Abstract
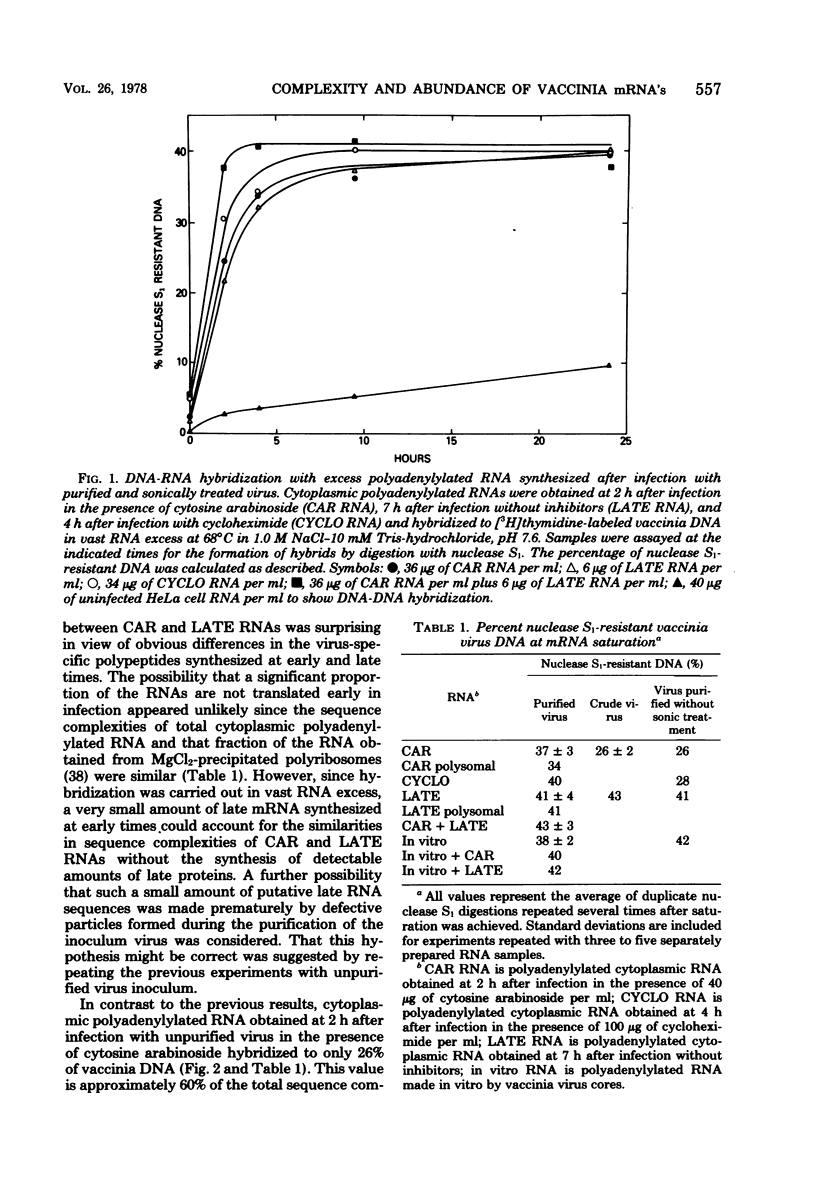
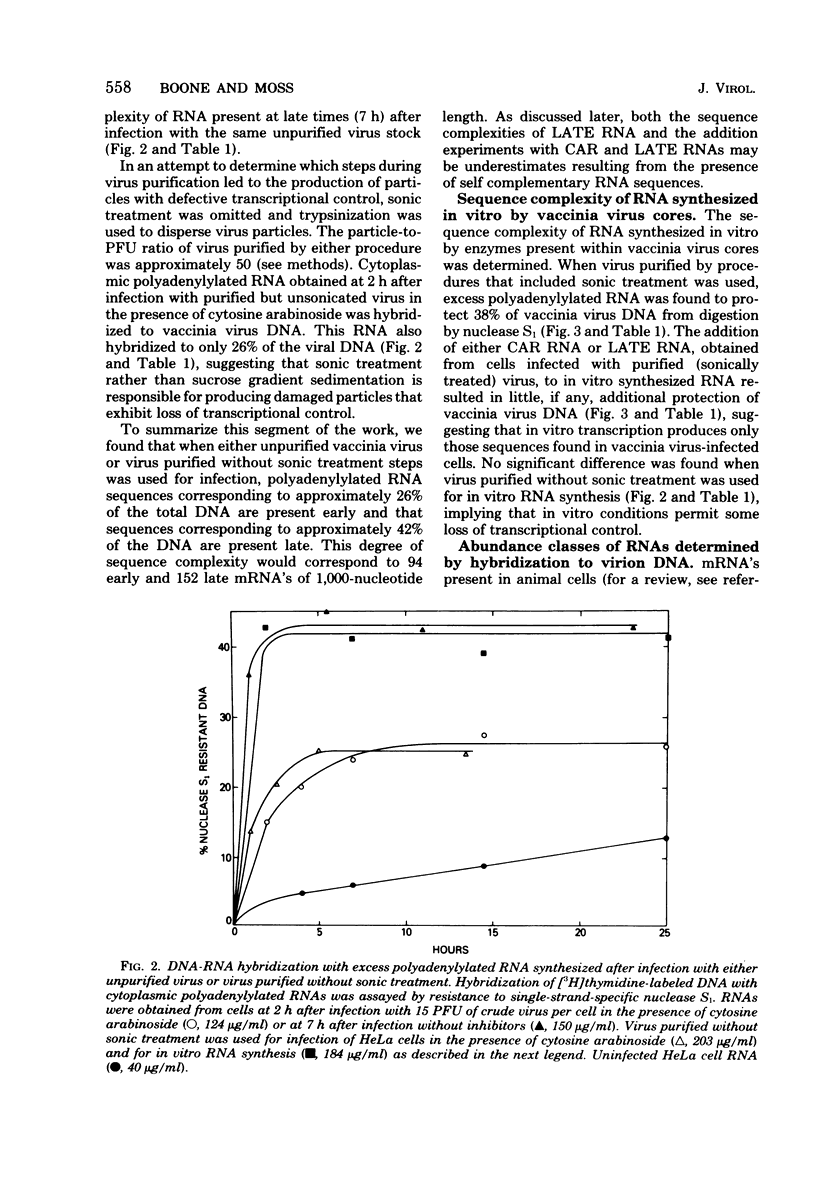
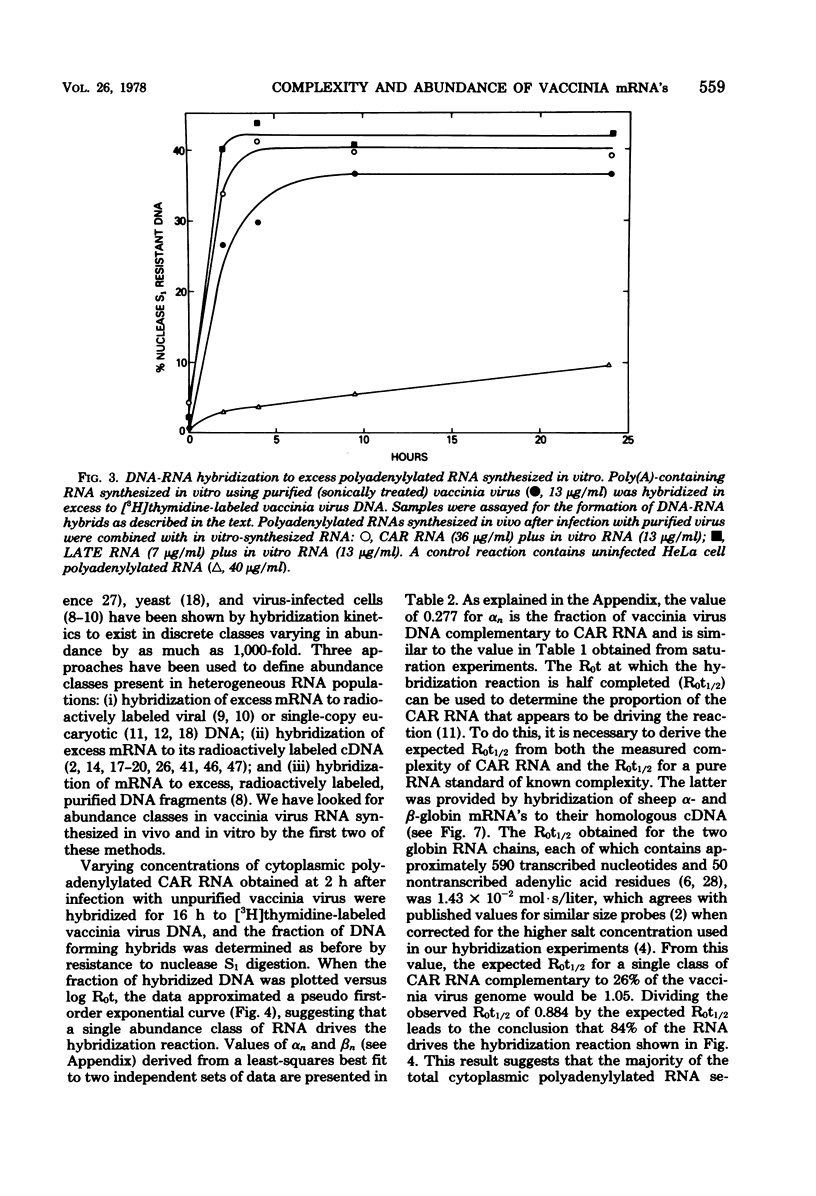
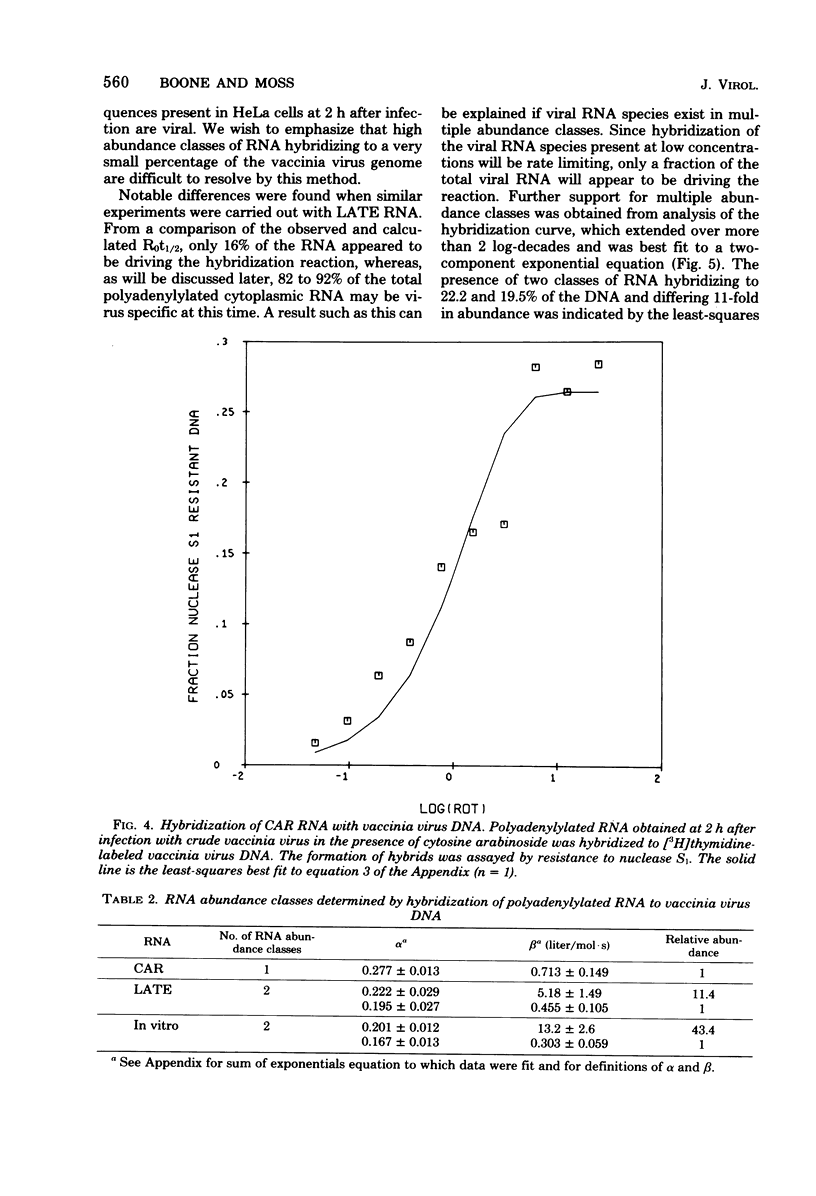
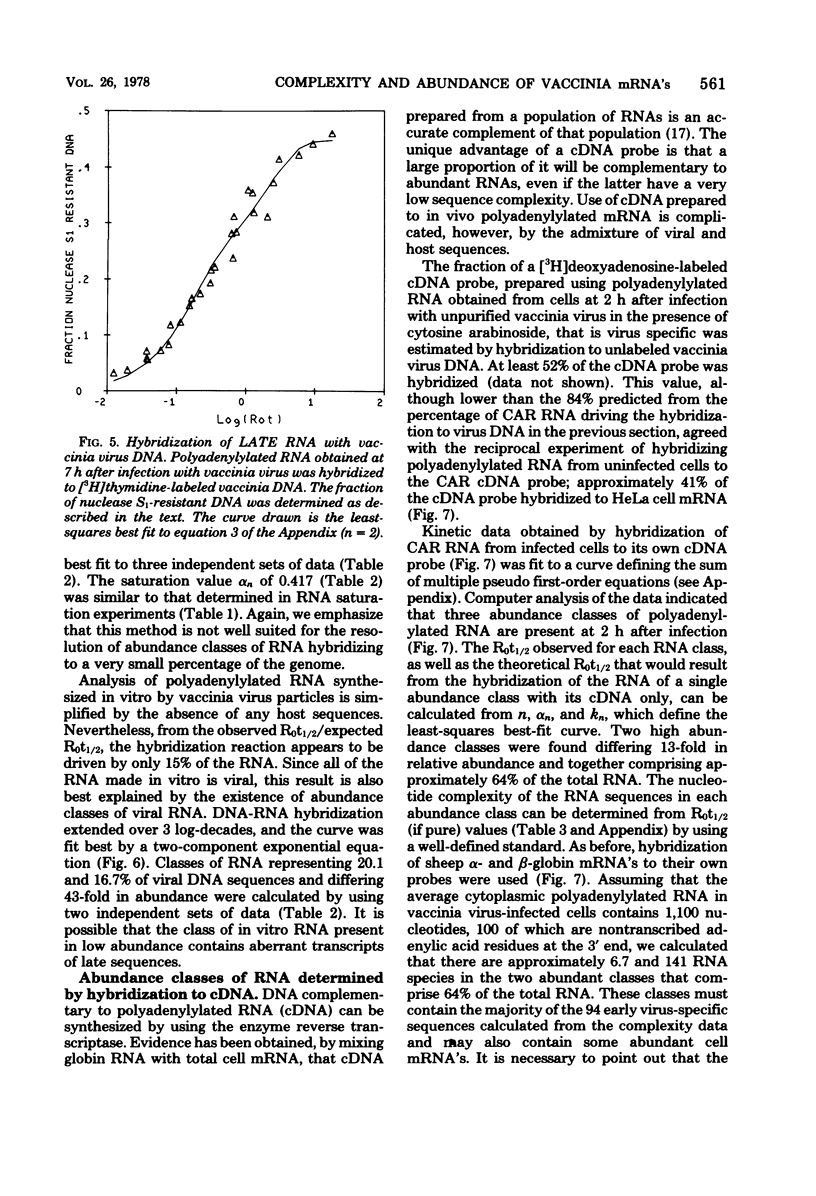
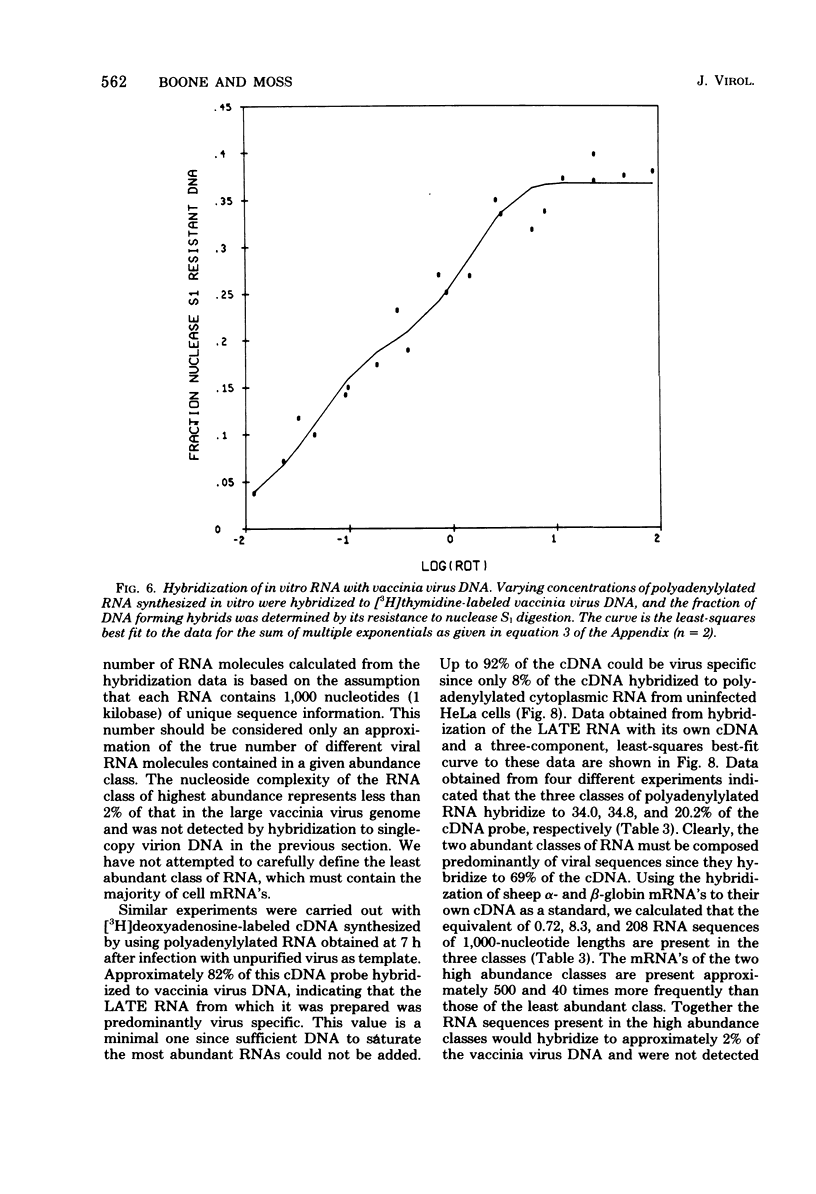
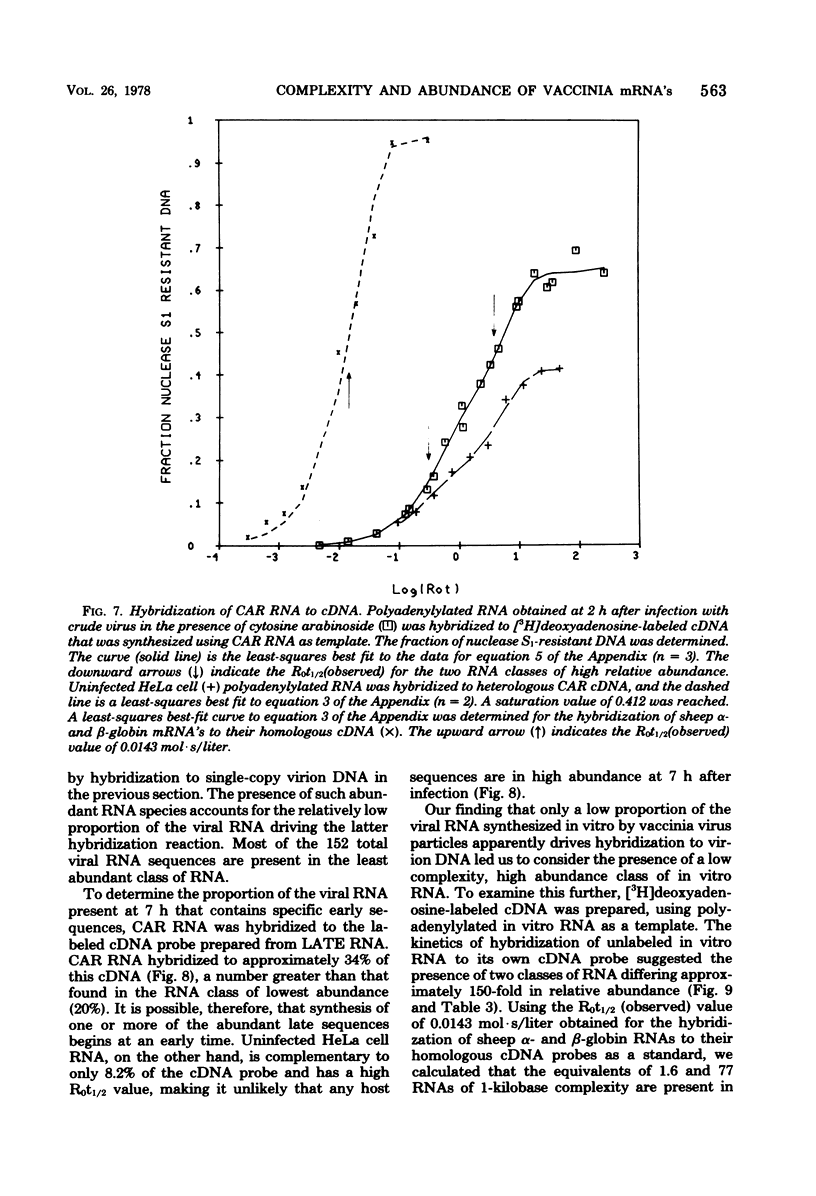
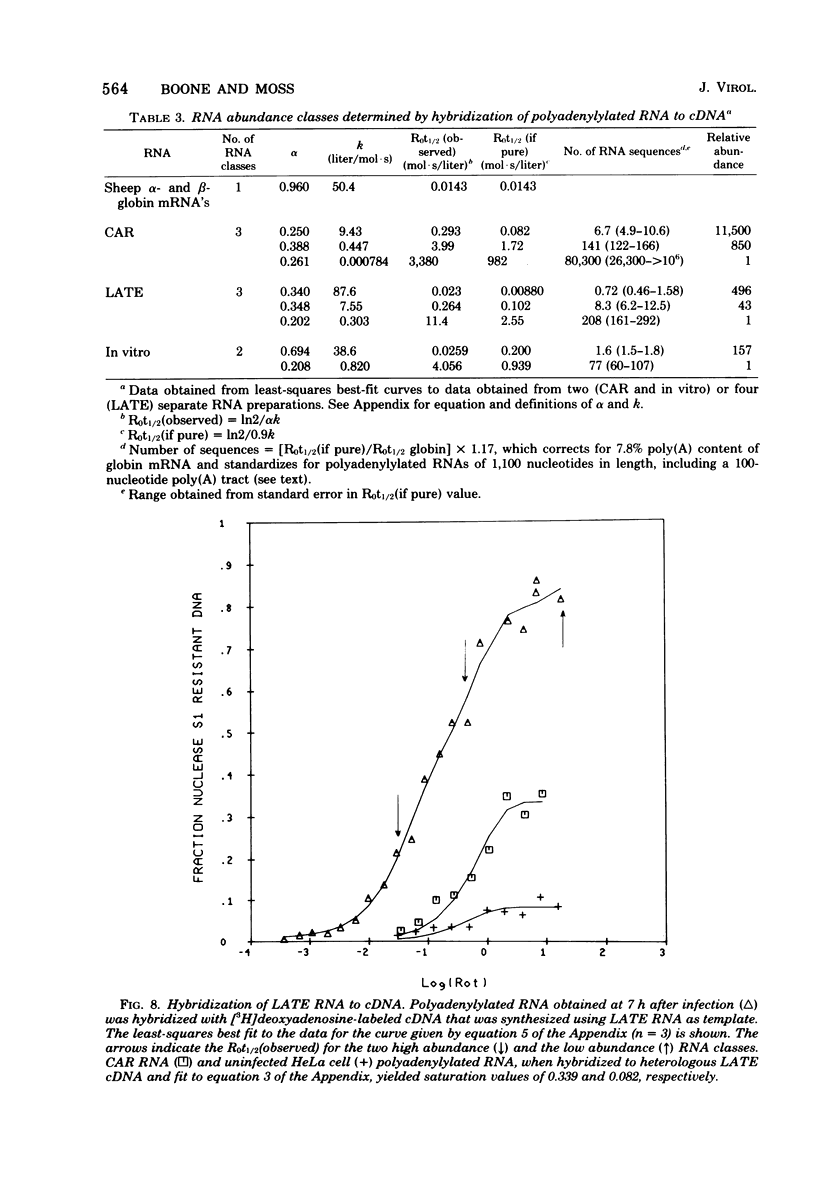
The sequence complexity and relative abundance of vaccinia virus mRNA's, synthesized in vivo and in vitro, have been measured by DNA-RNA hybridization. Up to 42% of [3H]thymidine-labeled virus DNA can be protected from digestion with nuclease S1, a single-strand specific nuclease, after annealing to excess polyadenylylated mRNA obtained at 7 h after infection. In contrast, only 26% of vaccinia virus DNA is protected when hybridized to polyadenylylated RNA obtained at 2 h after infection in the presence of an inhibitor of DNA synthesis. That the 94 kilobases transcribed early are a subset of the 152 kilobases present late was suggested by hybridization of DNA with a mixture of early and late RNAs. Some control of transcription is lost when virus purified by procedures that include sonic treatment is used for infection since under these conditions similar proportions of DNA are protected by either excess early or late RNA. Excess RNA, synthesized in vitro by enzymes within purified vaccinia virus particles, hybridized to approximately the same fraction of the DNA as did RNA present at late times in vivo. A second type of transcriptional control was demonstrated by kinetic analysis of the hybridization of polyadenylylated RNA to labeled DNA. With virion DNA used as the probe, a single abundance class for early RNA, two classes differing 11-fold in abundance for late RNA, and two classes differing 43-fold in abundance for in vitro RNA were found. To be able to detect high-abundance RNAs of very low sequence complexity, labeled complementary DNA probes to early, late, and in vitro polyadenylylated RNA were used. Evidence that, at late times, RNAs totaling 9 kilobases of sequence complexity are present 40 to 500 times more frequently than the bulk of the virus-specific RNA was obtained. In contrast, the highest abundance class of RNA present at 2 h after infection corresponded to 7 kilobases present in only a 13-fold molar excess over the majority of virus-specific sequences. RNA synthesized in vitro was found to contain a small amount of sequence information, approximately 2 kilobases, which occurred 150 times more frequently than the majority of viral sequences. Studies using hybridization of viral DNA to labeled complementary DNA probes also suggested that 52 to 59% of the polyadenylylated RNA present at 2 h after infection and 82 to 92% of that at 7 h are virus specific.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beatriz L. W., McCarthy B. J. Messenger RNA complexity in Drosophila melanogaster. Biochemistry. 1975 Jun 3;14(11):2440–2446. doi: 10.1021/bi00682a026. [DOI] [PubMed] [Google Scholar]
- Berns K. I., Silverman C. Natural occurrence of cross-linked vaccinia virus deoxyribonucleic acid. J Virol. 1970 Mar;5(3):299–304. doi: 10.1128/jvi.5.3.299-304.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop J. O., Morton J. G., Rosbash M., Richardson M. Three abundance classes in HeLa cell messenger RNA. Nature. 1974 Jul 19;250(463):199–204. doi: 10.1038/250199a0. [DOI] [PubMed] [Google Scholar]
- Flint S. J., Sharp P. A. Adenovirus transcription. V. Quantitation of viral RNA sequences in adenovirus 2-infected and transformed cells. J Mol Biol. 1976 Sep 25;106(3):749–774. doi: 10.1016/0022-2836(76)90263-1. [DOI] [PubMed] [Google Scholar]
- Frenkel N., Roizman B. Ribonucleic acid synthesis in cells infected with herpes simplex virus: controls of transcription and of RNA abundance. Proc Natl Acad Sci U S A. 1972 Sep;69(9):2654–2658. doi: 10.1073/pnas.69.9.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel N., Silverstein S., Cassai E., Roizman B. RNA synthesis in cells infected with herpes simplex virus. VII. Control of transcription and of transcript abundancies of unique and common sequences of herpes simplex virus 1 and 2. J Virol. 1973 Jun;11(6):886–892. doi: 10.1128/jvi.11.6.886-892.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galau G. A., Britten R. J., Davidson E. H. A measurement of the sequence complexity of polysomal messenger RNA in sea urchin embryos. Cell. 1974 May;2(1):9–20. doi: 10.1016/0092-8674(74)90003-8. [DOI] [PubMed] [Google Scholar]
- Galau G. A., Klein W. H., Davis M. M., Wold B. J., Britten R. J., Davidson E. H. Structural gene sets active in embryos and adult tissues of the sea urchin. Cell. 1976 Apr;7(4):487–505. doi: 10.1016/0092-8674(76)90200-2. [DOI] [PubMed] [Google Scholar]
- Geshelin P., Berns K. I. Characterization and localization of the naturally occurring cross-links in vaccinia virus DNA. J Mol Biol. 1974 Oct 5;88(4):785–796. doi: 10.1016/0022-2836(74)90399-4. [DOI] [PubMed] [Google Scholar]
- Getz M. J., Birnie G. D., Young B. D., MacPhail E., Paul J. A kinetic estimation of base sequence complexity of nuclear poly(A)-containing RNA in mouse Friend cells. Cell. 1975 Feb;4(2):121–129. doi: 10.1016/0092-8674(75)90118-x. [DOI] [PubMed] [Google Scholar]
- Glisin V., Crkvenjakov R., Byus C. Ribonucleic acid isolated by cesium chloride centrifugation. Biochemistry. 1974 Jun 4;13(12):2633–2637. doi: 10.1021/bi00709a025. [DOI] [PubMed] [Google Scholar]
- Grady L. J., Paoletti E. Molecular complexity of vaccinia DNA and the presence of reiterated sequences in the genome. Virology. 1977 Jun 15;79(2):337–341. doi: 10.1016/0042-6822(77)90361-0. [DOI] [PubMed] [Google Scholar]
- Hastie N. D., Bishop J. O. The expression of three abundance classes of messenger RNA in mouse tissues. Cell. 1976 Dec;9(4 Pt 2):761–774. doi: 10.1016/0092-8674(76)90139-2. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Herman R. C., Williams J. G., Penman S. Message and non-message sequences adjacent to poly(A) in steady state heterogeneous nuclear RNA of HeLa cells. Cell. 1976 Mar;7(3):429–437. doi: 10.1016/0092-8674(76)90173-2. [DOI] [PubMed] [Google Scholar]
- Hynes N. E., Groner B., Sippel A. E., Nguyen-Huu M. C., Schütz G. mRNA complexity and egg white protein mRNA content in mature and hormone-withdrawn oviduct. Cell. 1977 Aug;11(4):923–932. doi: 10.1016/0092-8674(77)90303-8. [DOI] [PubMed] [Google Scholar]
- JOKLIK W. K., BECKER Y. THE REPLICATION AND COATING OF VACCINIA DNA. J Mol Biol. 1964 Dec;10:452–474. doi: 10.1016/s0022-2836(64)80066-8. [DOI] [PubMed] [Google Scholar]
- Kates J. R., McAuslan B. R. Poxvirus DNA-dependent RNA polymerase. Proc Natl Acad Sci U S A. 1967 Jul;58(1):134–141. doi: 10.1073/pnas.58.1.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin B. Units of transcription and translation: sequence components of heterogeneous nuclear RNA and messenger RNA. Cell. 1975 Feb;4(2):77–93. doi: 10.1016/0092-8674(75)90113-0. [DOI] [PubMed] [Google Scholar]
- Moss B. Inhibition of HeLa cell protein synthesis by the vaccinia virion. J Virol. 1968 Oct;2(10):1028–1037. doi: 10.1128/jvi.2.10.1028-1037.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B., Rosenblum E. N., Paoletti E. Polyadenylate polymerase from vaccinia virions. Nat New Biol. 1973 Sep 12;245(141):59–63. doi: 10.1038/newbio245059a0. [DOI] [PubMed] [Google Scholar]
- Munyon W., Paoletti E., Grace J. T., Jr RNA polymerase activity in purified infectious vaccinia virus. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2280–2287. doi: 10.1073/pnas.58.6.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevins J. R., Joklik W. K. Poly (A) sequences of vaccinia virus messenger RNA: nature, mode of addition and function during translation in vitra and in vivo. Virology. 1975 Jan;63(1):1–14. doi: 10.1016/0042-6822(75)90365-7. [DOI] [PubMed] [Google Scholar]
- Ryffel G. U., McCarthy B. J. Complexity of cytoplasmic RNA in different mouse tissues measured by hybridization of polyadenylated RNA to complementary DNA. Biochemistry. 1975 Apr 8;14(7):1379–1385. doi: 10.1021/bi00678a006. [DOI] [PubMed] [Google Scholar]
- Sebring E. D., Salzman N. P. Metabolic properties of early and late vaccinia virus messenger ribonucleic acid. J Virol. 1967 Jun;1(3):550–558. doi: 10.1128/jvi.1.3.550-558.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibatani A. Precipitation and counting of minute quantities of labeled nucleic acids as cetyltrimethylammonium salt. Anal Biochem. 1970 Feb;33(2):279–285. doi: 10.1016/0003-2697(70)90298-8. [DOI] [PubMed] [Google Scholar]
- Wei C. M., Moss B. Methylation of newly synthesized viral messenger RNA by an enzyme in vaccinia virus. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3014–3018. doi: 10.1073/pnas.71.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. G., Penman S. The messenger RNA sequences in growing and resting mouse fibroblasts. Cell. 1975 Oct;6(2):197–206. doi: 10.1016/0092-8674(75)90010-0. [DOI] [PubMed] [Google Scholar]


