitro reconstitution of a highly processive recombinant human dynein complex. EMBO J. 2014;33:1855–1868. doi: 10.15252/embj.201488792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olenick MA, et al. Hook adaptors induce unidirectional processive motility by enhancing the dynein–dynactin interaction. J. Biol. Chem. 2016;291:18239–18251. doi: 10.1074/jbc.M116.738211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hutagalung AH, Novick PJ. Role of Rab GTPases in membrane traffic and cell physiology. Physiol. Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Johansson M, et al. Activation of endosomal dynein motors by stepwise assembly of Rab7–RILP–p150Glued, ORP1L, and the receptor betalll spectrin. J. Cell Biol. 2007;176:459–471. doi: 10.1083/jcb.200606077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cantalupo G, et al. Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 2001;20:683–693. doi: 10.1093/emboj/20.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cai Q, et al. Snapin-regulated late endosomal transport is critical for efficient autophagy–lysosomal function in neurons. Neuron. 2010;68:73–86. doi: 10.1016/j.neuron.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luiro K, et al. Interconnections of CLN3, Hook1 and Rab proteins link Batten disease to defects in the endocytic pathway. Hum. Mol. Genet. 2004;13:3017–3027. doi: 10.1093/hmg/ddh321. [DOI] [PubMed] [Google Scholar]
- 74.Bielska E, et al. Hook is an adapter that coordinates kinesin-3 and dynein cargo attachment on early endosomes. J. Cell Biol. 2014;204:989–1007. doi: 10.1083/jcb.201309022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maldonado-Baez L, Donaldson JG. Hook1, microtubules, and Rab22: mediators of selective sorting of clathrin-independent endocytic cargo proteins on endosomes. Bioarchitecture. 2013;3:141–146. doi: 10.4161/bioa.26638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao X, et al. FHIP and FTS proteins are critical for dynein-mediated transport of early endosomes in Aspergillus. Mol. Biol. Cell. 2014;25:2181–2189. doi: 10.1091/mbc.E14-04-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xu L, et al. An FTS/Hook/p107(FHIP) complex interacts with and promotes endosomal clustering by the homotypic vacuolar protein sorting complex. Mol. Biol. Cell. 2008;19:5059–5071. doi: 10.1091/mbc.E08-05-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Egan MJ, et al. Microtubule-based transport in filamentous fungi. Curr. Opin. Microbiol. 2012;15:637–645. doi: 10.1016/j.mib.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Penalva MA, et al. Searching for gold beyond mitosis: mining intracellular membrane traffic in Aspergillus nidulans. Cell Logist. 2012;2:2–14. doi: 10.4161/cl.19304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bielska E, et al. Long-distance endosome trafficking drives fungal effector production during plant infection. Nat. Commun. 2014;5:5097. doi: 10.1038/ncomms6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hynes MJ, et al. Genetic analysis of the role of peroxisomes in the utilization of acetate and fatty acids in Aspergillus nidulans. Genetics. 2008;178:1355–1369. doi: 10.1534/genetics.107.085795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Liu F, et al. Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J. Cell Biol. 2008;180:325–339. doi: 10.1083/jcb.200705049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zekert N, et al. Interaction of the Aspergillus nidulans microtubule-organizing center (MTOC) component ApsB with gamma-tubulin and evidence for a role of a subclass of peroxisomes in the formation of septal MTOCs. Eukaryot Cell. 2010;9:795–805. doi: 10.1128/EC.00058-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiang X, et al. Cytoplasmic dynein is involved in nuclear migration in Aspergillus nidulans. Proc. Natl. Acad. Sci. U.S.A. 1994;91:2100–2104. doi: 10.1073/pnas.91.6.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xiang X, et al. NudF, a nuclear migration gene in Aspergillus nidulans, is similar to the human LIS-1 gene required for neuronal migration. Mol. Biol. Cell. 1995;6:297–310. doi: 10.1091/mbc.6.3.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Welte MA. Expanding roles for lipid droplets. Curr. Biol. 2015;25:R470–R481. doi: 10.1016/j.cub.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]